1. Introduction
Aluminum (Al) stress mainly occurs in acidic soils (pH < 5.0) when the solubility of aluminum ions (Al
3+) in the soil increases significantly [
1]. The root system is the main site of damage from aluminum stress, and the root tip transition zone is the most sensitive to aluminum toxic stress [
2]. The cell wall plays a key role in aluminum sensing, the composition, structure, and modifications of the cell wall collectively influence plant responses to aluminum stress [
3]. Studies have shown that approximately 90% of Al
3+ ions accumulate in the cell wall [
4]. Pectin contains negatively charged carboxyl groups, making it a major site for aluminum binding [
5]. Aluminum binding to the cell wall triggers a series of undesirable consequences, as found in the groupia (
Neolamarckia cadamba), which enhances cell wall aluminum sequestration through activation of lignin synthesis while regulating Ca/Mg (calcium/magnesium) loss and phosphorus enrichment against aluminum toxicity [
6]; in sorghum (
Sorghum bicolor), aluminum stress significantly inhibits cell wall aluminum sequestration through enrichment of the root elongation zone, inducing callose deposition and disrupting the integrity of the plasma membrane, significantly inhibiting root growth in sorghum (
Sorghum bicolor) [
7]. In rice (
Oryza sativa), structural abnormalities were found in root tip epidermal and cortical cells after treatment with different concentrations of AlCl
3, resulting in blocked cell elongation, radial expansion, cell wall thickening, and enlarged vesicles [
8]. In both rice and tea plants (
Camellia sinensis), members of the PME (pectin methylesterase) gene family exhibiting elevated expression under aluminum stress have been identified. Notably, overexpression of
OsPME14 enhanced aluminum sensitivity in transgenic plants [
9]. Furthermore, transgenic rice overexpressing
OsXTH19 (
OsXTH19-OX) displayed promoted xyloglucan degradation. The resultant reduction in xyloglucan content conferred decreased root aluminum accumulation and enhanced root growth under aluminum stress conditions [
10].
The mechanisms of plant tolerance to aluminum toxicity fall into two main categories: internal tolerance and external rejection [
2,
11]. Internal tolerance mechanisms mitigate the toxicity of aluminum ions to cells through the compartmentalization of vesicles and chelation of organic acids [
12]. The maize root system significantly reduces cytoplasmic toxicity under aluminum stress by binding aluminum through citric acid within vesicles and forming a stabilizing complex [
13]. Aluminum ions absorbed by the rice root system can be segregated into vesicles through the vesicular membrane transport system, which uses the acidic environment of the vesicles to precipitate aluminum into aluminum phosphate or organic acid complexes, significantly reducing cytoplasmic toxicity [
14]. In contrast, the external exclusion mechanism reduces aluminum toxicity by chelating inter-root aluminum ions through the secretion of organic acids by the root system, reducing the entry of aluminum ions into the cell [
15,
16].
In addition to organic acids, plants have evolved antioxidant defense mechanisms to regulate and adapt to aluminum-induced oxidative stress. During reactive oxygen species (ROS)-mediated damage repair under aluminum stress [
2,
16], peroxidase (POD, EC1.11.1.7), superoxide dismutase (SOD, EC1.15.1.1), catalase (CAT, EC1.11.1.6), ascorbate peroxidase (APX, EC1.11.1.11), and several other enzymes are involved [
17,
18,
19,
20]. Low concentrations of aluminum promoted linear growth of rice roots but triggered ROS accumulation and cortical tissue damage with increasing doses [
21]; in cucumber (
Cucumis sativus), cucumber roots showed strong aluminum enrichment capacity, and aluminum stress significantly inhibited CAT activity in seedlings but activated APX activity, thus affecting cucumber’s aluminum tolerance [
22]; in peanut (
Arachis hypogaea), aluminum stress induces a mitochondria-dependent pathway ROS burst by inducing a decrease in mitochondrial membrane potential and cytochrome release, thereby triggering programmed cell death, which can be reversed by increasing CAT activity [
23]; salicylic acid significantly alleviates the effects of aluminum stress on tomato (
Solanum lycopersicum) by modulating the antioxidant system, decreasing aluminum accumulation and oxidative damage [
22]; and aluminum stress significantly alleviates the effects of aluminum stress on tomato (
Solanum lycopersicum) seedlings from aluminum toxicity [
24].
With the continuous development of molecular biology techniques, many genes closely associated with aluminum stress have been reported. In alfalfa, it has been found that
MsDHN1, a gene coding for dehydration proteins (DHNs), could improve aluminum tolerance by affecting the secretion of oxalate from the root tip, and its overexpression increased alfalfa growth rate under aluminum stress, whereas inhibiting the expression of
MsDHN1 decreased its aluminum tolerance [
25]. In soybean (
Glycine max), multidrug and toxic compound extrusion (MATE, multidrug and toxic compound efflux proteins) transporters
GmMATE13 and
GmMATE75 significantly enhanced plant aluminum tolerance by promoting citric acid secretion [
26]; overexpression of the
SbMATE gene in sugarcane (
Saccharum officinarum) improved citric acid secretion, maintained normal root growth under aluminum stress, and showed enhanced aluminum tolerance [
27]. Aluminum-activated malate transporter (ALMT) is closely associated with aluminum resistance in plants, and the activation of aluminum ions by the ALMT protein promotes the secretion of malate to the inter-root and chelates aluminum ions to reduce toxicity. Arabidopsis (
Arabidopsis thaliana)
AtALMT1 [
28], wheat (
Triticum aestivum)
TaALMT1 [
29], and maize
ZmALMT1 [
30] have been shown to be key genes for enhancing aluminum resistance in plants.
Plant endophytes (endophytes) are defined as bacterial taxa that colonize internal plant tissues, establishing a mutually beneficial symbiotic relationship with the host plant without causing symptomatic infection or detrimental effects. Typically, endophytes inhabit both the apoplast and symplast compartments of plants, and endophytes significantly enhance plant resistance to biotic (e.g., pathogens and pests) and abiotic stresses (e.g., drought, salinity, heavy metals, and temperature extremes) [
31,
32,
33]. Blueberry (
Vaccinium uliginosum), a typical acid-loving plant, can alleviate aluminum toxicity and nutrient imbalances in acidic soils by secreting iron carriers, organic acids, or hormone-like substances via its root endophytes [
34].
In China, pumpkin (
Cucurbita moschata) cultivation occupies approximately 600,000 hectares annually, with total production reaching ~20 million metric tons. Recent years have witnessed increasing global demand for pumpkin seeds. However, expanding areas of acidic soils have rendered aluminum stress a significant abiotic constraint on pumpkin production; under Al stress, root growth is severely impeded, resulting in reduced yield [
35]. In this study,
Bacillus sphaericus strain
J01, isolated from blueberry, was used to treat pumpkin seedlings under aluminum stress to investigate its physiological mechanisms and transcriptional regulation under aluminum stress to enrich the biological resources for acid and aluminum resistance in plants.
2. Materials and Methods
2.1. Materials and Stress Treatments
In this experiment, a mini-bebe pumpkin was used as the test material. Seeds were soaked in warm water at 50 °C for 10 min, wrapped in wet gauze, and placed in the dark at 30 °C for dewetting. They were cultured until two cotyledons unfolded, and then Hoagland nutrient solution was applied until three true leaves were grown for the aluminum stress treatment. For the control group, Hoagland (Solarbio Science & Technology Co. Ltd., Shanghai, China) culture solution with 750 μM of CaCl2 (pH = 4.0) was added for seedling culture.
For the aluminum-treated group, Hoagland nutrient solution was added with 750 μM of AlCl3 (pH = 4.0); for the strain J01 group, Hoagland nutrient solution was added with 750 μM of AlCl3 and 10 mL of OD600 = 1.0 J01 bacterial suspension. The nutrient solution was changed every four days, and equal amounts of aluminum solution and bacterial suspension were reintroduced for two weeks. Each treatment group contained 36 plants, and each index was determined using biological replicates.
2.2. Strain Isolation and Characterization
The blueberry branches were washed with demineralized water, and the water was drained. The leaf buds were cut, sequentially soaked in 75% alcohol and 0.1% NaClO (Solarbio Science & Technology Co. Ltd., Shanghai, China) for 2 min, and rinsed with sterile water. The preparation of endophytic bacterial isolation medium contained 3 g of beef paste, 5 g of sodium chloride, 10 g of peptone, and 20 g of agar; the pH was adjusted to 7.4–7.6, the above ingredients were added into deionized water. The volume was fixed to 1000 mL, and the ingredients were autoclaved at 121 °C for 30 min and cooled to room temperature. Sterilized leaf buds were divided into medium plates, and a sterile water-coated control was set up. The plates were incubated at a constant temperature of 30 °C. If no colonies grew on the control plate, the surface was sterilized thoroughly. After the other plates grew into colonies, single colonies were repeatedly picked and purified using plate delimitation. The purified strains were kept in a refrigerator at 4 °C for spare time, and microscopic observation and physiological and biochemical experiments were conducted. The dominant strains were sequenced using 16S rRNA and gyrB amplification, and the sequencing results were compared with the NCBI database to select 15 model strains with both 16S rDNA and gyrB sequences and other model strains. A phylogenetic tree was constructed by splicing 16S rDNA and gyrB sequences.
2.3. Comparative Analysis of Bacillus 16S rRNA Sequences
Phylogenetic tree construction: Characteristic gene sequences of J01 (16S rRNA gene sequences commonly used in bacteria) were extracted, and related homologous sequences were obtained from public databases as controls. Sequence quality was assessed using FastQC (v0.12.1) to remove low-quality, contaminated, or spliced sequences to ensure data reliability. The J01 16S rRNA gene sequence was compared with homologous sequences from Bacillus cereus ATCC 14579 (NR_114582.1), Bacillus subtilis subsp. subtilis ATCC 6051 (PQ482394.1), Bacillus thuringiensis strain ATCC 10792 (NR_114581.1 ), Bacillus amyloliquefaciens strain BCRC 11601 (NR_116022.1), Bacillus licheniformis strain BCRC 11702 (NR_116023.1), Bacillus pumilus strain ATCC 7061 (NR_043242.1), Bacillus mycoides strain ATCC 6462 (NR_115993.1), Bacillus vallismortis strain BCRC 17183 (EF433404.1), Bacillus malacitensis strain CECT 5687 (DQ993672.1), Bacillus axarquiensis strain CIP 108772 (DQ993670.1), Bacillus siamensis KCTC 13613 (PQ135913.1), Bacillus megaterium strain ATCC 14581 (JF749282.1), and Bacillus subtilis strain BCRC 10255 (NR_116017.1), and a phylogenetic tree was constructed using MEGA X v11.
2.4. Determination of Root Length and Root Vigor
Pumpkin root system was determined after 14 days of exposure to (1) aluminum-free control, (2) 750 μM of AlCl
3 stress, and (3) AlCl
3 stress with
J01 inoculation. Data were collected from 12 randomly selected plants per treatment group. The root system was thoroughly rinsed with deionized water for 5 min, and the residual water on the surface was blotted with filter paper. A straight edge was used to accurately determine the length of the primary roots, and the experiment was set up with three biological replicates, each containing three seedlings. Root vigor was determined using the triphenyltetrazolium chloride reduction method [
36].
2.5. Relative Conductivity Determination
The conductivity meter method was used for the determination of plant roots [
37].
2.6. Determination of Malondialdehyde Content and Antioxidant Enzyme Activities
The MDA content, SOD activity, POD activity, CAT activity, and APX activity were determined in accordance with the operation manual of Beijing Solepol Technology Co. (Shanghai, China) Following 14-day treatments (aluminum-free control, aluminum stress, and aluminum stress with J01 application), all parameters were quantified using three randomly selected plants per treatment group.
2.7. RNA Sequencing and Gene Expression Analysis
RNA was extracted from pumpkin root tips after 14 days of treatment using an RNA extraction kit from Wuhan Ekorui Biotechnology Co. (Wuhan, China). The quality of RNA was checked using agarose gel electrophoresis and a NanodroP microspectrophotometer. The RNA samples were sent to the Histogenics Biotechnology Company (Beijing, China) for the construction of strand-specific cDNA libraries and double-end transcriptome sequencing using an Illumina NovaSeq 6000 s-generation sequencer(Illumina, Inc., San Diego, CA, USA). Raw sequencing data underwent quality filtering to generate clean reads using the following criteria: removal of sequences containing >1 ambiguous base (N) per 20 bp window; exclusion of reads, including those lacking 5’/3’ adapters, those with zero length after processing, those with <17 bp or >30 bp post-adapter trimming, and those containing polyA tracts. Clean reads were aligned to the Cucurbitaceae family genome (
http://cucurbitgenomics.org/, accessed on 28 July 2025) using exact matching parameters. Perfectly matched tags were retained for degradome analysis. Sequences were cross-referenced against the Rfam database (v14.9;
http://rfam.sanger.ac.uk/, accessed on 28 July 2025) to identify structural RNAs. Processed reads were assembled using Trinity (v2.13.2) with default parameters. The longest isoform per gene locus was designated as a unigene for downstream analyses. rRNA, scRNA, snoRNA, tRNA, and snRNA sequences were systematically removed prior to expression quantification. Differentially expressed genes (DEGs) were identified using thresholds:|log
2(fold change)| > 1 and adjusted
p-value < 0.05 (Benjamini–Hochberg correction). The annotation of de novo splicing transcriptome is divided into two parts. One part is to predict the coding region (CDS) of UniGene and annotate the obtained protein sequence to multiple protein databases, including homology search known sequencing data (BLAST+/Uniprot), protein domain recognition (HMMER/PFAM), pathway action (GO/Kg databases), homologous protein clustering (eggNOG), etc. The other part is to annotate UniGene sequences without predicted CDS, including NCBI NR database (blastn, e-value ≤ 1 × 10
−5), Rfam covariance models, and plant-specific databases (
Arabidopsis TAIR10).
RNA was reverse transcribed into cDNA using a reverse transcription kit (Wuhan Acres Biotechnology Co., Ltd., Wuhan, China). Target genes were screened for key genes related to aluminum tolerance (transporter proteins
ALMT and
AQP, antioxidant genes
APX and
GST, and cell wall modification genes
XTH and
PME) based on transcriptome sequencing. qPCR was performed using the qPCR kit from Wuhan Acres Biotechnology Co. Stably expressed
GAPDH [
38] was selected as internal reference genes for data normalization. qPCR was performed using the qPCR kit from Wuhan Acres Biotechnology Co. The gene-specific primers are listed in
Table S1. The PCR program consisted of a preliminary step of 30 s at 95 °C followed by 40 cycles at 95 °C for 5 s and at 60 °C for 30 s.
2.8. Statistical Analysis of Data
All treatments were subjected to at least three independent biological replicates, which were organized and entered into Excel. Then, the following were used: one-way ANOVA using SPSS (26.0), post hoc multiple comparisons using SPSS, and plotting and correlation analyses using Origin 2019.
3. Results
3.1. Effects of Blueberry Endophytic Bacterial Strain J01 on the Growth and Development of Pumpkin Roots Under Aluminum Stress
A dominant bacterial strain, named
J01, was isolated from the leaf buds of blueberries (
Figure 1), and the kinship relationship was analyzed by constructing a phylogenetic tree. It was found that
J01 was closely clustered with several strains of the genus Bacillus, which proved that
J01 belonged to the bacterium
Bacillus subtilis (
Table 1).
As shown in
Figure 2, after 14 days of incubation, root elongation of pumpkin seedlings was significantly inhibited by aluminum treatment, whereas the treatment with endophyte J01 was able to improve the growth of pumpkin roots after aluminum treatment and promoted the growth of lateral roots. As shown in
Table 2, compared with the control group (Control), the plants in the Al
3+ treatment group demonstrated reduced root length of pumpkin seedlings to 0.82 times that of the control group and reduced root vigor to 0.16 times that of the control group. However, inoculation with the blueberry endophyte
J01 resulted in significantly higher root length and root vigor than the control group, but root vigor was still lower than that in the control group.
3.2. Effects of Blueberry Endophyte Strain J01 on MDA Content and Relative Conductivity of Pumpkin Roots Under Aluminum Stress
As shown in
Table 3, the MDA content of the AlCl
3-treated group increased by 22.6% compared with the control group, and the MDA content of the J01-treated group decreased by 9.4% compared with the AlCl
3 group but still increased by 11.1% compared with the control group; the relative electrical conductivity of the AlCl3-treated group increased by 362.5% compared with the control group, and the relative electrical conductivity of the J01-treated group decreased by 41.2% compared with the AlCl
3 group but was still 172.2% higher than the control group.
3.3. Effect of Blueberry Endophyte Strain J01 on Antioxidant Enzyme Activities in Pumpkin Roots Under Aluminum Stress
As shown in
Table 4, the SOD activity in the AlCl
3-treated group was significantly increased by 89.7% compared with the control group, 39.9% in the AlCl
3 + J01-treated group, and 165.4% compared with the control group; the POD activity in the AlCl
3-treated group was significantly decreased by 43.1% compared with the control group and increased by 58.0% in the AlCl
3 +
J01-treated group compared with AlCl
3, but it was still 10.1% lower than the control group; CAT activity in the AlCl
3-treated group was significantly reduced by 64.9% compared with the control group and increased by 93.7% in the AlCl
3 +
J01-treated group compared with the AlCl
3 group, but it was still 32.1% lower than the control group; the APX activity in the AlCl
3-treated group was significantly increased by 85.3% compared with the control group and reduced by 30.4% in the AlCl
3 + J01-treated group compared with the AlCl
3 group, but it was still increased by 29.0% compared with the control group.
3.4. Transcriptome Analysis of Blueberry Endophyte Strain J01 on Pumpkin Roots Under Aluminum Stress
The volcano plot shows 280 upregulated genes (red) and 729 downregulated genes (blue), and the remaining genes showed no significant difference in expression (gray) (
Figure 3A). GO enrichment analysis (
Figure 3B) was performed to analyze the functional classification and enrichment of differentially expressed genes in three dimensions: biological processes, cellular components, and molecular functions. The results showed that in biological processes, the functional categories related to metabolic processes and biological regulation enriched a large number of differentially expressed genes, whereas in cellular components, the categories related to cellular structure, such as cells and cell parts, enriched a larger number of differentially expressed genes. In molecular functions, functional categories such as binding and catalytic activity were the main functional directions of differentially expressed genes. Kyoto Encyclopedia of Genes and Genomes enrichment analysis (
Figure 3C) revealed that plant hormone signal transduction and phenylpropanoid biosynthesis pathways were highly enriched. Gene expression clustering analysis (
Figure 3D) showed that there were significant differences in the gene expression patterns between the AlCl
3-treated and AlCl
3 +
J01-treated groups, with most of the genes in the AlCl
3-treated group being expressed at lower levels, while some genes were significantly upregulated, and some genes were significantly downregulated in the AlCl
3 +
J01-treated group.
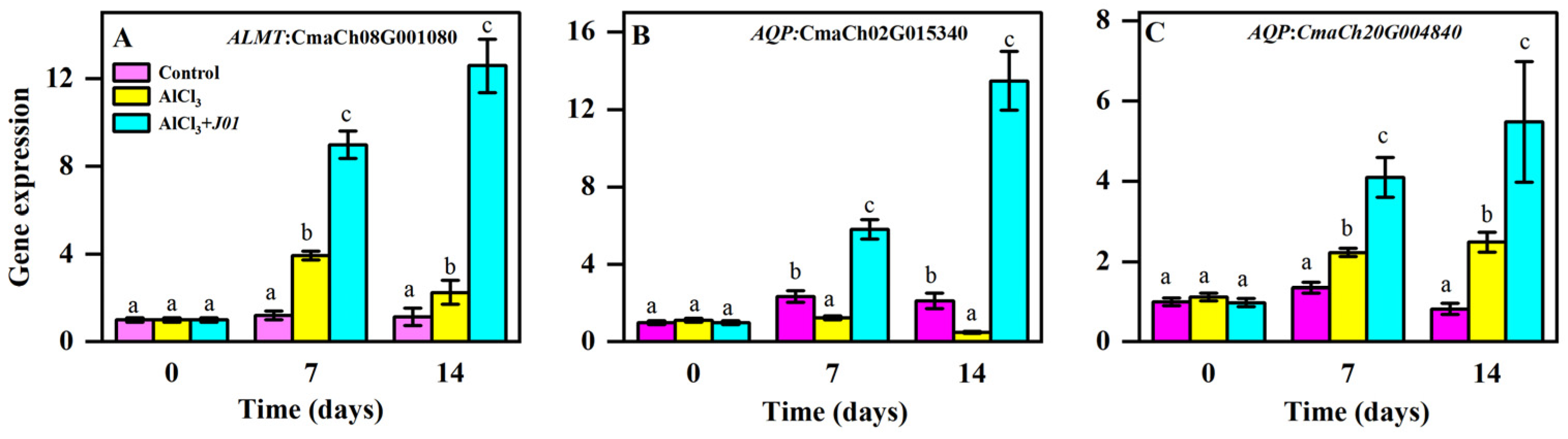
3.5. Effect of Blueberry Endophyte Strain J01 on the Expression of Transporter Protein-Related Genes Under Aluminum Stress
As shown in
Figure 4, the
ALMT gene (CmaCh08G001080), the expression of the control group at all time points, did not change significantly (
p > 0.05), indicating that the gene maintains the basal level of expression under normal conditions. The expression of the AlCl
3-treated group at seven days was significantly elevated, increasing by 294% compared with the control group (
p < 0.01). At 14 days, the expression declined, although it was still higher than that in the control group (
p < 0.05), but it decreased by 43% compared with seven days (
p < 0.01). The expression in the AlCl
3 +
J01 treatment group was elevated at seven days, which was significantly higher than that of the AlCl
3 group (
p < 0.01) and the control group (
p < 0.001) and was further elevated at 14 days, which was significantly increased by 40% compared with that at the seven days (
p < 0.05); the difference with the AlCl
3 group was highly significant (
p < 0.001).
For the AQP gene (CmaCh20G004840), there was no significant difference in the control group (p > 0.05), reflecting stable expression under the normal water transport requirement. The expression of the AlCl3-treated group increased by 65% at seven days compared with the control group (p < 0.05) and increased by 11.7% at 14 days compared with seven days (p > 0.05), which was higher than that of the control group (p < 0.05), but the increase was slower. The expression in the AlCl3 + J01 treatment group was higher than that in the AlCl3 group (p < 0.01) and the control group (p < 0.001) at seven days, and the expression at 14 days was significantly increased by 33.7% compared with that at 7 days (p < 0.05); the difference was highly significant compared with that in the AlCl3 group (p < 0.001).
The AQP gene (CmaCh02G015340), whose expression was significantly upregulated (p < 0.05) at 7 and 14 days in the control group compared with at 0 days, may be involved in specific water transport during the normal growth phase. The expression was decreased by 47.7% (p < 0.001) at seven days (p < 0.001) and 60.2% (p < 0.01) at 14 days (p < 0.001) in the AlCl3-treated group compared with that in the control group. The expression in the AlCl3 + J01 treatment group was significantly higher than that in the AlCl3 (p < 0.001) and control groups (p < 0.01) at seven days. The expression at 14 days was significantly increased by 131.6% compared with that at seven days (p < 0.001), and the difference with the AlCl3 group was highly significant (p < 0.001).
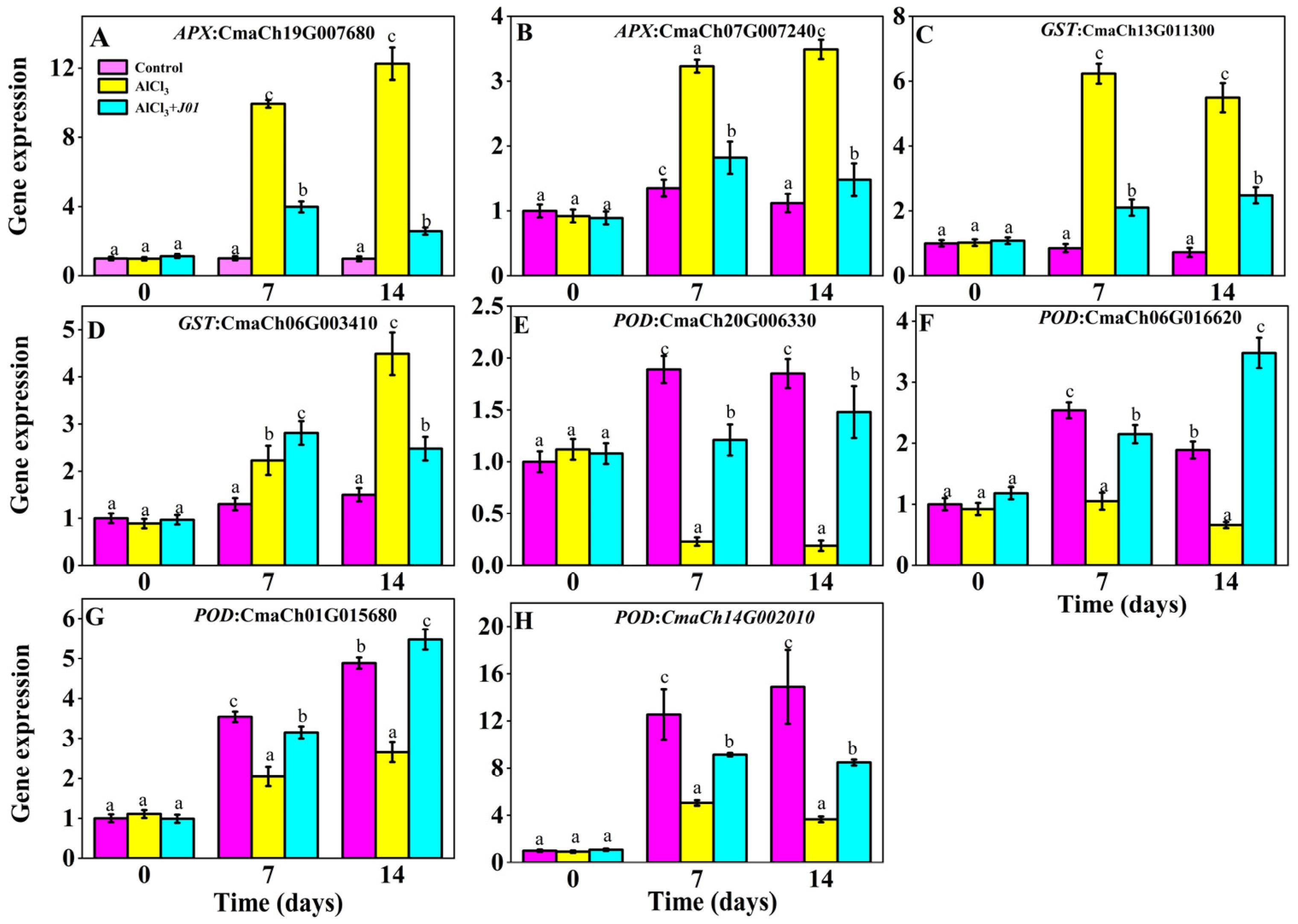
3.6. Effect of Blueberry Endophyte Strain J01 on the Expression of Antioxidant Defense-Related Genes in Pumpkin Roots Under Aluminum Stress
APX genes (CmaCh19G007680 and CmaCh07G007240) were significantly differentially expressed under aluminum stress (
Figure 5). For CmaCh19G007680, the expression was basically stable in the control group, and the expression in the AlCl
3 group was elevated approximately 9-fold compared with the control group at seven days (
p < 0.01) and further elevated at 14 days, whereas the expression in the AlCl
3 + J01 group was lower than that of the AlCl
3 group at seven days (
p < 0.01) but higher than that of the control group (
p < 0.05). Then, the expression declined at 14 days, approaching the control group’s level, indicating that
J01 could inhibit the overactivation of the
APX gene and avoid the overconsumption of ascorbic acid. The expression trend of CmaCh07G007240 was similar to that of CmaCh19G007680, but the response was lower.
The expression of GST genes (CmaCh13G011300 and CmaCh06G003410) showed different response patterns under aluminum stress, and the expression of the control group of CmaCh13G011300 decreased slightly with time; the expression of the AlCl3 group increased significantly at 7 and 14 days (p < 0.001), while the expression of the gene in the AlCl3 + J01-treated group increased gradually and was significantly lower than that of the AlCl3 group at 7 and 14 days (p < 0.01). The gene expression of the AlCl3 + J01 treatment group gradually increased and was significantly lower than that of the AlCl3 group at 7 and 14 days (p < 0.01), but it was still higher than that of the control group (p < 0.05). There was no significant change in the expression of the control group of CmaCh06G003410. The gene expression of the AlCl3 group gradually increased at 7 and 14 days (p < 0.05), and the gene expression of the J01 treatment group first increased and then decreased at 7 and 14 days, with the expression of the AlCl3 group being significantly higher than that of the AlCl3 group at seven days (p < 0.001). Expression was significantly higher than that in the AlCl3 group (p < 0.05).
The expression of the POD gene (CmaCh20G006330) in the control group remained stable at 7 and 14 days; the gene expression in the AlCl3 group showed a decreasing trend with time, decreasing by 87.8% and 89.7% compared with that of the control group at 7 and 14 days (p < 0.001), suggesting that aluminum stress severely inhibited the transcription of the POD gene. The expression of the AlCl3 + J01-treated group at 7 and 14 days gradually increased and was significantly higher than that of the AlCl3 group (p < 0.01) but was still lower than that of the control group (p < 0.05). Expression gradually increased and was significantly higher than that in the AlCl3 group (p < 0.01) but was still lower than that in the control group (p < 0.05). The expression of CmaCh06G016620 in the AlCl3 group was significantly lower than that of the control group at 7 and 14 days (p < 0.05); the expression of the gene was gradually increased and was significantly higher than that of the AlCl3 + J01-treated group at 7 and 14 days (p < 0.01), and the expression of the gene in the CmaCh06G016620 group was significantly lower than that of the control group (p < 0.05). p < 0.01) and exceeded that of the control group at 14 days (p < 0.05). The expression of the AlCl3 group of CmaCh01G015680 and CmaCh14G002010 was lower than that of the control group at 7 and 14 days, but the expression of the former group gradually increased with time and was significantly higher than that of the AlCl3 group at 14 days after treatment with J01 (p < 0.001). The latter expression amount first increased and then decreased and was significantly higher than that of the AlCl3 group at 7 and 14 days (p < 0.01).
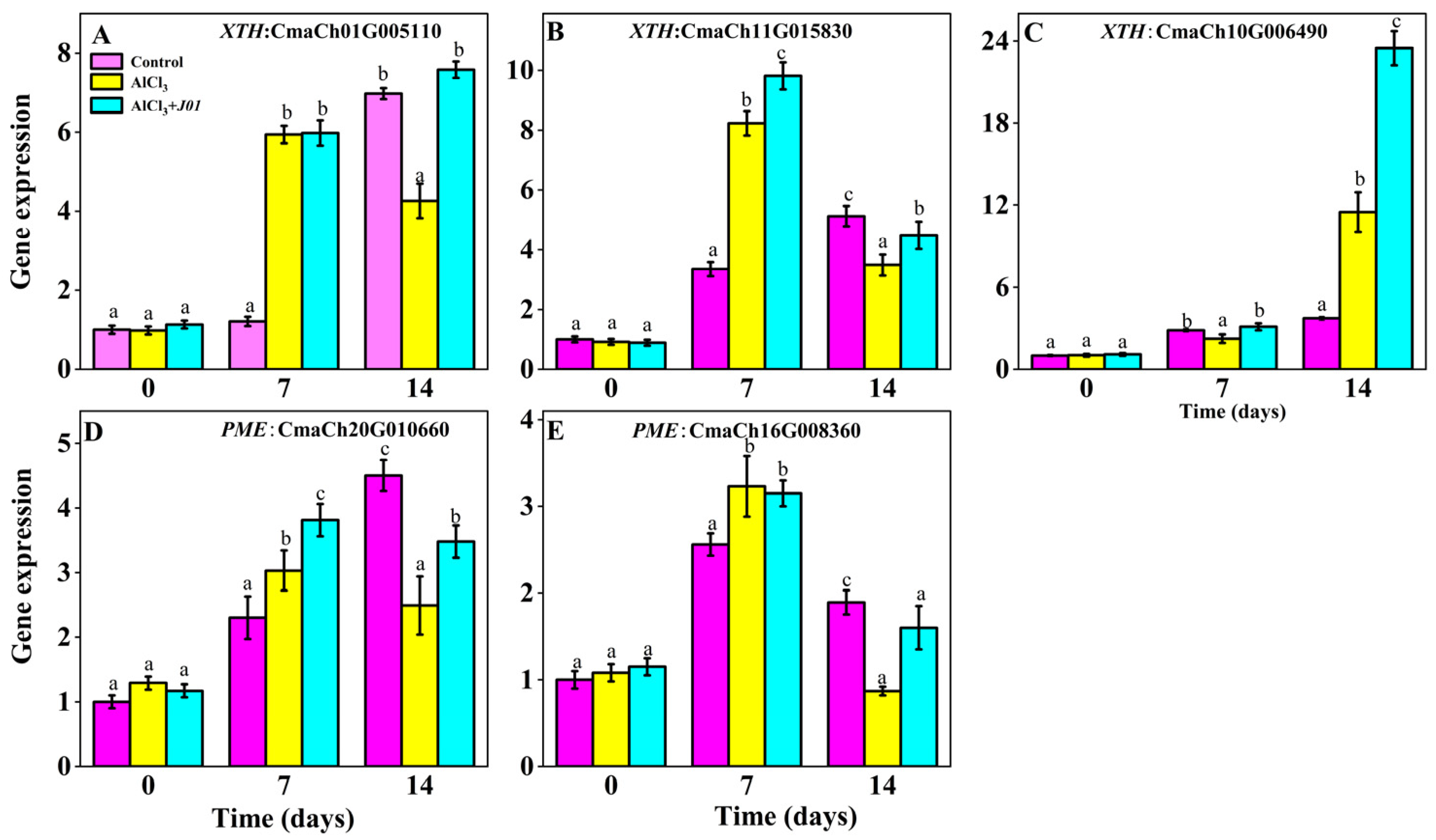
3.7. Effect of the Blueberry Endophyte Strain J01 on the Expression of Genes Related to Cell Wall Modification in Pumpkin Roots Under Aluminum Stress
The expression pattern of
XTH genes (CmaCh01G005110 and CmaCh11G015830) show (
Figure 6) that the expression of the control group gradually increased with time; in the AlCl
3-treated group, the expression of CmaCh01G005110 firstly increased and then decreased with time, and the expression increased by 490% compared with that of the control at seven days (
p < 0.01) and decreased to 4.26, which was still higher than the control (
p < 0.05). The expression of the AlCl
3 + J01 treatment group gradually increased with time, and at seven days, the expression was not significantly different from that of the AlCl
3 group (
p > 0.05). At 14 days, it increased by 78% compared with that of the AlCl
3 group (
p < 0.01). The expression of another
XTH gene (CmaCh11G015830) in the AlCl
3 group was 2.46 times higher than that of the control at seven days (
p < 0.001) and decreased to 3.49 at 14 days, which was significantly lower than that at seven days (
p < 0.01). The expression of the AlCl
3 +
J01-treated group increased by 19.3% at seven days compared with that of the AlCl
3 group (
p < 0.05), and although it decreased by 54.4%, it was still higher than that of the AlCl
3 group (
p < 0.05).
The expression of the PME gene (CmaCh20G010660) in the control group showed a trend of increasing and then decreasing with time; in the AlCl3 group, the expression of the PME gene increased by 31.7% at seven days compared with the control (p > 0.05) and decreased by 44.7% at 14 days (p < 0.01); the expression of the PME gene in the AlCl3 + J01-treated group gradually decreased but was higher than that in the AlCl3 group at 7 and 14 days (p < 0.05). The expression of CmaCh16G008360 was significantly downregulated under aluminum stress. The expression of the AlCl3 group was 1.26 times that of the control at seven days (p > 0.05) and then decreased to 0.87 at 14 days (53.9% decrease compared with the control, p < 0.001), whereas the expression of CmaCh16G008360 was decreased in the AlCl3 + J01-treated group at seven days (no significant difference from the control, p > 0.05) and increased by 83.7% at 14 days compared with the AlCl3 group. Expression increased by 83.9% (p < 0.01) compared with the AlCl3 group but was still lower than that of the control (p < 0.05).
4. Discussion
Aluminum stress, a major toxic factor in acidic soils, has become an important problem limiting crop production because of its significant inhibitory effect on root development [
17,
39]. Typical features of aluminum toxicity include decreased activity of apical meristematic tissues, reduced formation of lateral root primordia, and excessive aluminum incorporation into cell wall pectin, all of which are closely related to the high accumulation of aluminum ions in the apical transition zone [
5]. Aluminum stress not only destroys the cell wall structure through physical damage but also induces a burst of ROS, leading to lipid peroxidation, protein oxidation, and DNA damage [
19,
40,
41]. Low concentrations of aluminum transiently promote root elongation in some plants; however, oxidative damage triggered by ROS accumulation rapidly intensifies with increasing concentrations, ultimately inhibiting growth [
27].
Bacillus spp. strain Bt04 has been found to promote maize root development and alleviate aluminum stress through the production of growth hormones (IAA and PAA) and cytokinins via a mechanism that involves a reduction in aluminum accumulation, enhancement of antioxidant capacity, and a stress response dependent on the growth hormone transport pathway [
42]. In this study, we found that the blueberry endophyte strain
J01 alleviated the inhibition of pumpkin root growth by aluminum stress, as evidenced by enhanced root length and vigor (
Table 2,
Figure 2). The inhibition of root growth under aluminum stress mainly originates from the inhibition of root tip cell elongation and division by Al
3+ [
2], whereas
J01 may reduce the entry of inter-root Al
3+ into root cells by regulating the growth hormone signaling pathway or by secreting organic acids to chelate the inter-root Al
3+.
Aluminum stress can induce the formation of reactive oxygen species (ROS), leading to oxidative damage in plants. During long-term adaptation to acidic soil environments, plants have evolved an ROS scavenging system composed of enzymatic and non-enzymatic systems [
27].
J01 treatment reduced root MDA content and relative conductivity (
Table 3), suggesting that it mitigates aluminum toxicity injury by maintaining cell membrane integrity, which is similar to the mechanism by which endophytes in cucumbers maintain membrane stability by regulating antioxidant enzyme activity [
22]. Moreover, the significant enhancement of SOD and CAT activities by J01 indicated that it inhibited aluminum stress-induced oxidative damage by enhancing the scavenging capacity of superoxide anions (O
2−) and hydrogen peroxide (H
2O
2). This is similar to the mechanism of action of the antioxidant system in the aluminum tolerance of rice [
21] and peanuts [
23]. APX is a core component of the ascorbic acid–GSH cycle, and its overactivation may lead to the depletion of ascorbic acid [
17]. J01 inhibited the overactivation of
APX genes, maintained the expression of
GST genes, and restored
POD gene expression, thus optimizing the antioxidant defense system and preventing excessive depletion of ascorbic acid by oxidative stress. This indicates that, compared to aluminum stress treatment alone, the combined treatment of aluminum stress and
J01 significantly regulated numerous antioxidant enzyme-related genes in plants. Moreover, different members within the same gene family may play distinct roles in this process. Notably, during
J01-mediated alleviation of oxidative damage in pumpkin root tips, peroxidase genes and the ascorbate-glutathione cycle likely play more critical regulatory roles. This is evidenced by the significantly higher number of upregulated peroxidase genes versus downregulated genes, correlating with the enhanced hydrogen peroxide scavenging capacity observed under this condition.
Compared to aluminum stress alone, the combined treatment of aluminum stress and endophytic bacterium
J01 significantly upregulated multiple genes encoding malate transporters and aquaporins. The upregulation of the transporter protein genes
ALMT and
AQP indicated that
J01 enhanced the tolerance and resistance of pumpkin to aluminum ions by promoting malate efflux and water transport. The dynamic response of the
ALMT gene under aluminum stress was functionally similar to that of wheat
TaALMT1 [
29] and sugarcane SbMATE [
27]. Additionally, upregulation of the expression of the
AQP gene (CmaCh02G015340) suggests that
J01 might alleviate aluminum-induced osmotic stress by improving cellular water potential. These findings demonstrate that
J01-mediated malate secretion in pumpkin root tips likely constitutes a key mechanism by which the endophytic bacterium enhances aluminum tolerance. This increased malate secretion under
J01 treatment appears associated with significant upregulation of the malate transporter gene
ALMT and differential expression of specific aquaporin family members.
The cell wall, serving as the primary binding site for aluminum in plant roots, plays a critical role in regulating aluminum tolerance [
27]. The differential expression of cell wall modification-related genes indicated that
J01 remodeled cell wall extensibility by dynamically adjusting the activities of XTH and PME. For example, sustained high expression of the
XTH gene (CmaCh11G015830) under J01 treatment may enhance the elasticity of the cell wall and alleviate the mechanical damage induced by aluminum binding, whereas the phased downregulation of the expression of the
PME gene (CmaCh20G010660) may reduce pectin de-esterification and decrease the binding site of the cell wall to Al
3+, which is in line with the pectin modification in the mechanism of aluminum tolerance of the blueberry itself [
43]. These findings suggest that under aluminum stress, endophytic bacterium
J01 reduces aluminum accumulation in pumpkin root tips and cell walls, likely through enhancing pectin methylation levels, which consequently decreases aluminum-binding sites in the pectin matrix.
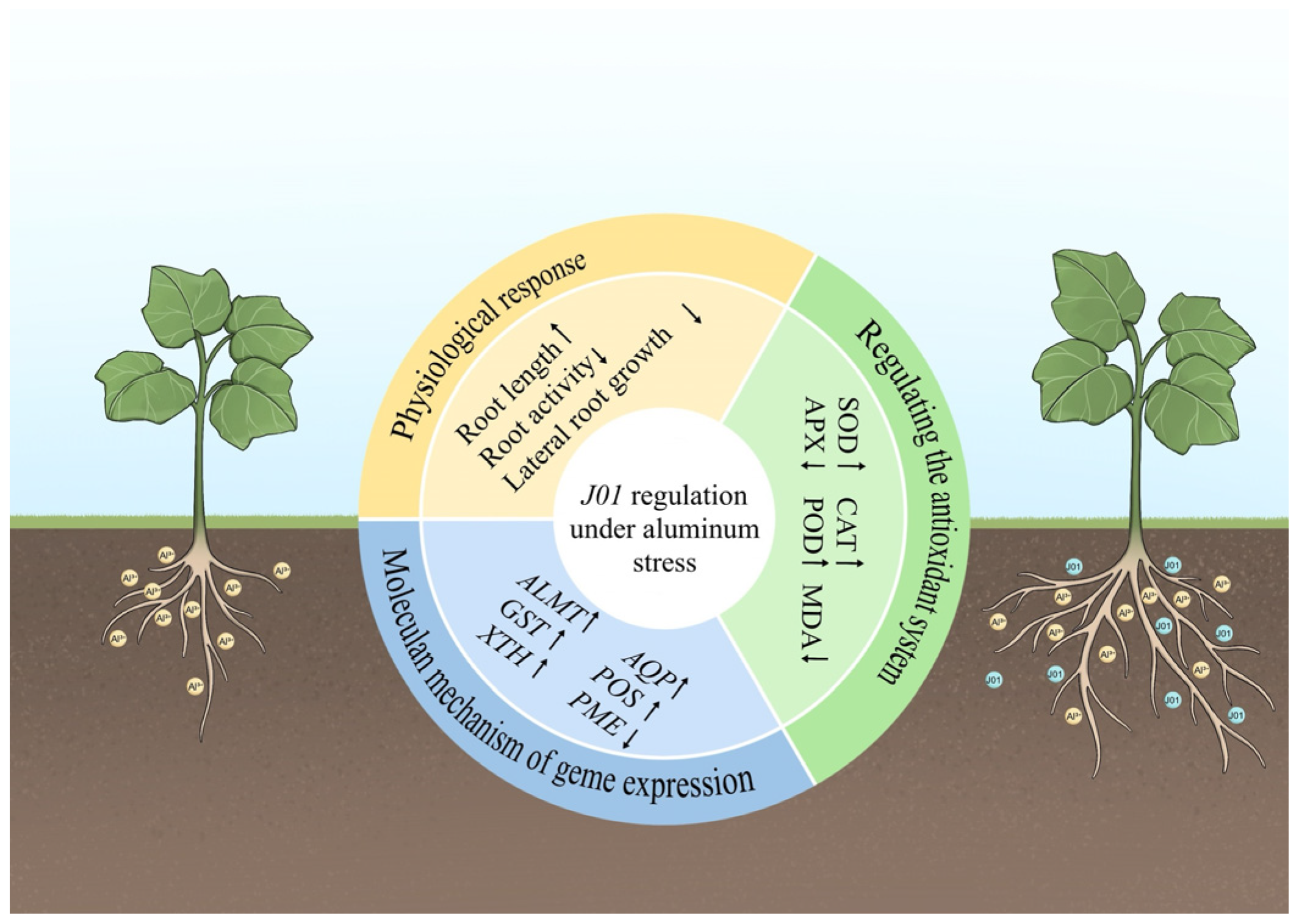
Transcriptome sequencing results indicated that J01 treatment activated phytohormone signaling (e.g., growth hormone and jasmonic acid pathway) and the phenylpropane biosynthesis pathway, with the former enhancing nutrient uptake by promoting lateral root proliferation, and the latter reinforcing cell wall fixation of aluminum through lignin deposition. However, it remains elusive which specific hormone signaling pathways and secondary metabolic pathways play dominant roles, and how these distinct hormonal pathways integrate into a coordinated regulatory network to collectively mediate aluminum tolerance in pumpkin—questions warranting further investigation. The spatiotemporal and temporal expression patterns of the differentially expressed genes further confirmed that J01 constructed a multilevel aluminum resistance regulatory network by coordinating the expression of genes related to antioxidant defense, organic acid transport, and cell wall modification.