Genetic Differentiation of Ornamental and Fruit-Bearing Prunus laurocerasus Revealed by SSR and S-Locus Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Extraction and PCR Analysis
2.3. Fragment Length Determination
2.4. Data Evaluation
3. Results and Discussion
3.1. Characterization of SSR Loci and the S-RNase Locus in Cherry Laurel Accessions
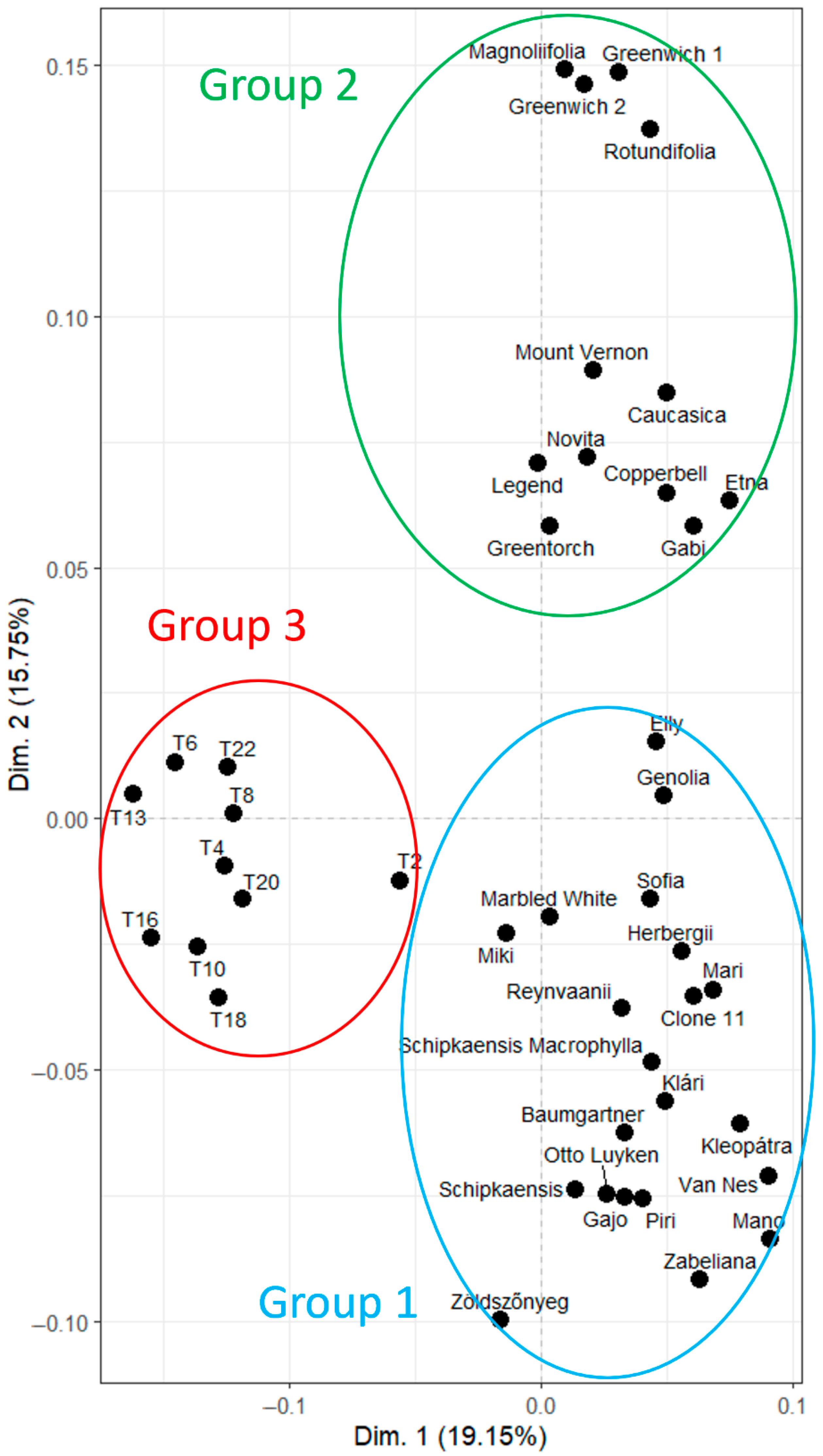
3.2. Genetic Distance of the Cherry Laurel Accessions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ercisli, S. A short review of the fruit germplasm resources of Turkey. Genet. Resour. Crop Evol. 2004, 51, 419–435. [Google Scholar] [CrossRef]
- Sulusoglu, M. The cherry laurel (Prunus laurocerasus L.) tree selection. Afr. J. Agric. Res. 2011, 6, 3574–3582. Available online: https://academicjournals.org/journal/AJAR/article-full-text-pdf/4AC6DB933771 (accessed on 8 June 2025).
- Akbulut, M.; Macit, I.; Ercisli, S.; Koc, A. Evaluation of 28 cherry laurel (Laurocerasus officinalis) genotypes in the Black Sea region, Turkey. N. Z. J. Crop Hortic. Sci. 2007, 35, 463–465. [Google Scholar] [CrossRef]
- Meurman, O. Prunus laurocerasus L., a species showing high polyploidy. J. Genet. 1929, 21, 85–94. [Google Scholar] [CrossRef]
- Zhao, L.; Jiang, X.W.; Zuo, Y.J.; Liu, X.L.; Chin, S.W.; Haberle, R.; Potter, D.; Chang, Z.Y.; Wen, J. Multiple events of allopolyploidy in the evolution of the racemose lineages in Prunus (Rosaceae) based on integrated evidence from nuclear and plastid data. PLoS ONE 2016, 11, e0157123. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, V.; Panchev, V. Effect of presowing treatment with ultrasound and stratification of Laurocerasus officinalis L. seeds on some growth behaviour of seedlings. Agro-Knowl. J. 2016, 17, 173–181. [Google Scholar] [CrossRef]
- Schmidt, G. Növények a Kertépítészetben; Mezőgazda Kiadó: Budapest, Hungary, 2003. [Google Scholar]
- Kolayli, S.; Kucuk, M.; Duran, C.; Candan, F.; Dincer, B. Chemical and antioxidant properties of Laurocerasus officinalis Roem. (cherry laurel) fruit grown in the Black Sea Region. J. Agric. Food Chem. 2003, 51, 7489–7494. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.L. Landscape Plants for Eastern North America: Exclusive of Florida and the Immediate Gulf Coast; John Wiley & Sons Inc.: New York, NY, USA, 1997; pp. 483–484. [Google Scholar]
- Schmidt, G. Növényházi Dísznövények Termesztése; Mezőgazda Kiadó: Budapest, Hungary, 2002. [Google Scholar]
- Chwil, M.; Kostryco, M.; Matraszek-Gawron, R. Comparative studies on structure of the floral nectaries and the abundance of nectar production of Prunus laurocerasus L. Protoplasma 2019, 256, 1705–1726. [Google Scholar] [CrossRef] [PubMed]
- Abrahamczyk, S.; Otto, J.; Weigend, M. The reproductive biology of the neophyte Prunus laurocerasus in Central Europe. Plant Species Biol. 2024, 40, 164–174. [Google Scholar] [CrossRef]
- Ustun, N.S.; Tosun, I. A research on composition of wild cherry laurel (Laurocerasus officinalis Roem). J. Food Technol. 2003, 1, 80–82. Available online: https://makhillpublications.co/view-article/1684-8462/jftech.2003.80.82 (accessed on 8 June 2025).
- Alasalvar, C. Cherry laurel syrup (Pekmez). In Handbook of Functional Beverages and Human Health; Watson, R.R., Preedy, V.R., Zibadi, S., Eds.; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2016; pp. 187–192. [Google Scholar]
- Alasalvar, C.; Al-Farsi, M.; Shahidi, F. Compositional characteristics and antioxidant components of cherry laurel varieties and pekmez. J. Food Sci. 2005, 70, S47–S52. [Google Scholar] [CrossRef]
- Celik, F.; Ercisli, S.; Yilmaz, S.O.; Hegedus, A. Estimation of certain physical and chemical fruit characteristics of various cherry laurel (Laurocerasus officinalis Roem.) genotypes. HortScience 2011, 46, 924–927. [Google Scholar] [CrossRef]
- Sütöriné Diószegi, M.; Schmidt, G. Prunus laurocerasus fajták télállóságának összehasonlító vizsgálata. In Növénynemesítés a Megújuló Mezőgazdaságban; Veisz, O., Ed.; A Magyar Tudományos Akadémia Agrártudományok Osztályának Növénynemesítési Tudományos Bizottsága: Budapest, Hungary, 2014; pp. 409–413. [Google Scholar]
- Van Laere, K.; Hokanson, S.C.; Contreras, R.; Van Huylenbroeck, J. Woody ornamentals of the temperate zone. In Ornamental Crops; Van Huylenbroeck, J., Ed.; Springer: Cham, Switzerland, 2018; Volume 11, pp. 803–887. [Google Scholar] [CrossRef]
- Ruter, J.M. Inducing sterility in Carolina laurel cherry using gamma irradiation. HortTechnology 2019, 29, 535–538. [Google Scholar] [CrossRef]
- Islam, A. ‘Kiraz’ cherry laurel (Prunus laurocerasus). N. Z. J. Crop Hortic. Sci. 2002, 30, 301–302. [Google Scholar] [CrossRef]
- Islam, A. A new cherry laurel cultivar: ‘Odü’. Akad. Ziraat Derg. 2024, 13, 219–226. [Google Scholar] [CrossRef]
- Gupta, P.K.; Balyan, I.S.; Sharma, P.C.; Ramesh, B. Microsatellites in plants: A new class of molecular markers. Curr. Sci. India 1996, 70, 45–54. [Google Scholar]
- Kalia, R.K.; Rai, M.K.; Kalia, S.; Singh, R.; Dhawan, A.K. Microsatellite markers: An overview of the recent progress in plants. Euphytica 2011, 177, 309–334. [Google Scholar] [CrossRef]
- Xu, Y.; Ma, R.C.; Xie, H.; Liu, J.T.; Cao, M.Q. Development of SSR markers for the phylogenetic analysis of almond trees from China and the Mediterranean region. Genome 2004, 47, 1091–1104. [Google Scholar] [CrossRef] [PubMed]
- Dirlewanger, E.; Cosson, P.; Tavaud, M.; Aranzana, M.; Poizat, C.; Zanetto, A.; Arús, P.; Laigret, F. Development of microsatellite markers in peach [Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.). Theor. Appl. Genet. 2002, 105, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Mnejja, M.; Garcia-Mas, J.; Howad, W.; Badenes, M.L.; Arús, P. Simple-sequence repeat (SSR) markers of Japanese plum (Prunus salicina Lindl.) are highly polymorphic and transferable to peach and almond. Mol. Ecol. Notes 2004, 4, 163–166. [Google Scholar] [CrossRef]
- Mnejja, M.; Garcias, J.; Howad, W.; Arús, P. Development and transportability across Prunus species of 42 polymorphic almond microsatellites. Mol. Ecol. Notes 2005, 5, 531–535. [Google Scholar] [CrossRef]
- Hajyzadeh, M.; Cavusoglu, A.; Sulusoglu, M.; Unver, T. DNA SSR fingerprinting analysis among cherry laurel (Prunus laurocerasus L.) types. J. Food Agric. Environ. 2013, 11, 630–638. [Google Scholar]
- Islam, A.; Orta, H.; Kaçar, Y.A.; Dönmez, D. Genetic diversity of cherry laurel (Laurocerasus officinalis Roemer) by SSR markers. J. Agric. Sci. 2023, 29, 239–248. [Google Scholar] [CrossRef]
- Gouta, H.; Ksia, E.; Buhner, T.; Moreno, M.A.; Zarrouk, M.; Mliki, A.; Gogorcena, Y. Assessment of genetic diversity and relatedness among Tunisian almond germplasm using SSR markers. Hereditas 2010, 147, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Makovics-Zsohár, N.; Tóth, M.; Surányi, D.; Kovács, S.; Hegedűs, A.; Halász, J. Simple sequence repeat markers reveal Hungarian plum (Prunus domestica L.) germplasm as a valuable gene resource. HortScience 2017, 52, 1655–1660. [Google Scholar] [CrossRef]
- Matsumoto, D.; Tao, R. Distinct self-recognition in the Prunus S-RNase-based gametophytic self-incompatibility system. Hortic. J. 2016, 85, 289–305. [Google Scholar] [CrossRef]
- McClure, B. S-RNase and SLF determine S-haplotype–specific pollen recognition and rejection. Plant Cell 2004, 16, 2840–2847. [Google Scholar] [CrossRef] [PubMed]
- Tao, R.; Iezzoni, A.F. The S-RNase-based gametophytic self-incompatibility system in Prunus exhibits distinct genetic and molecular features. Sci. Hortic. 2010, 124, 423–433. [Google Scholar] [CrossRef]
- Halász, J.; Molnár, A.B.; Ilhan, G.; Ercisli, S.; Hegedűs, A. Identification and molecular analysis of putative self-incompatibility ribonuclease alleles in an extreme polyploid species, Prunus laurocerasus L. Front. Plant Sci. 2021, 12, 715414. [Google Scholar] [CrossRef] [PubMed]
- Sulusoglu, M.; Cavusoglu, A. Pollination biology of cherry laurel and pollinizer effects on fruit characteristics. Yuzuncu Yıl Univ. J. Agric. Sci. 2014, 24, 280–289. [Google Scholar] [CrossRef][Green Version]
- Durul, M.S.; Unver, H. Cherry Laurel (P. laurocerasus L.) Flowering, Pollination and Fruit Set. In Proceedings of the IIIrd Balkan Agriculture Congress, Edirne, Turkey, 29 August–1 September 2021; pp. 1052–1056. [Google Scholar]
- Garkava-Gustavsson, L.; Kolodinska Brantestam, A.; Sehic, J.; Nybom, H. Molecular characterisation of indigenous Swedish apple cultivars based on SSR and S-allele analysis. Hereditas 2008, 145, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Gisbert, A.D.; Romero, C.; Martínez-Calvo, J.; Leida, C.; Llácer, G.; Badenes, M.L. Genetic diversity evaluation of a loquat (Eriobotrya japonica (Thunb) Lindl) germplasm collection by SSRs and S-allele fragments. Euphytica 2009, 168, 121–134. [Google Scholar] [CrossRef]
- Liu, C.; Qi, X.; Song, L.; Li, Y.; Li, M. Species identification, genetic diversity and population structure of sweet cherry commercial cultivars assessed by SSRs and the gametophytic self-incompatibility locus. Sci. Hortic. 2018, 237, 28–35. [Google Scholar] [CrossRef]
- Baraket, G.; Abdallah, D.; Ben Mustapha, S.; Ben Tamarzizt, H.; Salhi-Hannachi, A. Combination of simple sequence repeat, S-locus polymorphism and phenotypic data for identification of Tunisian plum species (Prunus spp.). Biochem. Genet. 2019, 57, 673–694. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, S.; Kitahara, K.; Maejima, T.; Komatsu, H. Parent-offspring relationships of apple cultivars’ Shinano Piccoro’ and ‘Komitsu’ by S-RNase and SSR markers. Acta Hort. 2007, 763, 303–308. [Google Scholar] [CrossRef]
- Sawamura, Y.; Takada, N.; Yamamoto, T.; Saito, T.; Kimura, T.; Kotobuki, K. Identification of parent-offspring relationships in 55 Japanese pear cultivars using S-RNase allele and SSR markers. Hort. J. 2008, 77, 364–373. [Google Scholar] [CrossRef][Green Version]
- Kim, H.T.; Robin, A.H.K.; Nou, I.S. Parentage confirmation of Korean bred pear cultivars by simple sequence repeat SSR genotyping and S-genotypes analysis. Plant Breed. Biotech. 2016, 4, 198–211. [Google Scholar] [CrossRef]
- Hrotkó, K.; Feng, Y.; Halász, J. Spontaneous hybrids of Prunus fruticosa Pall. in Hungary. Genet. Resour. Crop Evol. 2020, 67, 489–502. [Google Scholar] [CrossRef]
- Abrahamczyk, S.; Otto, J.; Böhnert, T.; Weigend, M. Naturalization of Prunus laurocerasus in a forest in Germany. Biol. Invasions 2024, 26, 2379–2386. [Google Scholar] [CrossRef]
- Foley, T.; Raulston, J.C. Prunus laurocerasus evaluations in the NCSU arboretum. Proc. South. Nursery Assoc. Res. Conf. 1994, 39, 364–368. [Google Scholar]
- Tóth, I. Lomblevelű Díszfák Díszcserjék Kézikönyve; Tarkavirág Kereskedelmi és Szolgáltató Kft.: Dunaharaszti, Hungary, 2012; 789p. [Google Scholar]
- Innoflora: Dr Józsa Miklós Fajtái. Available online: https://www.innoflora.hu/dr-jozsa-miklos-fajtai (accessed on 21 October 2024).
- Plantipp. Available online: https://plantipp.eu/uk/varieties (accessed on 21 October 2024).
- Safro Milan Havlis. Available online: https://www.havlis.cz (accessed on 21 October 2024).
- Pap, E. Magyar Siker a Zunderti Groot Groen Plus Kiállításon. Magyarmezőgazdaság.hu. 2019. Available online: https://magyarmezogazdasag.hu/2019/10/03/magyar-siker-zunderti-groot-groen-plus-kiallitason/ (accessed on 21 October 2024).
- Sonneveld, T.; Robbins, T.P.; Tobutt, K.R. Improved discrimination of self-incompatibility S-RNase alleles in cherry and high throughput genotyping by automated sizing of first intron polymerase chain reaction products. Plant Breed. 2006, 125, 305–307. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. iMEC: Online marker efficiency calculator. Appl. Plant Sci. 2018, 6, e01159. [Google Scholar] [CrossRef] [PubMed]
- Yeh, F.C.; Boyle, T.J.B. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg. J. Bot. 1997, 129, 157–163. [Google Scholar]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [PubMed]
- Dray, S.; Dufour, A.B. The ade4 package: Implementing the duality diagram for ecologists. J. Stat. Softw. 2007, 22, 1–20. [Google Scholar] [CrossRef]
- Gower, J.C. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 1966, 53, 325–338. [Google Scholar] [CrossRef]
- Cooke, R.S.; Eigenbrod, F.; Bates, A.E. Projected losses of global mammal and bird ecological strategies. Nat. Commun. 2019, 10, 2279. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Schliep, K.P. phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Shu, G.; Hu, Y.; Cao, G.; Wang, Y. Pattern and variation in simple sequence repeat (SSR) at different genomic regions and its implications to maize evolution and breeding. BMC Genom. 2023, 24, 136. [Google Scholar] [CrossRef] [PubMed]
- Gouta, H.; Ksia, E.; Buhner-Zaharieva, T.; Mliki, A.; Gogorcena, Y. Development of an SSR-based identification key for Tunisian local almonds. Sci. Agric. 2012, 69, 108–113. [Google Scholar] [CrossRef]
- Jalili, S.; Arzani, K.; Prudencio, A.S.; Salazar, J.A.; Martínez-García, P.J.; Bouzari, N.; Martínez-Gómez, P. Integrated morphological, physiological and molecular analysis of the drought response in cultivated and wild Prunus L. subgenera cerasus species. Plant Mol. Biol. Rep. 2023, 41, 440–453. [Google Scholar] [CrossRef]
- Halász, J.; Makovics-Zsohár, N.; Szőke, F.; Ercisli, S.; Hegedűs, A. Simple sequence repeat and S-locus genotyping to assist the genetic characterization and breeding of polyploid Prunus species, P. spinosa and P. domestica subsp. insititia. Biochem. Genet. 2021, 59, 1065–1087. [Google Scholar] [CrossRef] [PubMed]
- Urrestarazu, J.; Errea, P.; Miranda, C.; Santesteban, L.G.; Pina, A. Genetic diversity of Spanish Prunus domestica L. germplasm reveals a complex genetic structure underlying. PLoS ONE 2018, 13, e0195591. [Google Scholar] [CrossRef] [PubMed]
- Manco, R.; Chiaiese, P.; Basile, B.; Corrado, G. Comparative analysis of genomic- and EST-SSRs in European plum (Prunus domestica L.): Implications for the diversity analysis of polyploids. Biotech 2020, 10, 543. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Fickus, E.; Cregan, P. Characterization of trinucleotide SSR motifs in wheat. Theor. Appl. Genet. 2002, 104, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Gong, L.; Meng, R.; Li, S.; Dang, P.; Guo, B.; He, G. Development of trinucleotide (GGC)n SSR markers in peanut (Arachis hypogaea L.). Electron. J. Biotechnol. 2010, 13, 5–6. [Google Scholar] [CrossRef]
- Cipriani, G.; Spadotto, A.; Jurman, I.; Di Gaspero, G.; Crespan, M.; Meneghetti, S.; Testolin, R. The SSR-based molecular profile of 1005 grapevine (Vitis vinifera L.) accessions uncovers new synonymy and parentages, and reveals a large admixture amongst varieties of different geographic origin. Theor. Appl. Genet. 2010, 121, 1569–1585. [Google Scholar] [CrossRef] [PubMed]
- Sehic, J.; Garkava-Gustavsson, L.; Fernández-Fernández, F.; Nybom, H. Genetic diversity in a collection of European pear (Pyrus communis) cultivars determined with SSR markers chosen by ECPGR. Sci. Hortic. 2012, 145, 39–45. [Google Scholar] [CrossRef]
- Hong, J.H.; Chae, C.W.; Choi, K.J.; Kwon, Y.S. A database of simple sequence repeat (SSR) marker-based DNA profiles of citrus and related cultivars and germplasm. Hortic. Sci. Technol. 2016, 34, 142–153. [Google Scholar] [CrossRef]
- Serrote, C.M.L.; Reiniger, L.R.S.; Silva, K.B.; dos Santos Rabaiolli, S.M.; Stefanel, C.M. Determining the polymorphism information content of a molecular marker. Gene 2020, 726, 144175. [Google Scholar] [CrossRef] [PubMed]
- Antanyniené, R.; Šikšnianienė, J.B.; Stanys, V.; Frercks, B. Fingerprinting of plum (Prunus domestica) genotypes in Lithuania using SSR markers. Plants 2023, 12, 1538. [Google Scholar] [CrossRef] [PubMed]
- Tamarzizt, H.B.; Walker, D.; Mustapha, S.B.; Abdallah, D.; Baraket, G.; Hannachi, A.S.; Azzouzi, S.Z. DNA variation and polymorphism in Tunisian plum species (Prunus spp.): Contribution of flow cytometry and molecular markers. Genet. Mol. Res. 2015, 14, 18034–18046. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, B.H. DNA fingerprints of 51 sweet and sour Prunus accessions using simple sequence repeats. J. Hortic. Sci. Biotechnol. 2006, 81, 118–124. [Google Scholar] [CrossRef]
- Arismendi, M.J.; Hinrichsen, P.; Almada, R.; Pimentel, P.; Pinto, M.; Sagredo, B. Characterization of genetic diversity of stone fruit rootstocks used in Chile by means of microsatellite markers. J. Am. Soc. Hortic. Sci. 2012, 137, 302–310. [Google Scholar] [CrossRef]
- Urbanovich, O.Y.; Kuzmitskaya, P.V.; Kilchevsky, A.V. Identification and genetic diversity of plum cultivars grown in Belarus. Russ. J. Genet. 2017, 53, 775–784. [Google Scholar] [CrossRef]
- Hayashi, K.; Shimazu, K.; Yaegaki, H.; Yamaguchi, M.; Iketani, H.; Yamamoto, T. Genetic diversity in fruiting and flower-ornamental Japanese apricot (Prunus mume) germplasms assessed by SSR markers. Breed. Sci. 2008, 58, 401–410. [Google Scholar] [CrossRef]
- Hasnaoui, N.; Buonamici, A.; Sebastiani, F.; Mars, M.; Zhang, D.; Vendramin, G.G. Molecular genetic diversity of Punica granatum L. (pomegranate) as revealed by microsatellite DNA markers (SSR). Gene 2012, 493, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Gill, K.; Kumar, P.; Sharma, V.; Negi, S.; Sharma, M.; Sharma, R.; Joshi, A.K. Unraveling phenotypic ambiguities of kagzi and ornamental lime accessions: A comprehensive exploration through morpho-biochemical and DNA marker profiling. Genet. Resour. Crop Evol. 2024, 71, 3765–3790. [Google Scholar] [CrossRef]
- Schulze, J.A.; Contreras, R.N.; Scagel, C.F. Comparing vegetative propagation of two ‘Schipkaensis’ common cherry laurel ploidy levels. HortTechnology 2017, 27, 69–72. [Google Scholar] [CrossRef]
- Dirr, M.A. Manual of Woody Landscape Plants: Their Identification, Ornamental Characteristics, Culture, Propagation and Uses; Stipes Pub. Co.: Champaign, IL, USA, 1990; pp. 654–656. [Google Scholar]
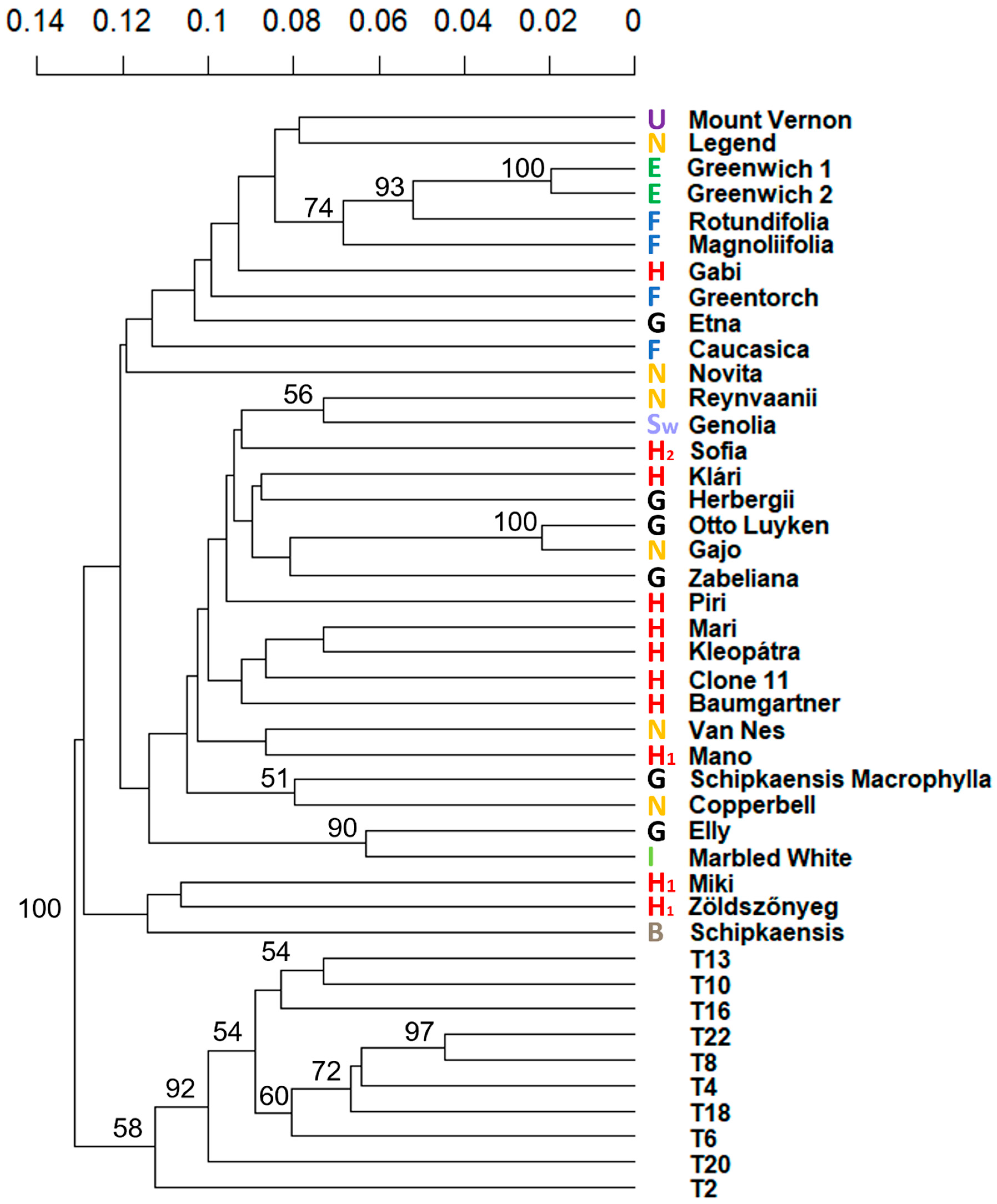
| Name | Type a | Sample Collection Site b | Origin | Breeder (Release Year) |
|---|---|---|---|---|
| Clone 11 | O | Sz | Hungary | Miklós Józsa |
| ‘Baumgartner’ | O | Sz | Hungary | Géza Baumgartner and Miklós Józsa (1990) |
| ‘Caucasica’ | O | Sz | France | n.d. (1852) |
| ‘Copperbell’ | O | Sz | The Netherlands | Frans Muysers Boomkwekerijen vof |
| ‘Elly’ | O | Sz | Germany | Adrian Straver, Straver Gbr |
| ‘Etna’ | O | Sz | Germany | Adrian Straver, Straver Gbr |
| ‘Gabi’ | O | Sz | Hungary | Miklós Józsa (2007) |
| ‘Gajo’ | O | Sz | The Netherlands | Wilhelmus P.C. Nouws (1996) |
| ‘Genolia’ | O | Sz | Switzerland | Pépinières de Genolier (2002) |
| ‘Greentorch’ | O | Sz | France | Pépinières Minier (2009) |
| Greenwich 1 | O | ROG | England | n.d. |
| Greenwich 2 | O | ROG | England | n.d. |
| ‘Herbergii’ | O | Sz | Germany | Herberg (1930) |
| ‘Klári’ | O | Sz | Hungary | Miklós Józsa (1998) |
| ‘Kleopátra’ | O | Sz | Hungary | Miklós Józsa (2007) |
| ‘Legend’ | O | Sz | The Netherlands | Guido Rouwette |
| ‘Magnoliifolia’ | O | BA | France | n.d. (1869) |
| ‘Mano’ | O | Sz | Hungary | Elemér Barabits (2004) |
| ‘Marbled White’ | O | BA | Ireland | n.d. (1811) |
| ‘Mari’ | O | Sz | Hungary | Miklós Józsa (1989) |
| ‘Miki’ | O | BA | Hungary | Elemér Barabits (2004) |
| ‘Mount Vernon’ | O | Sz | USA | Wells Nursery (1967) |
| ‘Novita’ | O | Sz | The Netherlands | n.d. |
| ‘Otto Luyken’ | O | Sz | Germany | Hermann Albrecht Hesse (1940) |
| ‘Piri’ | O | Sz | Hungary | Miklós Józsa (1989) |
| ‘Reynvaanii’ | O | Sz | The Netherlands | A.J. Reynvaan (1913) |
| ‘Rotundifolia’ | O | Sz | France | L.C.B.Billard, Barre (1865) |
| ‘Schipkaensis’ | O | BA | Bulgaria | Franz Ludwig Späth (1889) |
| ‘Schipkaensis Macrophylla’ | O | Sz | Germany | G. D. Böhlje (1940) |
| ‘Sofia’ | O | Sz | Hungary | Gábor Németh |
| ‘Van Nes’ | O | Sz | The Netherlands | P. van Nes (1935) |
| ‘Zabeliana’ | O | Sz | Germany | Franz Ludwig Späth (1898) |
| ‘Zöldszőnyeg’ | O | BA | Hungary | Elemér Barabits Jr. (2004) |
| T2 | F | BSR | Türkiye | selection |
| T4 | F | BSR | Türkiye | selection |
| T6 | F | BSR | Türkiye | selection |
| T8 | F | BSR | Türkiye | selection |
| T10 | F | BSR | Türkiye | selection |
| T13 | F | BSR | Türkiye | selection |
| T16 | F | BSR | Türkiye | selection |
| T18 | F | BSR | Türkiye | selection |
| T20 | F | BSR | Türkiye | selection |
| T22 | F | BSR | Türkiye | selection |
| Primer Name | Sequence (5′→3′) | Amplicon Size in the Original Study (bp) | Linkage Group | Repeat Motif | Reference |
|---|---|---|---|---|---|
| ASSR63 | F: CACCAATTTATGTTGCAAGATTATATG R: GTTTTAGATTTCACAGTACTATG | 154 | G8 | (GAT) 5 | [24] |
| BPTCT 007 | F: TCATTGCTCGTCATCAGC R: CAGATTTCTGAAGTTAGCGGTA | 149 | G3 | (AG) 22(CG) 2(AG) 4 | [25] |
| BPPCT 025 | F: TCCTGCGTAGAAGAAGGTAGC R: CGACATAAAGTCCAAATGGC | 197 | G6 | (GA) 29 | [25] |
| BPPCT 037 | F: CATGGAAGAGGATCAAGTGC R: CTTGAAGGTAGTGCCAAAGC | 155 | G5 | (GA) 25 | [25] |
| BPPCT 039 | F: ATTACGTACCCTAAAGCTTCTGC R: GATGTCATGAAGATTGGAGAGG | 154 | G3 | (GA) 20 | [25] |
| BPPCT 040 | F: ATGAGGACGTGTCTGAATGG R: AGCCAAACCCCTCTTATACG | 135 | G4 | (GA) 14 | [25] |
| CPSCT018 | F: AGGACATGTGGTCCAACCTC R: GGGTTCCCCGTTACTTTCAT | 162 | G8 | (CA) 5 (CT) 20 | [26] |
| CPSCT 021 | F: GCCACTTCGGCTAAAAGAGA R: TCCATATCTCCTCCTGCTTGA | 139 | G2 | (GA) 15 | [26] |
| CPDCT 044 | F: ACATGCCGGGTAATTAGCAA R: AAAATGCACGTTTCGTCTCC | 175 | G2 | (GA) 15 | [27] |
| CPDCT 045 | F: TGTGGATCAAGAAAGAGAACCA R: AGGTGTGCTTGCACATGTTT | 142 | G4 | (GA) 16 | [27] |
| PaConsI | F: MCTTGTTCTTGSTTTYGCTTTCTTC R2: GCCATTGTTGCACAAATTGA | 234–458 | G6 | - | [53] |
| Na | ANa | Na/Accession | Size Range (bp) | Gu | PIC | Rp | Dp | |
|---|---|---|---|---|---|---|---|---|
| ASSR 63 | 5 | 3.44 | 2–4 | 156–167 | 9 | 0.19 | 1.95 | 0.53 |
| BPPCT007 | 27 | 7.58 | 4–12 | 99–192 | 41 | 0.20 | 9.77 | 0.92 |
| BPPCT025 | 52 | 9.67 | 5–13 | 147–268 | 42 | 0.24 | 18.84 | 0.96 |
| BPPCT037 | 46 | 9.49 | 7–14 | 106–231 | 41 | 0.23 | 12.93 | 0.96 |
| BPPCT039 | 65 | 12.21 | 7–17 | 100–242 | 42 | 0.24 | 21.07 | 0,97 |
| BPPCT040 | 38 | 8.02 | 5–12 | 113–170 | 43 | 0.23 | 13.07 | 0.96 |
| CPDCT044 | 77 | 6.88 | 1–15 | 105–283 | 41 | 0.28 | 13.40 | 0.99 |
| CPDCT045 | 23 | 6.07 | 3–9 | 117–159 | 37 | 0.21 | 6.84 | 0.93 |
| CPSCT018 | 18 | 2.33 | 1–5 | 101–162 | 29 | 0.26 | 4.33 | 0.86 |
| CPSCT021 | 58 | 11.02 | 8–15 | 117–185 | 43 | 0.24 | 19.63 | 0.96 |
| S-RN-ase 1st intron | 89 | 11.49 | 2–18 | 193–536 | 42 | 0.26 | 21.35 | 0.98 |
| Mean | 45.27 | 8.30 | 37 | 0.23 | 13.02 | 0.91 |
| Number of Accessions | Total Allele Number | Average Allele Number | Number of Unique Alleles | Average Number of Unique Alleles | Polymorphic Alleles (%) | |
|---|---|---|---|---|---|---|
| Group 1 | 21 | 371 | 18 | 92 | 4.4 | 73.5 |
| Group 2 | 12 | 302 | 25 | 41 | 3.4 | 59.2 |
| Group 3 | 10 | 243 | 24 | 68 | 6.8 | 48.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hegedűs, A.; Honfi, P.; Ercisli, S.; Ilhan, G.; Tóth, E.G.; Halász, J. Genetic Differentiation of Ornamental and Fruit-Bearing Prunus laurocerasus Revealed by SSR and S-Locus Markers. Horticulturae 2025, 11, 854. https://doi.org/10.3390/horticulturae11070854
Hegedűs A, Honfi P, Ercisli S, Ilhan G, Tóth EG, Halász J. Genetic Differentiation of Ornamental and Fruit-Bearing Prunus laurocerasus Revealed by SSR and S-Locus Markers. Horticulturae. 2025; 11(7):854. https://doi.org/10.3390/horticulturae11070854
Chicago/Turabian StyleHegedűs, Attila, Péter Honfi, Sezai Ercisli, Gulce Ilhan, Endre György Tóth, and Júlia Halász. 2025. "Genetic Differentiation of Ornamental and Fruit-Bearing Prunus laurocerasus Revealed by SSR and S-Locus Markers" Horticulturae 11, no. 7: 854. https://doi.org/10.3390/horticulturae11070854
APA StyleHegedűs, A., Honfi, P., Ercisli, S., Ilhan, G., Tóth, E. G., & Halász, J. (2025). Genetic Differentiation of Ornamental and Fruit-Bearing Prunus laurocerasus Revealed by SSR and S-Locus Markers. Horticulturae, 11(7), 854. https://doi.org/10.3390/horticulturae11070854









