Abstract
In this study, the molecular mechanism underlying melanin synthesis within the fermentation broth of Auricularia heimuer (A. heimuer) was explored. The absorbance at 500 nm, melanin yield, and tyrosinase activity in the fermentation broth were assessed across different fermentation durations. Mycelia samples were gathered on the 4th (R1), 8th (R2), and 10th (R3) days of liquid fermentation for transcriptome sequencing. As fermentation progressed, the absorbance, melanin yield, and tyrosinase activity in the broth rose. A total of 5915 differentially expressed genes (DEGs) were detected, with 1136 DEGs between R2 and R1, 3717 between R3 and R1, and 2950 between R3 and R2. Gene Ontology (GO) analysis showed that DEGs were significantly enriched in oxidoreductase activity and ribosome structural constituent terms. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed significant enrichment of DEGs in ribosome and amino acid metabolism pathways. Key DEGs, including transcription factors, glycosidases, P450 enzymes, laccases, and glutamate dehydrogenase, were identified during melanin production in the fermentation broth. These DEGs may play crucial roles in melanin synthesis. This study offers a foundation for further exploring the melanin synthesis mechanism in A. heimuer fermentation broth.
1. Introduction
Auricularia heimuer (A. heimuer) [1], commonly known as black wood ear and wood jellyfish, is a traditional edible and medicinal mushroom with high nutritional and pharmacological value. It is rich in various bioactive compounds [2,3,4,5], such as melanin, polysaccharides, and adenosine, which possess multiple biological functions, including antioxidant [6], lipid-lowering [7], and anti-aging properties [8]. As one of the five types of black foods, A. heimuer is an important source for melanin preparation.
Melanin is a class of chemically complex, heterogeneous polymers of phenolic or indolic substances. It is one of the most abundant natural pigments in nature, appearing black or brown and widely distributed in animals, plants, and microorganisms [9]. It possesses various biological activities, including antiviral [10], antibacterial [11], radioprotective [12], and antioxidant properties [13]. As a secondary metabolite, melanin is not essential for the growth and development of organisms. However, it can effectively enhance their survival and competitive abilities. Its widespread presence is thus considered the result of long-term adaptation between life and the environment in the evolutionary process [14]. Melanin from A. heimuer can be extracted from the fruiting bodies of A. heimuer. However, artificial cultivation of A. heimuer has issues, such as long production cycles, high labor intensity, and yields and quality being affected by climate and other factors. These problems restrict the application and commercialization of A. heimuer melanin. Our team’s previous research found that melanin from A. heimuer can also be obtained using liquid fermentation technology. Compared with traditional fruiting body cultivation, liquid fermentation has the advantages of short cultivation cycles, convenient inoculation, and suitability for large-scale production. Currently, many researchers have conducted related research on A. heimuer melanin. They mainly focused on the extraction, physicochemical properties, and biological functions of A. heimuer melanin. However, there has been little research on A. heimuer melanin synthesis and metabolism [15,16].
At the molecular level, the mechanism of melanin synthesis is still not fully understood. Fan [17] confirmed that G11815 (glutaminyl transferase) is a key enzyme gene for pigment synthesis in Auricularia auricula-judae. Du et al. [18] found that genes related to melanin synthesis, such as tyrosinase, polyketide synthase, and laccase, are also closely associated with the color-changing process during the cultivation of Pleurotus ostreatus. There are relatively few reports on genes related to melanin synthesis in Auricularia polytricha. Wang et al. [19] discovered that dopachrome tautomerase is an important rate-limiting enzyme in the melanin synthesis pathway and conducted cloning and bioinformatics analysis of the dopachrome tautomerase gene.
Transcriptomics is a means of studying the transcription of genes and the regulatory mechanisms of transcription at the cellular level on a holistic scale. It primarily investigates gene expression at the RNA level, and transcriptional regulation is one of the most widespread forms of biological regulation known to date [20]. As a holistic research approach, transcriptomics can provide comprehensive expression information for specific cells or tissues under particular conditions. By comparing this information with the genomes of model organisms, it is possible to infer the functions of unknown genes and further elucidate the mechanisms of their regulatory factors. Most studies on the genes related to melanin synthesis in A. heimuer have focused on the fruiting bodies, with fewer molecular studies on liquid fermentation. Qiu et al. [21] have revealed the mechanism underlying light intensity-induced melanin synthesis of A. heimuer through transcriptome analysis.
In this study, to examine the changing trends of physiological and biochemical indicators and the genes related to melanin synthesis during the liquid fermentation of A. heimuer, A. heimuer strain 1703, selected as the experimental strain, was used for liquid fermentation experiments. The color and tyrosinase activities of the fermentation broth under different fermentation times were measured. Transcriptome sequencing was used to explore genes related to A. heimuer melanin synthesis. The findings contribute to a broader understanding of the mechanisms of melanin synthesis in A. heimuer under different times.
2. Materials and Methods
2.1. Auricularia Heimuer Strain
The A. heimuer strain 1703 used in this study was isolated from a wild strain that was collected in the suburbs of Harbin City. It was preserved in a cryovial containing PDA medium in the Institute of Microbiology, Heilongjiang Academy of Sciences, and a partial ITS sequence has been deposited in GenBank as accession number OR527936.1. The pure culture was inoculated on potato dextrose agar (PDA, 200 g·L−1 diced potatoes, 20 g·L−1 glucose, and 18 g·L−1 agar) medium at 25 °C.
2.2. Physiological Assay
The A. heimuer strain 1703 was inoculated into a 250 mL conical flask containing 100 mL of liquid medium. The liquid medium consisted of 200 g·L−1 potato, 21.40 g·L−1 sucrose, 5.34 g·L−1 peptone, 0.56 g·L−1 MgSO4, with natural pH, based on Yao et al. [22]. The fermentation broth was cultured at 25 °C and 160 r/min in the dark for 10 days in a constant temperature shaker. The fermentation broth was centrifuged at 10,000 rpm for 10 min, and the supernatant was collected every day. The absorbance of the supernatant at 500 nm was measured using a UV757CRT UV-Vis spectrophotometer (Tianmei Shanghai, Shanghai, China). In addition, the tyrosinase activities of the supernatant were determined using a tyrosinase activity assay kit (Solrbio, Beijing, China) according to the manufacturer’s instructions.
2.3. Transcriptome Analysis
The mycelia of A. heimuer strain 1703 cultured on liquid media at 25 °C were collected on the 4th day (R1), the 8th day (R2), and the 10th day (R3) by centrifugation at 10,000 rpm for 10 min. The mycelia under three different times were taken for three biological replicates, which were frozen in liquid nitrogen and stored at −80 °C for further use. Total RNA was extracted using the Omega Fungal RNA Kit (Cambridge, MA, USA) according to the manufacturer’s instructions. The total RNA was assessed by 1% agarose gel electrophoresis. The purity of total RNA was evaluated using a microvolume spectrophotometer (Kairo, Beijing, China), with an acceptable standard of OD260/OD280 ≥ 1.8. After passing the quality control, cDNA libraries were constructed, and RNA-Seq was performed by OE Biotech, Inc., Shanghai, China.
DESeq2 software 1.46.0 was employed to normalize the counts of genes in each sample (using BaseMean values to estimate expression levels), calculate fold changes, and perform significance tests using the negative binomial (NB) distribution. Differential protein-coding genes were screened based on fold changes and significance test results. The criteria for identifying differentially expressed genes (DEGs) were set as p-value < 0.05 and fold change > 2. Following the identification of DEGs, GO (Gene Ontology) functional enrichment analysis and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis were conducted.
2.4. Real-Time-Quantitative PCR (RT-qPCR) Analysis
Seven DEGs from the transcriptome sequencing were randomly selected for RT-qPCR analysis to verify the reliability of the transcriptome data. The RT-qPCR primers were designed using Primer Premier 5 software and synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) (Table 1). The total RNA samples were reverse-transcribed into cDNA using the TaKaRa Reverse Transcription Kit (Takara Biotechnology Co., Ltd., Dalian, China). The 18s rRNA gene was used as the internal reference. The RT-qPCR amplifications were performed using the 2 × Universal Blue SYBR Green qPCR Master Mix (Servicebio, Wuhan, China) and the Applied Biosystems 7500 Real-Time PCR System (Foster City, CA, USA). Each sample was set up in triplicate for biological replication. The relative gene expression levels were calculated using the 2−∆∆CT method. [23,24].

Table 1.
RT-qPCR primers.
2.5. Statistical Analysis
All statistics were evaluated using SPSS software (version 20, IBM Corp., Armonk, NY, USA). The data normality and homogeneity of variance were checked. Data were presented as mean ± SD.
3. Results
3.1. Physiological Indicators
The color of the A. heimuer fermentation broth, the absorbance, the melanin yield, and the tyrosinase activities of A. heimuer supernatant are shown in Figure 1. The results indicate that the color of the fermentation broth gradually deepened with the extension of fermentation time. The color changes of the fermentation broth were not obvious from the 1st day to the 4th day, and it turned from yellow to black on the 9th day of fermentation. The absorbance of A. heimuer supernatant at 500 nm increased with the extension of fermentation time. The absorbance of A. heimuer supernatant began to increase after the 4th day of fermentation. After 10 days of fermentation, the absorbance of A. heimuer supernatant remained almost unchanged. The melanin yield varied in a manner basically consistent with the changes in absorbance. The yield of melanin began to increase after the 4th day of fermentation. After 10 days of fermentation, the yield of melanin remained almost unchanged. The tyrosinase activities of A. heimuer supernatant increased with the extension of fermentation time. The tyrosinase activities began to rise after 4 days of fermentation and showed a slight increase but no significant change after 8 days of fermentation. Therefore, the mycelia from the fermentation broth on the 4th day, 8th day, and 10th day were selected for subsequent experiments.
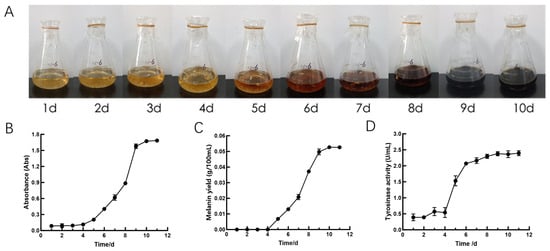
Figure 1.
The physiological indicators of A. heimuer fermentation broth: (A) Change in coloration in fermentation broth. (B) Absorbance of A. heimuer supernatant. (C) Melanin yield of A. heimuer supernatant. (D) Tyrosinase activities of A. heimuer supernatant.
3.2. RNA-Seq Data Quality Statistics
Based on physiological indicators, mycelia on the 4th day (R1), the 8th day (R2), and the 10th day (R3) were selected as materials for transcriptome sequencing. Through the sequencing platform, a large amount of paired-end sequencing data was obtained for the samples. Considering the impact of data error rates on the results, fastp software (version 0.20.1) was used to perform quality preprocessing on the raw data, and the number of reads throughout the quality control process was statistically summarized (Table 2). The average number of raw bases for all samples was >6 G, and the number of clean bases was also >6 G. After filtering, the base count for all samples in this experiment was >6 G, indicating sufficient sequencing depth to meet the requirements for subsequent data analysis. The Q30 values for all samples were greater than 95%, indicating that the base quality of the sequencing results was satisfactory and that the sequencing results for the nine samples were stable.

Table 2.
Data filtering analysis results.
3.3. DEGs Analysis
To further explore the molecular mechanisms underlying melanin synthesis in the fermentation broth of A. heimuer, transcriptomic sequencing was employed to identify genes potentially involved in melanin synthesis. Samples from the 4th day (R1), 8th day (R2), and 10th day (R3) were compared pairwise. The results showed that a total of 5915 differentially expressed genes (DEGs) were finally identified. A total of 2081 DEGs were identified compared to R2 and R1, including 1136 upregulated DEGs (54.59%, 1136/2081) and 945 downregulated DEGs (45.41%, 945/2081). A total of 3717 DEGs were detected compared to R3 and R1, including 1824 upregulated DEGs (49.07%, 1824/3717) and 1893 downregulated DEGs (50.93%, 1893/3717). A total of 2950 DEGs were detected compared to R3 and R2, including 1361 upregulated DEGs (46.13%, 1361/2950 and 1598 downregulated DEGs (54.17%, 1598/2950). A total of 289 DEGs were shared among R1, R2, and R3 (Figure 2).
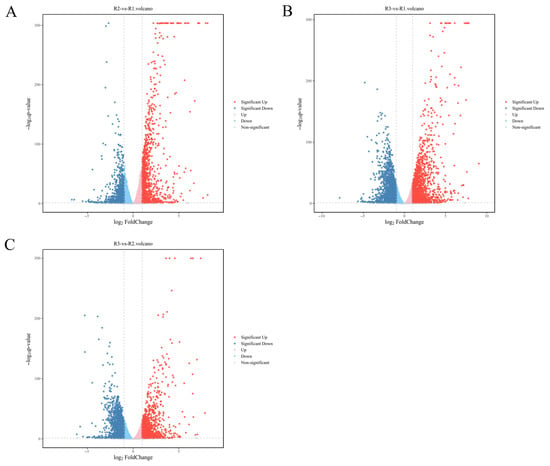
Figure 2.
Statistical analysis of DEGs in A. heimuer melanin synthesis at different fermentation times. (A) DEGs between R2 and R1. (B) DEGs between R3 and R1. (C): DEGs between R3 and R2.
3.4. GO and KEGG Classification
GO analysis was applied in accordance with an international standardized gene functional classification system to gain an in-depth comprehension of the functional classification of annotated genes. The GO enrichment analysis and functional annotation of DEGs during the melanin production process of A. heimuer were conducted at three time points. The successfully annotated DEGs were categorized into the three major classes of GO: Biological Process (BP), Cellular Component (CC), and Molecular Function (MF) (Figure 3, Green represents Biological Process, orange represents Cellular Component, and blue represents Molecular Function.). The GO enrichment analysis between R2 and R1 revealed that DEGs were significantly enriched in subcategories such as oxidoreductase activity, protein aggregate center, unfolded protein binding, and aminoacyl-tRNA editing activity (Figure 3A). The GO enrichment analysis between R3 and R1 showed that DEGs were significantly enriched in subcategories related to cytoplasmic translation, cytoplasmic large ribosomal subunit, and structural constituent of ribosome (Figure 3B). The GO enrichment analysis between R3 and R2 indicated that DEGs were significantly enriched in subcategories such as cytoplasmic translation, cytosolic small ribosomal subunit, cytosolic large ribosomal subunit, structural constituent of ribosome, and oxidoreductase activity (Figure 3C). Through GO enrichment analysis, it was found that the enriched functional categories in the melanin production process of A. heimuer are mainly related to oxidoreductase activity and structural constituent of ribosome.
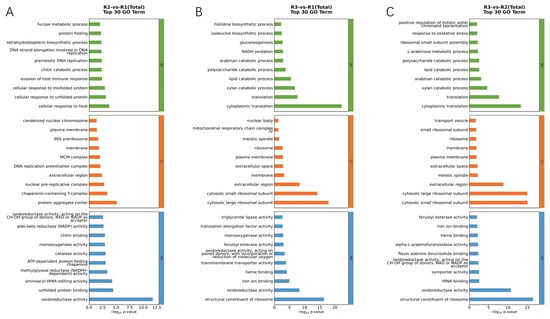
Figure 3.
Histogram of GO pathways enriched by DEGs: (A) GO analysis of DEGs between R2 and R1. (B) GO analysis of DEGs between R3 and R1. (C) GO analysis of DEGs between R3 and R2.
An enriched pathway analysis of DEGs was performed to further understand the biochemical pathways. The top 20 enriched pathways from KEGG enrichment analysis in each comparison at each developmental stage are displayed in Figure 4 (The color indicates the significance level of enrichment. The darker the color, the smaller the p-value, which means higher significance of enrichment. The size of the bubble represents the number of differentially expressed genes or metabolites enriched in a particular pathway. The larger the bubble, the more differentially expressed genes or metabolites are included in that pathway). The representative KEGG pathways included between R2 and R1 revealed significant enrichment in pathways such as Aminoacyl-tRNA biosynthesis, Mismatch repair, Homologous recombination, Valine, leucine, and isoleucine biosynthesis, DNA replication, and Nucleotide excision repair (Figure 4A). The representative KEGG pathways included between R3 and R1 showed significant enrichment in pathways including ribosome; glycolysis/gluconeogenesis; valine, leucine, and isoleucine biosynthesis; pyruvate metabolism; alanine, aspartate, and glutamate metabolism; and arginine biosynthesis. The representative (Figure 4B) KEGG pathways included between R3 and R2 indicated significant enrichment in pathways such as ribosome; glycine, serine, and threonine metabolism; pyruvate metabolism; steroid biosynthesis; glycosphingolipid biosynthesis-globo and isoglobo series; and phenylalanine, tyrosine, and tryptophan biosynthesis (Figure 4C). Through KEGG enrichment analysis, it was found that the main metabolic pathways enriched in the melanin production process of A. heimuer are ribosome and amino acid metabolism.
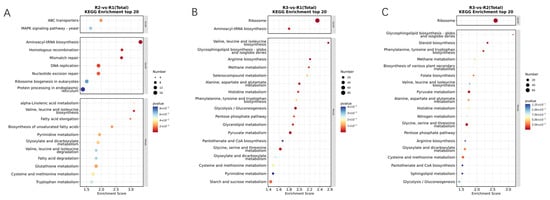
Figure 4.
Bubble chart of top 20 KEGG pathways enriched by DEGs: (A) KEGG analysis of DEGs between R2 and R1. (B) KEGG analysis of DEGs between R3 and R1. (C) KEGG analysis of DEGs between R3 and R2.
3.5. Transcriptomic Responses to Melanin Synthesis
3.5.1. Analysis of Transcription Factor-Related Genes
Transcription factors (TF) are a class of protein molecules that can specifically bind to the promoter regions or upstream specific sequences of genes. They regulate gene transcription by modulating the activity of RNA polymerase, thereby either promoting or inhibiting transcription. TF play a central role in gene expression regulation and are crucial for the growth, development, and environmental response of fungi [25]. In the analysis of DEGs between R2 and R1, the transcription factor RIM101 (gene-03035) was found to be upregulated by 1.2-fold. In the comparison between R3 and R1, the transcription factor CON7 (gene-06153) was upregulated by 1.0-fold, while transcription factor hapX (gene-03008) was downregulated by 2.7-fold. In the comparison between R3 and R2, RIM101 (gene-03035) was upregulated by 1.3-fold, and hapX (gene-03008) was downregulated by 2.0-fold.
3.5.2. Analysis of Glycosidase-Related Genes
Glycosidases (GH) are a class of enzymes that hydrolyze glycosidic bonds and are widely present in almost all living organisms. They play significant roles in carbohydrate metabolism and the hydrolysis and synthesis of glycoconjugates. In the analysis of DEGs between R2 and R1, glycoside hydrolase family 18 members CHI1 (gene-01307, gene-07291) and chiB1 (gene-09531) were significantly upregulated. Other glycosidases (gene-02294, gene-04255), ARB-02077 (gene-03408), and An02g11890 (gene-04961) were also upregulated. In the comparison between R3 and R1, CHI1 (gene-01307, gene-07291, gene-15507) were significantly upregulated. Other glycosidases exgB (gene-02294, gene-06850, gene-09512) and An02g11890 (gene-04961, gene-09587) were also upregulated. In the comparison between R3 and R2, CHI1 (gene-15507) was significantly upregulated. For other glycosidases, four exgB genes were identified, with two genes (gene-06850, gene-09512) upregulated and two genes (gene-04189, gene-04255) downregulated. An02g11890 (gene-04961, gene-06649) were both upregulated.
3.5.3. Analysis of Metabolism-Related Genes
Cytochrome P450 (CYP450) is a large superfamily of enzymes, with heme as a prosthetic group, involved in the oxidative degradation of both endogenous and exogenous compounds [26]. In the DEGs between R2 and R1, R3 and R1, and R3 and R2, 20, 50, and 32 genes were annotated as cytochrome P450, respectively, with most of these genes showing upregulated expression.
Laccase is a polyphenol oxidase that oxidizes phenolic substrates to quinone intermediates through single-electron oxidation. These intermediates subsequently undergo self-coupling or cross-coupling reactions to form complex polymeric products. In the DEGs between R2 and R1, lcc2 (gene-08210, gene-10345) and lcc3 (gene-09500) were both downregulated. In the DEGs between R3 and R1, lcc2 (gene-08210) and lcc3 (gene-09500) were downregulated. In the DEGs between R3 and R2, lcc2 (gene-08210) was downregulated. Studies have shown that laccase promotes melanin formation through the oxidative polymerization of phenolic substrates [27]. However, the laccase genes screened in this study were all downregulated.
Secondary metabolism regulator LaeA (CC1G_00498) is a global regulator involved in the biosynthesis of secondary metabolites in fungi [27,28,29]. In the DEGs between R2 and R1, CC1G_00498 (gene-00760, gene-04512, gene-07190) was downregulated. Similarly, in the DEGs between R3 and R1, CC1G_00498 (gene-04512, gene-07190) was also downregulated.
3.5.4. Analysis of Genes Related to Growth and Development
Serine/threonine protein kinase (srk1) is an enzyme that catalyzes the transfer of phosphate groups to serine or threonine residues on proteins, regulating fungal growth, development, and sexual reproduction [30,31]. During melanin production, srk1 (gene-16234) was significantly upregulated in the comparison between R2 and R1, with a 2.1-fold increase. Similarly, srk1 (gene-16234) was upregulated by 2.3-fold in the comparison between R3 and R1, and by 1.2-fold in the comparison between R3 and R2.
Tyrosine protein phosphatase (Yvh1) primarily regulates ribosome biogenesis and cellular stress responses in fungi. During melanin production, Yvh1 genes (gene-07078, gene-08992, gene-16295) were downregulated in the comparison between R2 and R1. In the comparison between R3 and R1, Yvh1 (gene-10614) was upregulated, while Yvh1 (gene-07078, gene-08992, gene-13991, gene-16295) were downregulated. In the comparison between R3 and R2, Yvh1 (gene-10614) was upregulated, and Yvh1 (gene-16295) was downregulated. This indicates that the expression levels of Yvh1 vary at different fermentation times, and the synthesis of melanin under different fermentation times is regulated by different genes.
3.5.5. Analysis of Nitrogen Utilization-Related Genes
Glutamate dehydrogenase (GLDH or GDH) is an important mitochondrial enzyme that catalyzes the interconversion between glutamate and α-ketoglutarate, while reducing NAD(P)+ to NAD(P)H. It is mainly involved in amino acid metabolism and energy metabolism processes. In the DEGs between R2 and R1, gdh-1 (gene-02256) was significantly upregulated, with an increase of 2.5-fold. In the DEGs between R3 and R1, gdh-1 (gene-02256) was also significantly upregulated.
NmrA-like proteins are a class of negative transcriptional regulators that are primarily involved in nitrogen metabolism regulation. They interact with nitrogen metabolism-related enzymes or transcription factors to regulate the absorption, assimilation, and utilization of nitrogen. In the DEGs between R2 and R1, 14 genes were annotated as NmrA-like proteins, with 11 upregulated and 3 downregulated. In the DEGs between R3 and R1, 13 genes were annotated as NmrA-like proteins, with 9 upregulated and 4 downregulated. In the DEGs between R3 and R2, 18 genes were annotated as NmrA-like proteins, with 3 upregulated and 15 downregulated.
3.6. RT-qPCR Verification of DEGs
To verify the reliability of the transcriptome sequencing results, a total of seven DEGs were selected from the transcriptome data to assess the relative expression levels through RT-qPCR on the 4th day, 8th day, and 10th day during the liquid fermentation of A. heimuer. The validated DEGs included aes1 (gene-3653), CYP075 (gene-04749), srk1 (gene-16234), yvh1 (gene-16295), ustYa (gene-14060), mdlA (gene-06647), and adh-1 (gene-08645). The results demonstrated that relative expression levels of these seven DEGs detected by RT-qPCR on the 4th day, 8th day, and 10th day were consistent with the transcriptome sequencing data (Figure 5), indicating that the transcriptomic sequencing data are reliable.
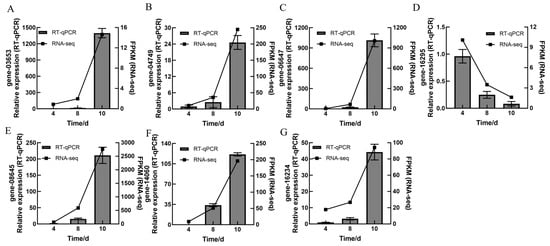
Figure 5.
RT-qPCR results and transcriptome data of seven DEGs.: (A) aes1 (gene-3653). (B) CYP075 (gene-04749). (C) mdlA (gene-06647). (D) yvh1 (gene-16295). (E) adh-1 (gene-08645). (F) ustYa (gene-14060). (G) srk1 (gene-16234).
4. Discussion
In this study, transcriptome sequencing was performed on mycelial samples collected at different fermentation times. The sequencing data were aligned and analyzed against the reference genome to identify DEGs. Additionally, GO and KEGG enrichment analyses were conducted. These analyses helped reveal the mechanisms underlying melanin synthesis at the transcriptional level. In the DEGs identified between R2 and R1, R3 and R1, and R3 and R2, we preliminarily screened genes related to transcriptional regulation, nutrient accumulation, metabolism, growth and development, and nitrogen utilization. These DEGs might be involved in or regulate the melanin synthesis of A. heimuer.
Eukaryotic transcription factors, as proteins that regulate gene expression, play important roles in growth, development, and responses to environmental stress [32,33]. The pH-responsive transcription factor PacC (RIM101) is a key transcription factor in fungi, widely involved in regulating the response to external pH changes to help fungi adapt to different acidic and alkaline environments. Feng et al. [34] analyzed the expression levels of PacC (the homolog of RIM101) in Penicillium italicum (P. italicum) cultured for different durations and found that the expression levels of PacC changed relatively mildly and remained at a high level during the early, middle, and late stages of P. italicum growth. It is speculated that during the growth of P. italicum, PacC maintains the pH required for the growth, thereby maintaining the acid–base balance necessary for its growth. CON7 is a C2H2 zinc finger domain transcription factor that is important for fungal hyphal growth, metabolic regulation, and stress responses. Zhang [35] found that upregulation of CON7 may help fungi enhance their survival capabilities in adverse environments by regulating the expression of related genes. HapX is a key bZIP transcription factor involved in iron uptake, storage, and utilization [36]. The downregulation of HapX may help fungi reduce iron-dependent reactions, thereby decreasing the generation of reactive oxygen species. This mechanism helps fungi survive in oxidative stress environments. Therefore, transcription factors RIM101, CON7, and HapX play important roles in the process of melanin production.
Fungi utilize carbohydrate-active enzymes and redox enzymes to degrade renewable lignocellulose, thereby obtaining carbohydrates such as monosaccharides and oligosaccharides to meet their nutritional requirements for growth, development, and reproduction [37]. Glycoside hydrolases represent the largest family of carbohydrate-active enzymes and play a significant role in fungal carbohydrate metabolism and development. For instance, chitinases within the glycoside hydrolase family are involved in various physiological processes such as nutrition, parasitism, morphogenesis, and immunity [38]. Chu et al. [25] conducted a transcriptomic analysis of Lentinula edodes mycelium under light-induced pigmentation and initial unpigmented conditions and found that genes of the glycoside hydrolase family were significantly upregulated, which is consistent with the findings of this study. This indicates that these enzymes play a crucial role in providing nutrition for melanin formation.
CYP450 is a class of enzymes with redox functions, widely involved in metabolic processes within organisms. In fungi, the upregulation of CYP450 genes is closely related to the synthesis of secondary metabolites. Jiang et al. [39] found that CYP450 genes in Ganoderma lucidum play an important role in the synthesis of triterpenes, a type of secondary metabolite in Ganoderma lucidum. They can catalyze the downstream oxidation reactions in the synthesis of triterpenes and modify the carbon ring skeleton. In a study on Arnebia euchroma by Li et al. [40], the upregulation of CYP450 genes related to the synthesis of shikonin, a secondary metabolite in Arnebia euchroma, suggests that the upregulation of CYP450 genes is closely related to the biosynthetic regulation of secondary metabolites. Laccase is a polyphenol oxidase that can catalyze the oxidation of phenolic or amine substrates, thereby promoting the synthesis of melanin. However, the laccase gene found in this study was downregulated, which is inconsistent with the findings of Chu et al. [25]. LaeA can regulate the secondary metabolism and growth and development of filamentous fungi, thereby affecting fungal morphogenesis, toxin synthesis, and the expression of silent genes, which have important impacts on the biological characteristics of fungi [38]. Studies have shown that the secretion of lipase is significantly reduced after the deletion of LaeA, which in turn affects the absorption of nutrients by hyphae and the formation of aflatoxin [41]. The differential expression of CYP450, laccase, and LaeA may affect the yield of melanin in the fermentation broth of A. heimuer.
Srk1 is a serine/threonine protein kinase that can regulate the activity of other splicing factors through phosphorylation, thereby affecting the developmental processes of fungi. Wang et al. [42] found that in Fusarium graminearum, the upregulation of srk1 is closely related to the processes of sexual reproduction and hyphal morphogenesis of the fungus. Yvh1 is a dual-specificity protein phosphatase, belonging to the atypical dual-specificity phosphatase subfamily. The downregulation of Yvh1 may be associated with the inhibition of the cell cycle. Under certain growth-inhibiting conditions, fungi may downregulate Yvh1 to slow down the cell cycle process, thereby reducing cell division and growth [43]. This regulatory mechanism may help fungi conserve energy in nutrient-poor or stressful environments.
Nitrogen sources, as essential nutrients for living organisms, are crucial for the synthesis of amino acids, proteins, nucleic acids, and secondary metabolites. They have an impact on the growth and development, pathogenicity, secondary metabolite synthesis, and sporulation capacity of fungi. In Candida albicans, knockout of the GDH gene, which is involved in nitrogen metabolism, has confirmed that nitrogen metabolism is associated with hyphal growth and morphogenesis [44]. NmrA plays an important regulatory role in the activity of GATA transcription factors during nitrogen metabolism. Pan et al. [45] discussed the impact of nitrogen sources on secondary metabolism in filamentous fungi, especially GATA-type regulatory proteins. These proteins regulate the synthesis of secondary metabolites by binding to the promoter regions of nitrogen utilization-related genes. The upregulation of GDH and NmrA genes can indirectly indicate that the formation of melanin requires the active participation of nitrogen sources.
5. Conclusions
In this study, the Illumina second-generation high-throughput sequencing platform was utilized, and a reference-based transcriptome analysis was conducted using the genome of A. heimuer to investigate DEGs during the mycelial pigmentation process of shiitake mushrooms. The study successfully identified differentially expressed transcription factors and preliminarily screened DEGs related to glycosidases, laccases, and serine/threonine protein kinases. These DEGs are closely associated with fungal growth, development, and nutrient metabolism. In the future, some exogenous expression of the screened melanin genes should be conducted to verify their functions. For the functional genes, the impact of their overexpression on melanin production should be verified by the overexpression method.
Author Contributions
Conceptualization, P.Z. and Y.M.; methodology, software, X.Y.; validation, C.S., B.Y. and Y.G.; formal analysis, X.Y., C.H. and Y.M.; investigation, X.Y. and S.T.; resources, P.Z.; data curation, J.Z. (Jianan Zhu); writing—original draft preparation, X.Y.; writing—review and editing, Y M., J.Z. (Jiechi Zhang) and S.Z.; visualization, J.L. and X.Z.; supervision, X.D.; project administration, P.Z.; funding acquisition, Y.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the earmarked fund for CARS-20 (Edible Mushroom).
Data Availability Statement
The original contributions presented in the study are included in the article; further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Dai, Y.; Li, X.; Song, B.; Sun, L.; Yang, C.; Zhang, X.; Wang, Y.; Zhang, Z.; Fu, Y.; Li, Y. Genomic Analyses Provide Insights Into the Evolutionary History and Genetic Diversity of Auricularia Species. Front. Microbiol. 2019, 10, 2255. [Google Scholar] [CrossRef]
- Chen, N.; Zhang, H.; Zong, X.; Li, S.; Wang, J.; Wang, Y.; Jin, M. Polysaccharides from Auricularia auricula: Preparation, structural features and biological activities. Carbohydr. Polym. 2020, 247, 116750. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, L.; Xue, B.; Zhao, D.; Zhang, Y.; Yan, X. A New Lectin from Auricularia auricula Inhibited the Proliferation of Lung Cancer Cells and Improved Pulmonary Flora. BioMed Res. Int. 2021, 2021, 5597135. [Google Scholar] [CrossRef]
- Ma, Z.; Liu, X.; Liu, Y.; Chen, W.; Wang, C. Studies on the biosynthetic pathways of melanin in Auricularia auricula. J. Basic Microbiol. 2022, 62, 843–856. [Google Scholar] [CrossRef]
- Zong, X.; Zhang, H.; Zhu, L.; Deehan, E.C.; Fu, J.; Wang, Y.; Jin, M. Auricularia auricula polysaccharides attenuate obesity in mice through gut commensal Papillibacter cinnamivorans. J. Adv. Res. 2023, 52, 203–218. [Google Scholar] [CrossRef]
- Shi, Q.; Yang, Z.; Fan, R.; Chu, J.; Fang, C.; Zhang, Y.; Shi, W.; Zhang, Y. Isolation, Characterization, and Antioxidant Activity of Melanin from Auricularia auricula (Agaricomycetes). Int. J. Med. Mushrooms 2023, 25, 55–73. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Chen, S.; Huang, Q.; Tan, J.; Zeng, J.; Yao, J.; Feng, T.; Wang, G.; Zhang, Y. The lipid lowering and antioxidative stress potential of polysaccharide from Auricularia auricula prepared by enzymatic method. Int. J. Biol. Macromol. 2021, 187, 651–663. [Google Scholar] [CrossRef]
- Han, Q.; Li, H.; Zhao, F.; Gao, J.; Liu, X.; Ma, B. Auricularia auricula Peptides Nutritional Supplementation Delays H2O2-Induced Senescence of HepG2 Cells by Modulation of MAPK/NF-κB Signaling Pathways. Nutrients 2023, 15, 3731. [Google Scholar] [CrossRef]
- Cordero, R.J.B.; Casadevall, A. Melanin. Curr. Biol. 2020, 30, R142–R143. [Google Scholar] [CrossRef]
- Montefiori, D.C.; Zhou, J.Y. Selective antiviral activity of synthetic soluble L-tyrosine and L-dopa melanins against human immunodeficiency virus in vitro. Antivir. Res. 1991, 15, 11–25. [Google Scholar] [CrossRef]
- Pascoe, M.J.; Maillard, J.Y. The role of melanin in Aspergillus tolerance to biocides and photosensitizers. Lett. Appl. Microbiol. 2021, 72, 375–381. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, P.; Dai, X.; Yao, X.; Zhou, S.; Ma, Q.; Liu, J.; Tian, S.; Zhu, J.; Zhang, J.; et al. Extraction, physicochemical properties, and antioxidant activity of natural melanin from Auricularia heimuer fermentation. Front. Nutr. 2023, 10, 1131542. [Google Scholar] [CrossRef]
- Yin, C.M.; Yao, F.; Wu, W.; Fan, X.Z.; Chen, Z.; Ma, K.; Shi, D.F.; Gao, H. Physicochemical Properties and Antioxidant Activity of Natural Melanin Extracted from the Wild Wood Ear Mushroom, Auricularia auricula (Agaricomycetes). Int. J. Med. Mushrooms 2022, 24, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Chen, X.D.; Yu, L.S. Biosynthesis, function and applications of melanin. Biot. Resour. 2020, 42, 652–659. [Google Scholar] [CrossRef]
- Sun, S.; Zhang, X.; Chen, W.; Zhang, L.; Zhu, H. Production of natural edible melanin by Auricularia auricula and its physicochemical properties. Food Chem. 2016, 196, 486–492. [Google Scholar] [CrossRef]
- Wang, J.; Ma, Z.; Wang, C.; Chen, W. Melanin in Auricularia auricula: Biosynthesis, production, physicochemical characterization, biological functions, and applications. Food Sci. Biotechnol. 2024, 33, 1751–1758. [Google Scholar] [CrossRef]
- Fan, X. The study of melanin property and color evaluation of Auricularia cornea and the expression of its key enzymes gene for pigment synthesis. Master’s Thesis, Jilin Agricultural University, Jilin, China, 2019. [Google Scholar] [CrossRef]
- Du, J.; Wang, S.; Yan, D.; Gao, Q.; Fan, Y.; Yu, Z.; Liu, Y. Analyses of melanin synthesis pathway and the related genes in Lentinula edodes based on whole genome sequence comparison. Mycosystema 2023, 42, 1114–1128. [Google Scholar] [CrossRef]
- Wang, G.; Li, D.; Zhu, B.; Ma, H.; Mu, Y.; Lv, F.; Jiang, M. Cloning and bioinformatics analysis of dopachrome tautomerase gene from Auricularia auricula. Hubei Agric. Sci. 2022, 61, 153–156. [Google Scholar] [CrossRef]
- He, M.; Wang, T.; Tang, C.; Xiao, M.; Pu, X.; Qi, J.; Li, Y.; Li, X. Metabolomics and transcriptomics reveal the effects of different fermentation times on antioxidant activities of Ophiocordyceps sinensis. J. Fungi. 2025, 11, 51. [Google Scholar] [CrossRef]
- Qiu, Z.; Gao, Y.; Wang, S.; Wang, J.; Wang, X.; Cai, N.; Zhao, J.; Li, T.; Li, H.; Li, T.; et al. Mechanism Underlying Light Intensity-Induced Melanin Synthesis of Auricularia heimuer Revealed by Transcriptome Analysis. Cells 2022, 12, 56. [Google Scholar] [CrossRef]
- Yao, X.G.; Guo, Y.; Han, C.; Tian, S.; Zhu, J.N.; Zhou, S.Y.; Liu, J.N.; Dai, X.D.; Zhang, P.Q.; Ma, Q.F.; et al. Optimization of fermentation medium formula for increasing melanin yield and physicochemical properties of melanin of Auricularia heimuer. Mycosystema 2024, 43, 241–254. [Google Scholar] [CrossRef]
- Fu, R.; Sun, W.; Liu, B.; Sun, J.; Wu, Q.; Liu, X.; Xiang, M. Genome and transcriptome reveal lithophilic adaptation of Cladophialophora brunneola, a new rock-inhabiting fungus. Mycology 2024, 14, 326–343. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Chu, T.; Shang, J.J.; Guan, W.; Yang, R.H.; Bao, D.P.; Tang, L.H. Transcriptome analysis of Lentinula edodes during brown mycelium-film formation. Mycosystema 2022, 41, 260–273. [Google Scholar] [CrossRef]
- Omura, T. Forty years of cytochrome P450. Biochem. Biophys. Res. Commun. 1999, 266, 690–698. [Google Scholar] [CrossRef]
- Chen, H.L.; Zhang, Q.Y.; Sun, K. Laccase-Mediated Oxidative Coupling of Phenolic Compounds in vivo: From Fundamentals to Multifunctional Applications in Green Synthesis. Biotechnol. Bull. 2020, 36, 193–204. [Google Scholar] [CrossRef]
- Bok, J.W.; Balajee, S.A.; Marr, K.A.; Andes, D.; Nielsen, K.F.; Frisvad, J.C.; Keller, N.P. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot. Cell 2005, 4, 1574–1582. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Wang, M.; Li, L.; Si, J.; Song, B.; Zhou, C.; Yu, M.; Wang, X.; Zhang, Y.; Ding, G.; et al. Overexpression of the Global Regulator LaeA in Chaetomium globosum Leads to the Biosynthesis of Chaetoglobosin Z. J. Nat. Prod. 2016, 79, 2487–2494. [Google Scholar] [CrossRef]
- Luo, Q.; Li, N.; Xu, J.W. A methyltransferase LaeA regulates ganoderic acid biosynthesis in Ganoderma lingzhi. Front. Microbiol. 2022, 13, 1025983. [Google Scholar] [CrossRef]
- Rispail, N.; Soanes, D.M.; Ant, C.; Czajkowski, R.; Grünler, A.; Huguet, R.; Perez-Nadales, E.; Poli, A.; Sartorel, E.; Valiante, V.; et al. Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet. Biol. 2009, 46, 287–298. [Google Scholar] [CrossRef]
- Golson, M.L.; Kaestner, K.H. Fox transcription factors: From development to disease. Development 2016, 143, 4558–4570. [Google Scholar] [CrossRef]
- Lu, Z.M.; Zhang, R.T.; Huang, X.B.; Cao, X.T.; Shen, X.Y.; Fan, L.; Hou, C.L. Optimisation of hypocrellin production in Shiraia-like fungi via genetic modification involving a transcription factor gene and a putative monooxygenase gene. Mycology 2023, 15, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zhang, M.H.; Li, X.Y.; Li, Q.R.; Peng, L.T. Cloning and expression analysis of the PacC gene of the postharvest pathogen Penicillium italicum in citrus. J. Food Sci. Technol. 2022, 43, 145–152. [Google Scholar] [CrossRef]
- Zhang, S.J. Functional studies of the thiol-disulfide reductase Trx in Magnaporthe oryzae and a unique secreted protein in Fusarium graminearum. Unpublished doctoral dissertation, Northwest A&F University, Yangling, China, 2015. [Google Scholar]
- Sun, K.; Li, Y.; Gai, Y.; Wang, J.; Jian, Y.; Liu, X.; Wu, L.; Shim, W.B.; Lee, Y.W.; Ma, Z.; et al. HapX-mediated H2B deub1 and SreA-mediated H2A.Z deposition coordinate in fungal iron resistance. Nucleic Acids Res. 2023, 51, 10238–10260. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Shen, D.; Wang, J.; Dong, Y.; Zhang, M.; Tang, Z.; Xia, Q.; Nyawira, K.T.; Jing, M.; Dou, D.; Xia, A. The glycoside hydrolase 18 family chitinases are associated with development and virulence in the mosquito pathogen Pythium guiyangense. Fungal Genet. Biol. 2009, 135, 103290. [Google Scholar] [CrossRef]
- Jiang, X.Y.; Han, W.; Liu, Y.F.; Tang, C.H.; Feng, J.; Zhang, J.S. Identification of key factors affecting liquid fermentation of Ganoderma lucidum for triterpenes: A review. Microbiol. Bull. 2023, 50, 2155–2172. [Google Scholar] [CrossRef]
- Li, X.W.; Liu, Z.Y.; Xu, Y.J.; Zhu, J.B.; Wu, Y.M. Explore of Molecular Mechanism on Fungal Elicitors Regulating Shikonin Synthesis. Chin. Agric. Sci. Bull. 2024, 26, 78–88. [Google Scholar] [CrossRef]
- Hu, Y.S.; Lv, A.; Zhang, J.Y.; LEI, Y.; Wang, L.; Lv, Y.Y. Research progresses of aflatoxin biosynthetic regulation by LaeA. J. Henan Univ. Technol. 2018, 39, 127–132. [Google Scholar] [CrossRef]
- Wang, G.; Sun, P.; Gong, Z.; Gu, L.; Lou, Y.; Fang, W.; Zhang, L.; Su, L.; Yang, T.; Wang, B.; et al. Srk1 kinase, a SR protein-specific kinase, is important for sexual reproduction, plant infection and pre-mRNA processing in Fusarium graminearum. Environ. Microbiol. 2018, 20, 3261–3277. [Google Scholar] [CrossRef]
- Zang, H.; Shackelford, R.; Bewley, A.; Beeser, A.E. Mutational Analyses of the Cysteine-Rich Domain of Yvh1, a Protein Required for Translational Competency in Yeast. Biology 2022, 11, 1246. [Google Scholar] [CrossRef] [PubMed]
- Han, T.L.; Cannon, R.D.; Gallo, S.M.; Villas-Bôas, S.G. A metabolomic study of the effect of Candida albicans glutamate dehydrogenase deletion on growth and morphogenesis. NPJ Biofilms Microbiomes 2019, 5, 13. [Google Scholar] [CrossRef]
- Pan, Y.Y.; Liu, G. Research advances on molecular regulation of filamentous fungal secondary metabolism in China. Hereditas 2018, 40, 874–887. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).