Genome-Wide Identification of Cucumber Lhc Genes’ Family and Their Expression Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Chromosomal Localization of Lhc Family in Cucumber
2.2. Characterization and Phylogenetic Analysis of Cucumber Lhc Family
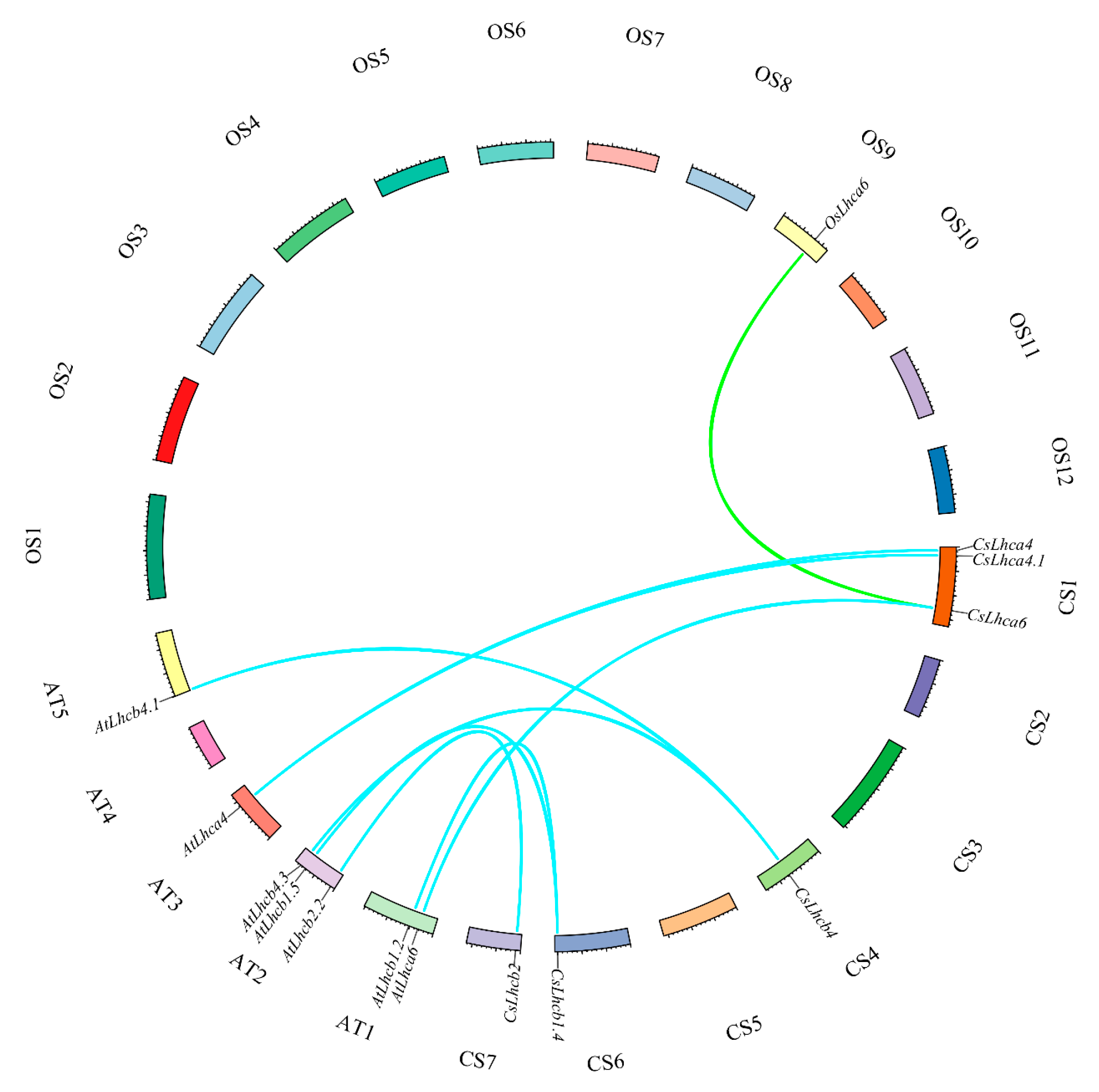
2.3. Collinearity Analysis of Cucumber Lhc Family
2.4. Re-Analysis of Cucumber Transcriptome Sequencing Big Data Based on RNA-Seq
2.5. Expression Analysis of Lhc Family in Cucumber
2.6. qRT-PCR Analysis of Lhc Family in Cucumber Under High Temperature and Salt Stress
3. Results
3.1. Basic Information of Lhc Family Members in Cucumber
3.2. Chromosomal Localization of Lhc Family in Cucumber
3.3. Clustering Analysis of Lhc Family in Cucumber, Arabidopsis, and Rice
3.4. Structure and Conserved Sequence Analysis of Lhc Family in Cucumber
3.5. Collinearity Analysis of Lhc Family in Cucumber, Arabidopsis, and Rice
3.6. Cis Elements Analysis of the Promoter Sequence of Cucumber Lhc Family
3.7. Tissue-Specific Expression Analysis of Lhc Family in Cucumber
3.8. Expression Pattern Analysis of Cucumber Lhc Family Under Abiotic Stress
3.9. Expression Pattern Analysis of Cucumber Lhc Family Under Biotic Stress
3.10. Regulatory Patterns Analysis of Cucumber Lhc in Response to Abiotic and Biotic Stresses
3.11. Expression Profiles of Cucumber Lhc Family in Response Low Temperature and Salt Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kirilovsky, D.; Büchel, C. Evolution and function of light-harvesting antenna in oxygenic photosynthesis advances in botanical research. Elsevier 2019, 91, 247–293. [Google Scholar] [CrossRef]
- Wang, P.; Grimm, B. Connecting chlorophyll metabolism with accumulation of the photosynthetic apparatus. Trends Plant Sci. 2021, 26, 484–495. [Google Scholar] [CrossRef]
- Zhao, S.; Gao, H.; Luo, J.; Wang, H.; Dong, Q.; Wang, Y.; Yang, K.; Mao, K.; Ma, F. Genome-wide analysis of the light-harvesting chlorophyll a/b-binding gene family in apple (Malus domestica) and functional characterization of MdLhcb4. 3, which confers tolerance to drought and osmotic stress. Plant Physiol. Biochem. 2020, 154, 517–529. [Google Scholar] [CrossRef]
- Negi, S.; Perrine, Z.; Friedland, N.; Kumar, A.; Tokutsu, R.; Minagawa, J.; Berg, H.; Barry, A.N.; Govindjee, G.; Sayre, R. Light regulation of light-harvesting antenna size substantially enhances photosynthetic efficiency and biomass yield in green algae(†). Plant J. Cell Mol. Biol. 2020, 103, 584–603. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.E.; Ma, J.; Wu, N.; Su, Y.Q.; Zhang, Z.W.; Yuan, M.; Zhang, H.Y.; Zeng, X.Y.; Yuan, S. The roles of Arabidopsis proteins of Lhcb4, Lhcb5 and Lhcb6 in oxidative stress under natural light conditions. Plant Physiol. Biochem. 2018, 130, 267–276. [Google Scholar] [CrossRef] [PubMed]
- De Bianchi, S.; Dall’Osto, L.; Tognon, G.; Morosinotto, T.; Bassi, R. Minor antenna proteins CP24 and CP26 affect the interactions between photosystem II subunits and the electron transport rate in grana membranes of Arabidopsis. Plant Cell 2008, 20, 1012–1028. [Google Scholar] [CrossRef]
- Zou, Z.; Li, M.; Jia, R.; Zhao, H.; He, P.; Zhang, Y.; Guo, A. Genes encoding light-harvesting chlorophyll a/b-binding proteins in papaya (Carica papaya L.) and insight into lineage-specific evolution in Brassicaceae. Gene 2020, 748, 144685. [Google Scholar] [CrossRef]
- Jansson, S. A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 1999, 4, 236–240. [Google Scholar] [CrossRef]
- Zhang, Q.; Ma, C.; Wang, X.; Ma, Q.; Fan, S.; Zhang, C. Genome-wide identification of the light-harvesting chlorophyll a/b binding (Lhc) family in Gossypium hirsutum reveals the influence of GhLhcb2. 3 on chlorophyll a synthesis. Plant Biol. 2021, 23, 831–842. [Google Scholar] [CrossRef]
- Vass, I.; Cser, K.; Cheregi, O. Molecular mechanisms of photosynthe. Ann. N. Y. Acad. Sci. 2010, 1113, 114–122. [Google Scholar] [CrossRef]
- Gao, J.; Wang, H.; Yuan, Q.; Feng, Y. Structure and function of the photosystem supercomplexes. Front. Plant Sci. 2018, 9, 357. [Google Scholar] [CrossRef] [PubMed]
- Fristedt, R.; Herdean, A.; Blaby-Haas, C.E.; Mamedov, F.; Merchant, S.S.; Last, R.L.; Lundin, B. Phtosystem II Protein33, a protein conserved in the plastid lineage, is associated with the chloroplast thylakoid membrane and provides stability to photosystem II supercomplexes in Arabidopsis. Plant Physiol. 2014, 167, 481–492. [Google Scholar] [CrossRef]
- Wang, M.; Sun, K.; Qin, X.; Gong, S.; Li, Z.; Fan, K. Comparative transcriptome analysis reveals a tissue-specific pathway involved in nitrogen utilization between genotypes with different nitrogen use efficiencies in tea plants (Camellia sinensis). Agronomy 2024, 14, 2824. [Google Scholar] [CrossRef]
- Umate, P. Genome-wide analysis of the family of light-harvesting chlorophyll a/b-binding proteins in Arabidopsis and rice. Plant Signal. Behav. 2010, 5, 1537–1542. [Google Scholar] [CrossRef]
- Zou, Z.; Yang, J. Genomics analysis of the light-harvesting chlorophyll a/b-binding (Lhc) superfamily in cassava (Manihot esculenta Crantz). Gene 2019, 702, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Huang, Q.; An, F. Genome-wide identification, classification and expression analysis of Lhc supergene family in castor bean (Ricinus communis L.). Agric. Biotechnol. 2013, 2, 44–48. [Google Scholar]
- Luo, J.; Abid, M.; Tu, J.; Gao, P.; Wang, Z.; Huang, H. Genome-wide identification of the LHC gene family in kiwifruit and regulatory role of AcLhcb3. 1/3.2 for chlorophyll a content. Int. J. Mol. Sci. 2022, 23, 6528. [Google Scholar] [CrossRef]
- Xu, Y.H.; Liu, R.; Yan, L.; Liu, Z.Q.; Jiang, S.C.; Shen, Y.Y.; Wang, X.F.; Zhang, D.P. Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 2012, 63, 1095–1106. [Google Scholar] [CrossRef]
- Peterson, R.B.; Schultes, N.P. Light-harvesting complex B7 shifts the irradiance response of photosynthetic light-harvesting regulation in leaves of Arabidopsis thaliana. J. Plant Physiol. 2014, 171, 311–318. [Google Scholar] [CrossRef]
- Wang, S.S.; Song, Z.B.; Sun, Z.; Zhang, J.; Mei, Y.; Nian, H.J.; Li, K.Z.; Chen, L.M. Effects of formaldehyde stress on physiological characteristics and gene expression associated with photosynthesis in Arabidopsis thaliana. Plant Mol. Biol. Report. 2012, 30, 1291–1302. [Google Scholar] [CrossRef]
- Pietrzykowska, M.; Suorsa, M.; Semchonok, D.A.; Tikkanen, M.; Boekema, E.J.; Aro, E.-M.; Jansson, S. The light-harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis. Plant Cell 2014, 26, 3646–3660. [Google Scholar] [CrossRef]
- Yadavalli, V.; Neelam, S.; Rao, A.S.; Reddy, A.R.; Subramanyam, R. Differential degradation of photosystem I subunits under iron deficiency in rice. J. Plant Physiol. 2012, 169, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Xu, Z.S.; Wang, F.; Li, M.Y.; Ma, J.; Xiong, A.S. Effects of abiotic stresses on the expression of Lhcb1 gene and photosynthesis of Oenanthe javanica and Apium graveolens. Biol. Plant. 2014, 58, 256–264. [Google Scholar] [CrossRef]
- Wen, C.L.; Cheng, Q.; Zhao, L.; Mao, A.; Yang, J.; Yu, S.; Weng, Y.; Xu, Y. Identification and characterisation of Dof transcription factors in the cucumber genome. Sci. Rep. 2016, 6, 23072. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Li, R.; Zhang, Z.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P. The genome of the cucumber, Cucumis sativus L. Nature Genetics 2009, 41, 1275–1281. Nature Genetics 2009, 41, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Jiang, W.; Zhang, Y.; Yu, H.; Mao, Z.; Gu, X.; Huang, S.; Xie, B. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011, 12, 471. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Liu, S. Genome-wide analysis of the MADS-box gene family in cucumber. Genome 2012, 55, 245–256. [Google Scholar] [CrossRef]
- Wan, H.; Yuan, W.; Bo, K.; Shen, J.; Pang, X.; Chen, J. Genome-wide analysis of NBS-encoding disease resistance genes in Cucumis sativus and phylogenetic study of NBS-encoding genes in Cucurbitaceae crops. BMC Genom. 2013, 14, 109. [Google Scholar] [CrossRef]
- Baloglu, M.C.; Eldem, V.; Hajyzadeh, M.; Unver, T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE 2014, 9, e96014. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.-B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.-H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE. A database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. Stringtie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2-and EdgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Z.; Yan, P.; Huang, S.; Fei, Z.; Lin, K. RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genom. 2011, 12, 540. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, X.; Han, J.; Lu, W.; Ren, Z. Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol. 2020, 20, 443. [Google Scholar] [CrossRef]
- Zhu, Y.; Yin, J.; Liang, Y.; Liu, J.; Jia, J.; Huo, H.; Wu, Z.; Yang, R.; Gong, H. Transcriptomic dynamics provide an insight into the mechanism for silicon-mediated alleviation of salt stress in cucumber plants. Ecotoxicol. Environ. Saf. 2019, 174, 245–254. [Google Scholar] [CrossRef]
- Zhu, Y.X.; Yang, L.; Liu, N.; Yang, J.; Zhou, X.K.; Xia, Y.C.; He, Y.; He, Y.Q.; Gong, H.J.; Ma, D.F. Genome-wide identification, structure characterization, and expression pattern profiling of aquaporin gene family in cucumber. BMC Plant Biol. 2019, 19, 345. [Google Scholar] [CrossRef]
- Kęska, K.; Szcześniak, M.W.; Makałowska, I.; Czernicka, M. Long-term waterlogging as factor contributing to hypoxia stress tolerance enhancement in cucumber: Comparative transcriptome analysis of waterlogging sensitive and tolerant accessions. Genes 2021, 12, 189. [Google Scholar] [CrossRef]
- Tian, Z.; Jahn, M.; Qin, X.; Obel, H.O.; Yang, F.; Li, J.; Chen, J. Genetic and transcriptomic analysis reveal the molecular basis of photoperiod-regulated flowering in Xishuangbanna cucumber (Cucumis sativus L. var. xishuangbannesis Qi et Yuan). Genes 2021, 12, 1064. [Google Scholar] [CrossRef] [PubMed]
- Burkhardt, A.; Day, B. Transcriptome and small RNAome dynamics during a resistant and susceptible interaction between cucumber and downy mildew. Plant Genome 2016, 9, plantgenome2015-08. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Xu, X.; Shi, Y.; Qi, X.; Chen, X. Elucidation of the molecular responses of a cucumber segment substitution line carrying Pm5. 1 and its recurrent parent triggered by powdery mildew by comparative transcriptome profiling. BMC Genom. 2017, 18, 21. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, C.; Zhang, K.; Tian, Z.; Xu, J.; Yang, S.; Lou, Q.; Li, J.; Chen, J.F. Comparative transcriptomics reveals suppressed expression of genes related to auxin and the cell cycle contributes to the resistance of cucumber against Meloidogyne incognita. BMC Genom. 2018, 19, 583. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Wang, Y.; Xian, Q.; Chen, X.; Xu, J. Transcriptome analysis reveals ethylene-mediated defense responses to Fusarium oxysporum f. sp. cucumerinum infection in Cucumis sativus L. BMC Plant Biol. 2020, 20, 334. [Google Scholar] [CrossRef]
- Słomnicka, R.; Olczak-Woltman, H.; Sobczak, M.; Bartoszewski, G. Transcriptome profiling of cucumber (Cucumis sativus L.) early response to Pseudomonas syringae pv. Lachrymans. Int. J. Mol. Sci. 2021, 22, 4192. [Google Scholar] [CrossRef] [PubMed]
- Mansfeld, B.N.; Colle, M.; Kang, Y.; Jones, A.D.; Grumet, R. Transcriptomic and metabolomic analyses of cucumber fruit peels reveal a developmental increase in terpenoid glycosides associated with age-related resistance to Phytophthora capsici. Hortic. Res. 2017, 4, 17022. [Google Scholar] [CrossRef]
- Slavokhotova, A.; Korostyleva, T.; Shelenkov, A.; Pukhalskiy, V.; Korottseva, I.; Slezina, M.; Istomina, E.; Odintsova, T. Transcriptomic analysis of genes involved in plant defense response to the cucumber green mottle mosaic virus infection. Life 2021, 11, 1064. [Google Scholar] [CrossRef]
- Hamilton, J.P.; Buell, C.R. Advances in plant genome sequencing. Plant J. 2012, 70, 177–190. [Google Scholar] [CrossRef]
- Wang, L.; Wei, J.; Shi, X.; Qian, W.; Mehmood, J.; Yin, Y.; Jia, H. Identification of the light-harvesting chlorophyll a/b binding protein gene family in peach (Prunus persica L.) and their expression under drought stress. Genes 2023, 14, 1475. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Zhou, Y.; Sun, P.; Cao, M.; Li, H.; Mao, Y. Identification of light-harvesting chlorophyll a/b-binding protein genes of Zostera marina L. and their expression under different environmental conditions. J. Ocean. Univ. China 2016, 15, 152–162. [Google Scholar] [CrossRef]
- Jiang, L.; Yun, M.; Ma, Y.; Qu, T. Melatonin mitigates water deficit stress in Cenchrus alopecuroides (L.) Thunb through up-regulating gene expression related to the photosynthetic rate, flavonoid synthesis, and the assimilatory sulfate reduction pathway. Plants 2024, 13, 716. [Google Scholar] [CrossRef] [PubMed]










| Family Member | Gene ID | CDS Size | Number of Amino Acids | Molecular Weight | pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Prediction of Subcellular Location |
|---|---|---|---|---|---|---|---|---|---|
| CsLhca1 | CsaV3_5G025740.1 | 681 | 226 | 24.43 | 6.74 | 42.39 | 87.21 | 0.034 | chloroplast |
| CsLhca2 | CsaV3_5G030270.1 | 816 | 271 | 29.09 | 6.42 | 34.76 | 84.02 | 0.050 | chloroplast |
| CsLhca3 | CsaV3_1G043710.1 | 822 | 273 | 29.42 | 8.67 | 29.66 | 80.48 | −0.063 | chloroplast |
| CsLhca4 | CsaV3_1G002410.1 | 759 | 252 | 27.67 | 6.90 | 49.82 | 84.01 | −0.08 | chloroplast |
| CsLhca4.1 | CsaV3_1G005600.1 | 753 | 250 | 27.79 | 6.43 | 41.15 | 81.92 | −0.182 | chloroplast |
| CsLhca4.2 | CsaV3_3G012890.1 | 579 | 192 | 20.21 | 9.56 | 44.84 | 88.96 | 0.143 | cell membrane |
| CsLhca4.3 | CsaV3_6G034480.1 | 819 | 272 | 29.05 | 6.76 | 24.70 | 102.21 | 0.272 | chloroplast |
| CsLhca5 | CsaV3_2G027790.1 | 780 | 259 | 28.48 | 7.17 | 40.57 | 92.66 | −0.022 | cell membrane |
| CsLhca6 | CsaV3_1G041230.1 | 804 | 267 | 29.35 | 5.86 | 43.04 | 77.53 | −0.131 | cell membrane |
| CsLhcb1.1 | CsaV3_1G032510.1 | 798 | 265 | 28.27 | 5.29 | 26.89 | 78.08 | −0.036 | chloroplast |
| CsLhcb1.2 | CsaV3_3G031580.1 | 798 | 265 | 28.31 | 5.47 | 25.43 | 78.83 | −0.045 | chloroplast |
| CsLhcb1.3 | CsaV3_6G013800.1 | 804 | 267 | 28.35 | 5.15 | 25.67 | 79.36 | −0.024 | chloroplast |
| CsLhcb1.4 | CsaV3_6G051520.1 | 798 | 265 | 28.19 | 5.14 | 25.53 | 78.49 | −0.033 | chloroplast |
| CsLhcb1.5 | CsaV3_6G051530.1 | 798 | 265 | 28.14 | 5.14 | 25.53 | 78.87 | −0.013 | chloroplast |
| CsLhcb2 | CsaV3_7G002620.1 | 798 | 265 | 28.54 | 5.46 | 28.71 | 79.58 | −0.036 | chloroplast |
| CsLhcb3 | CsaV3_3G005950.1 | 795 | 264 | 28.54 | 4.89 | 20.73 | 86.14 | 0.011 | chloroplast |
| CsLhcb4 | CsaV3_4G026100.1 | 855 | 284 | 31.09 | 5.67 | 37.71 | 88.66 | −0.082 | chloroplast |
| CsLhcb5 | CsaV3_5G039350.1 | 867 | 288 | 30.93 | 5.33 | 32.42 | 88.19 | −0.016 | chloroplast |
| CsLhcb6 | CsaV3_1G019610.1 | 618 | 205 | 21.96 | 10.70 | 34.92 | 88.54 | 0.001 | cell membrane |
| CsLhcb6.1 | CsaV3_6G005340.1 | 768 | 255 | 27.22 | 6.75 | 23.24 | 86.94 | 0.089 | chloroplast |
| CsLhcb7 | CsaV3_4G037740.1 | 816 | 271 | 30.10 | 4.86 | 35.56 | 96.49 | 0.086 | cell membrane |
| Motif | Sequence | Number of Amino Acids | Pfam Annotation |
|---|---|---|---|
| motif 1 | EJKNGRLAMLAMLGFFVPEILT | 22 | Chlorophyll a/b binding protein |
| motif 2 | RNGVKFGEAVWFKAGSQIFSEGGLDYLGNPSLVHAQSILAIWACQVVLMGAVEGYRIAG | 59 | Chlorophyll a/b binding protein |
| motif 3 | PSYLDGELPGDYGFDPLGLSADPE | 24 | Chlorophyll a/b binding protein |
| motif 4 | GKGPLENLADHLADPVHNNAW | 21 | - |
| motif 5 | GSFDPLGLADDPEAFAELKVK | 21 | - |
| motif 6 | IGIINVPSWYDAGKAEYFADSSTLFVIEFILFGWVEGRRWQDIKNPGSVNQDPIFPQYKLPPNDVGYPGG | 70 | Chlorophyll a/b binding protein |
| motif 7 | SGSPWYGPDRVKYLGPFSGEP | 21 | - |
| motif 8 | AASSMALSSPSFAGQAVKLSPTAPEJQGNAKFTMRKTASKS | 41 | - |
| motif 9 | AYATNFVPGK | 10 | - |
| motif 10 | PLGEVTDPIYP | 11 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miao, Y.; Zhang, K. Genome-Wide Identification of Cucumber Lhc Genes’ Family and Their Expression Analysis. Horticulturae 2025, 11, 736. https://doi.org/10.3390/horticulturae11070736
Miao Y, Zhang K. Genome-Wide Identification of Cucumber Lhc Genes’ Family and Their Expression Analysis. Horticulturae. 2025; 11(7):736. https://doi.org/10.3390/horticulturae11070736
Chicago/Turabian StyleMiao, Yongmei, and Kaijing Zhang. 2025. "Genome-Wide Identification of Cucumber Lhc Genes’ Family and Their Expression Analysis" Horticulturae 11, no. 7: 736. https://doi.org/10.3390/horticulturae11070736
APA StyleMiao, Y., & Zhang, K. (2025). Genome-Wide Identification of Cucumber Lhc Genes’ Family and Their Expression Analysis. Horticulturae, 11(7), 736. https://doi.org/10.3390/horticulturae11070736







