Cloning and Functional Analysis of Flavonol Synthase Gene ZjFLS from Chinese Jujube (Ziziphus jujuba Mill.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. ZjFLS Cloning and Sequence Analysis
2.3. qRT-PCR Expression Analysis of ZjFLS
2.4. Subcellular Localization of ZjFLS
2.5. Prokaryotic Expression Analysis of ZjFLS
2.6. Acquisition of Arabidopsis thaliana Strains Overexpressed by ZjFLS
2.7. qPCR Detection of ZjFLS Gene Expression Level and Determination of Total Flavonoid Content in Positive Arabidopsis thaliana Strains
3. Results
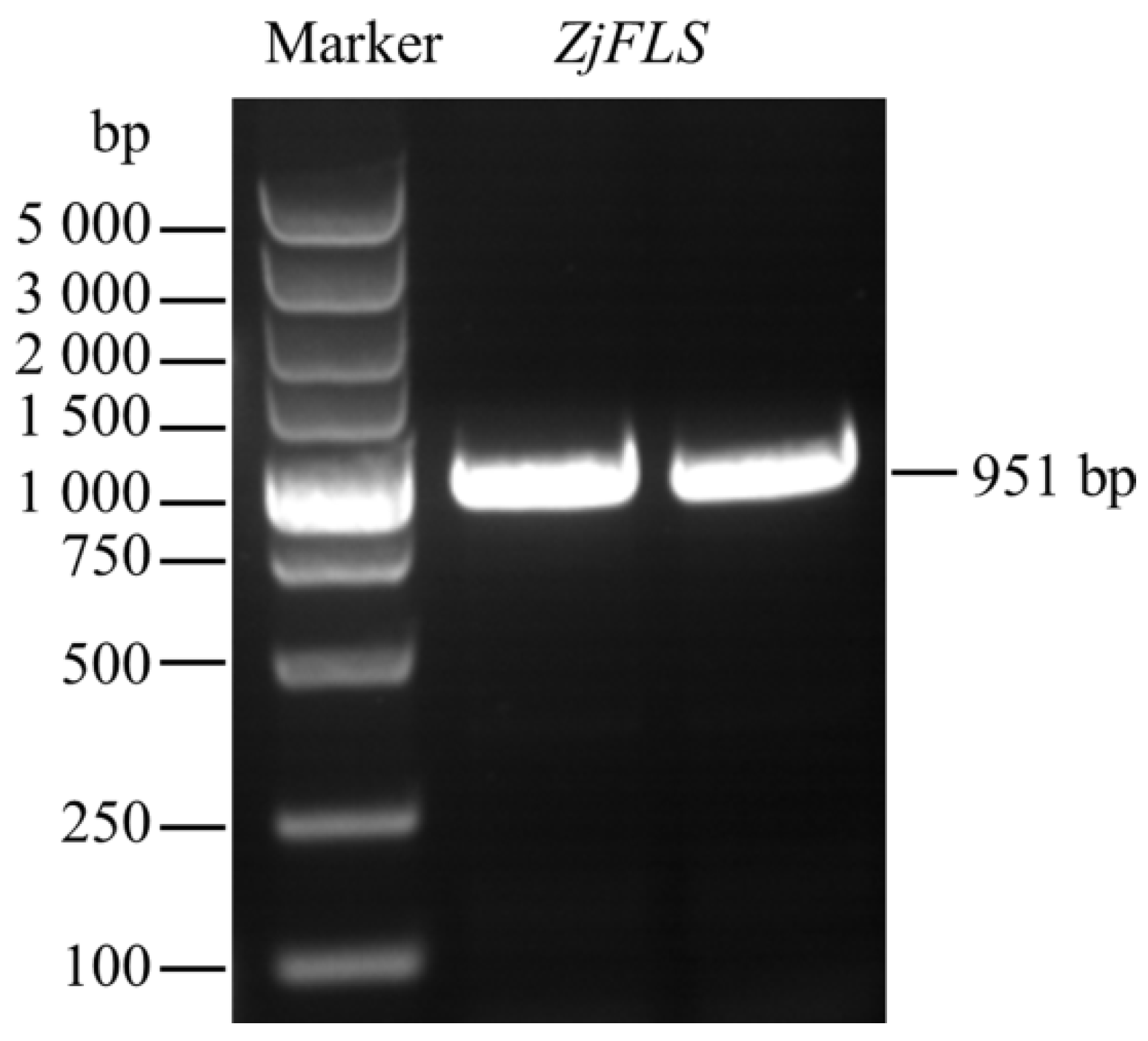
3.1. Cloning and Bioinformatics Analyses of the ZjFLS Gene
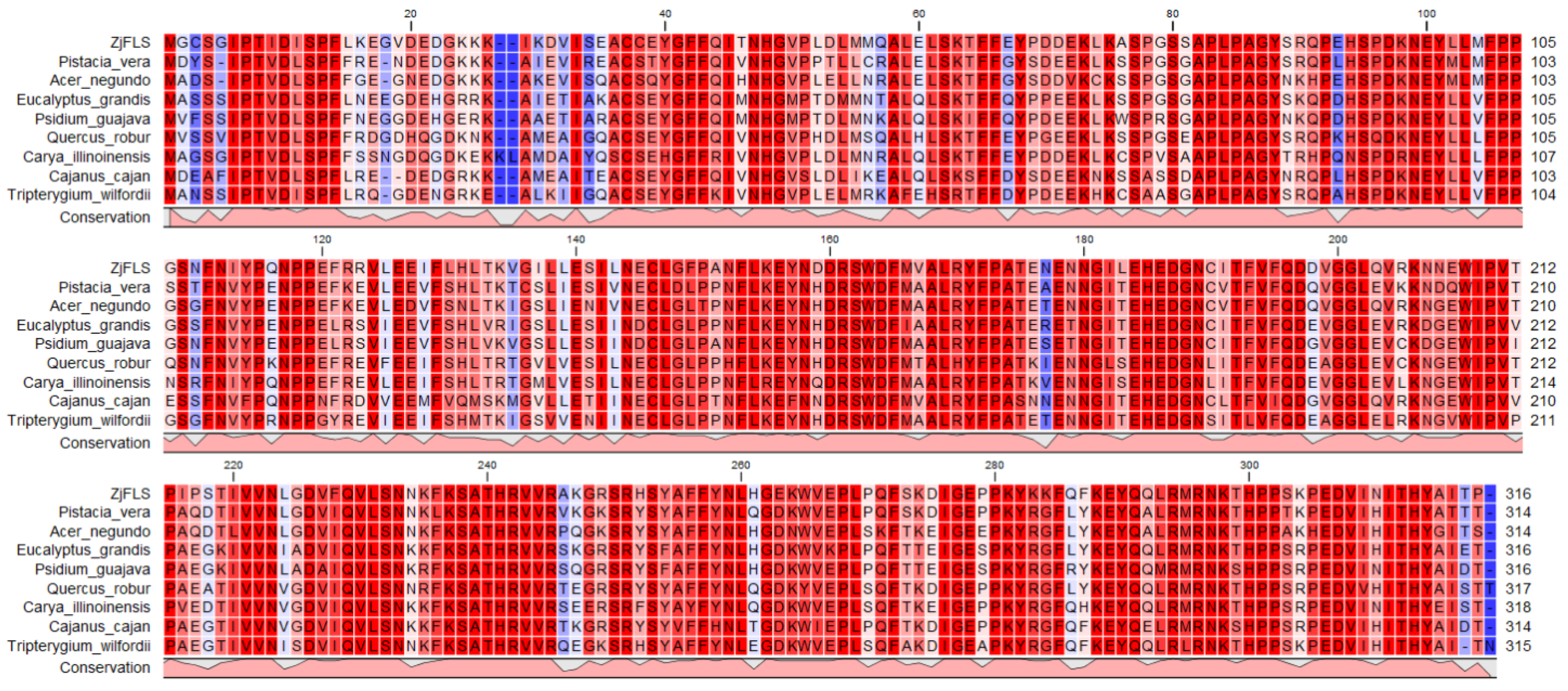
3.2. Analysis of ZjFLS Sequence Characteristics
3.3. Tissue and Organ Expression Characteristics of ZjFLS
3.4. Subcellular Localization of ZjFLS Protein
3.5. Prokaryotic Expression Analysis of ZjFLS
3.6. Identification of Arabidopsis thaliana Strains with Heterologous Overexpression of the ZjFLS Gene
3.7. Analysis of the Expression Level of ZjFLS and Total Flavonoid Content in Positive Arabidopsis thaliana Strains
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qu, Z.Z.; Wang, Y.H. Fruit records. In Chinese jujube (Ziziphus jujuba Mill.); China Forestry Publishing House: Beijing, China, 1993; pp. 1–2. [Google Scholar]
- Liu, M.J.; Wang, M. Germplasm Resources of Chinese jujube; China Forestry Publishing House: Beijing, China, 2009; pp. 23–24. [Google Scholar]
- Li, D.K.; Niu, X.W.; Tian, J.B. The Illustrated Germplasm Resources of Chinese jujube; China Agriculture Press: Beijing, China, 2013; pp. 523–526. [Google Scholar]
- Zhu, D.Q.; Jiang, N.; Wang, N.; Zhao, Y.F.; Liu, X.M. A literature review of the pharmacological effects of jujube. Foods 2024, 13, 193. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.F.; Zhao, A.L.; Wang, Y.K.; Ren, H.Y.; Li, Y.; Li, D.K.; Du, J.J. Metabolomics-based analysis of flavonoid metabolites in Chinese jujube and sour jujube fruits from different harvest periods. J. Food Sci. 2022, 87, 3752–3765. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.L.; Zhang, S.L.; Jin, X.Y.; Yin, C.X.; Zhang, Y.; Guo, X.D.; Lin, X.M. Systematic comparison of structural characterization of polysaccharides from Ziziphus jujuba cv. Muzao. Molecules 2023, 28, 562. [Google Scholar] [CrossRef]
- Yang, M.; Han, L.; Zhang, S.F.; Dai, L.; Li, B.; Han, S.K.; Zhao, J.; Liu, P.; Zhao, Z.H.; Liu, M.J. Insights into the evolution and spatial chromosome architecture of jujube from an updated gapless genome assembly. Plant Commun. 2023, 4, 100662. [Google Scholar] [CrossRef]
- Liu, Z.G.; Yuan, Y.; Wang, L.L.; Zhao, X.; Wang, L.X.; Wang, L.H.; Zhao, Z.H.; Zhao, X.; Chu, Y.T.; Gao, Y.N.; et al. Three novel adenylate cyclase genes show significant biological functions in plant. J. Agric. Food Chem. 2023, 71, 1149–1161. [Google Scholar] [CrossRef]
- Pan, F.X.; Zhao, Z.H.; Liu, F.W.; Liu, F.W.; Luo, Z.; Chen, S.J.; Liu, Z.G.; Zhao, Z.H.; Liu, M.J.; Wang, L.L. Triterpenoids in jujube: A review of composition, content diversity, pharmacological effects, synthetic pathway, and variation during domestication. Plants 2023, 12, 1501. [Google Scholar] [CrossRef]
- Cai, W.T.; Zhuang, H.N.; Wang, X.Y.; Fu, X.; Chen, S.; Yao, L.Y.; Sun, M.; Wang, H.T.; Yu, C.; Feng, T. Functional nutrients and jujube-based processed products in Ziziphus jujuba. Molecules 2024, 29, 3437. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.F.; Zhao, A.L.; Wang, Y.K.; Ren, H.Y.; Du, J.J.; Li, D.K.; Li, Y. Composition and content of phenolic acids and flavonoids among the different varieties, development stages, and tissues of Chinese jujube (Ziziphus jujuba Mill.). PLoS ONE 2021, 16, e0254058. [Google Scholar] [CrossRef]
- Choi, S.S.; Park, S.R.; Lee, K.A. A comparative study of rutin and rutin glycoside: Antioxidant activity, anti-inflammatory effect, effect on platelet aggregation and blood coagulation. Antioxidants 2021, 10, 1696. [Google Scholar] [CrossRef]
- Wu, Q.; Needs, P.W.; Lu, Y.L.; Kroon, P.A.; Ren, D.Y.; Yang, X.B. Correlation: Different antitumor effects of quercetin, quercetin-3′-sulfate and quercetin-3-glucuronide in human breast cancer MCF-7 cells. Food Funct. 2019, 10, 4452. [Google Scholar] [CrossRef]
- Brenda, W.S. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar]
- Xu, F.; Li, L.L.; Zhang, W.W.; Cheng, H.; Sun, N.N.; Cheng, S.Y.; Wang, Y. Isolation, characterization, and function of a flavonol synthase gene from Ginkgo biloba. Mol. Biol. Rep. 2012, 39, 2285–2296. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Vos, R.; Bartelniewoehner, L.; Ishihara, H.; Sagasser, M.; Martens, S.; Weisshaar, B. Metabolomic and genetic analyses of flavonol synthesis in Arabidopsis thaliana support the in vivo involvement of leucoanthocyanidin dioxygenase. Planta 2009, 229, 427–445. [Google Scholar] [CrossRef]
- Holton, T.A.; Brugllera, F.; Tanaka, Y. Cloning and expression of flavonol synthase from Petunia hybrida. Plant J. 1993, 4, 1003–1010. [Google Scholar] [CrossRef]
- Liu, X.; Lu, J.; Hao, Y.J.; You, C.X. Cloning, bioinformatics analysis and catalytic activity identification of MdFLS1 gene in apple. J. Fruit. Sci. 2018, 35, 905–916. [Google Scholar]
- Xiao, W.J.; Hu, Y.L.; Wang, Q.Y.; Duan, Y.J.; Hu, H.G. Cloning, bioinformatics and functional analysis of MaFLS1 in banana. J. Fruit. Sci. 2024, 41, 229–240. [Google Scholar]
- Ouyang, Y.N.; Liu, Y.; Peng, J.S.; Chen, Y.Y.; Zhao, H.; He, Y.T.; Zhao, X.Y.; Tang, X.K.; Zhou, M.L. Function and genetic diversity analysis of FtFLS1 from tartary buckwheat. J. Plant Genet. Resour. 2023, 24, 1401–1412. [Google Scholar]
- Wang, W.H.; Li, D.B.; He, F.; Wang, W.J.; Wu, Y.Y. Cloning and functional analysis of RhFLS gene of Rhododendron. Mol. Plant Breed. 2022, 20, 4243–4250. [Google Scholar]
- Liu, X.L.; Sun, T.T.; Yang, J.; He, H.B. Cloning and expression analysis of FLS gene of flavonol synthetase in Lilium auratum and L. speciosum var. gloriosoides. Acta Agric. Zhejiangensis 2024, 36, 344–357. [Google Scholar]
- Li, H.H.; Liu, Y.L.; Liu, H.L.; Lou, Q. Cloning and expression analysis of FLS gene in Muscari armeniacum. J. Northwest For. Univ. 2019, 34, 116–121. [Google Scholar]
- Liu, H.L.; Su, B.B.; Zhang, H.; Gong, J.X.; Zhang, B.X.; Liu, Y.L.; Du, L.J. Identification and functional analysis of a flavonol synthase gene from grape hyacinth. Molecules 2019, 24, 1579. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.L.; Shi, Y.F.; Fu, Z.P.; Li, W.W.; Lai, S.Y.; Wu, Y.H.; Wang, Y.S.; Liu, Y.J.; Gao, L.P.; Xia, T. Functional characterization of three flavonol synthase genes from Camellia sinensis: Roles in flavonol accumulation. Plant Sci. 2020, 300, 110632. [Google Scholar] [CrossRef]
- Tian, J.; Han, Z.Y.; Zhang, J.; Hu, Y.J.; Song, T.T.; Yao, Y.C. The balance of expression of dihydroflavonol 4-reductase and flavonol synthase regulates flavonoid biosynthesis and red foliage coloration in Crabapples. Sci. Rep. 2015, 5, 122228. [Google Scholar] [CrossRef]
- Lei, T.; Huang, J.; Ruan, H.X.; Qian, W.; Fang, Z.; Gu, C.Y.; Zhang, N.N.; Liang, Y.X.; Wang, Z.Y.; Gao, L.P.; et al. Competition between FLS and DFR regulates the distribution of flavonols and proanthocyanidins in Rubus chingii Hu. Front. Plant Sci. 2023, 14, 1134993. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, N.; Pucker, B. Conserved amino acid residues and gene expression patterns associated with the substrate preferences of the competing enzymes FLS and DFR. PLoS ONE 2024, 19, e0305837. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Huang, W.D. Salicylic acid modulated flavonol biosynthesis in three key phases during grape berry development. Eur. Food Res. Technol. 2013, 237, 441–448. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, X.Y.; Miao, Y.N.; Wang, D.X.; Zhang, Z.H.; Liu, Y.X. Genome-wide identification and expression profile analysis of the WUSCHEL-related homeobox (WOX) genes in woodland strawberry (Fragaria vesca). Horticulturae 2022, 8, 1043. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhao, A.L.; Li, D.K.; Wang, Y.K.; Sui, C.L.; Du, X.M.; Cao, Y.Q. Study on the content of triterpene acids and flavones for jujubes of different varieties, different growing periods and different parts. Acta Hortic. 2009, 840, 533–540. [Google Scholar] [CrossRef]
- Li, W. Study on Mechanisms of TaFLS1 and TaANS1 of Triticum aestivum in Abiotic Stress Response. Ph.D. Thesis, Shandong University, Jinan, China, 2011. [Google Scholar]
- Wu, C.; Dai, M.Y.; Zhang, C.; Shi, C.G.; Ren, M.J.; Ma, J.J.; Shen, Y.M. Study on the mechanism of the FLS gene regulating flower color formation in Yulania denudata and Y. liliiflora. J. Nucl. Agric. Sci. 2023, 37, 1947–1956. [Google Scholar]
- Wang, H.J.; Li, T.J.; Luo, J.; Qu, Y. Cloning and expression analysis of FLS gene in three different color Meconopsis species. Plant Phys. J. 2023, 59, 2063–2073. [Google Scholar]
- Sun, M.; Wu, L.Y.; Gong, Y.J.; Bao, R.; Gui, M.; Li, Z.B.; Du, G.H. Gene cloning and its expression analysis of SsFLS2 in Solanum sisymbriifolium. Southwest China J. Agric. Sci. 2024, 37, 1669–1676. [Google Scholar]
- Zhang, C.Y.; Liu, H.C.; Jia, C.G.; Liu, Y.J.; Wang, F.T.; Wang, J.Y. Cloing, characterization and functional analysis of a flavonol synthase from Vaccinium corymbosum. Trees 2016, 30, 1595–1605. [Google Scholar] [CrossRef]
- Prelich, G. Gene overexpression: Uses, mechanisms, and interpretation. Genetics 2012, 190, 841–854. [Google Scholar] [CrossRef]
- Gao, M.; Zhao, H.Y.; Zheng, L.T.; Zhang, L.H.; Peng, Y.J.; Ma, W.F.; Tian, R.; Yuan, Y.Y.; Ma, F.W.; Li, M.J.; et al. Overexpression of apple Ma12, a mitochondrial pyrophosphatase pump gene, leads to malic acid accumulation and the upregulation of malate dehydrogenase in tomato and apple calli. Hortic. Res. 2022, 9, uhab053. [Google Scholar] [CrossRef]
- Chen, G.; Wang, Y.P.; Liu, X.L.; Duan, S.Y.; Jiang, S.H.; Zhu, J.; Zhang, Y.G.; Hou, H.M. The MdmiR156n regulates tolerance and flavonoid synthesis in apple calli and Arabidopsis. Int. J. Mol. Sci. 2023, 24, 6049. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.C.; Li, M.F.; Zhang, Z.; Han, F.L.; Yan, W.; Liu, Y.L.; Li, M.R.; Xia, Y.N.; Yang, J.; Xu, X.F.; et al. Enhancement of grape tolerance through VvbZIP36-mediated quercetin production. Int. J. Bio. Macromol. 2025, 297, 139826. [Google Scholar] [CrossRef]
- Wei, L.L.; Wang, W.J.; Gao, X.Q.; Yao, S.X.; Deng, L.L.; Zeng, K.F. Alterations in glucose metabolism and shikimate pathway affected by transient overexpression of CsPAL gene in postharvest citrus fruit. Postharvest Biol. Technol. 2024, 213, 112936. [Google Scholar] [CrossRef]
- Yin, Y.C.; Hou, J.M.; Tian, S.K.; Yang, L.; Zhang, Z.X.; Li, W.D.; Liu, Y. Overexpressing chalcone synthase (CHS) gene enhanced flavonoids accumulation in Glycyrrhiza uralensis hairy roots. Bot. Lett. 2020, 167, 219–231. [Google Scholar] [CrossRef]
- Ma, X.T.; Hou, Y.Y.; Umar, A.W.; Wang, Y.H.; Yu, L.L.; Ahmad, N.; Yao, N.; Zhang, M.; Liu, X.M. Safflower CtFLS1-induced drought tolerance by stimulating the accumulation of flavonols and anthocyanins in Arabidopsis thaliana. Int. J. Mol. Sci. 2024, 25, 5546. [Google Scholar] [CrossRef]
- Fu, J.; Lu, Y.; Zhang, J.L.; Ni, R.; Liu, X.Y.; Hu, M.W.; Li, J.H.; Zhang, J.Z.; Jiang, N.; Xu, D.D.; et al. Functional characterization of promiscuous 2-ODD enzymes sheds light on the molecular basis for flavone and flavonol biosynthesis in ferns. Plant J. 2025, 122, e70189. [Google Scholar] [CrossRef] [PubMed]
- Kou, M.; Li, C.; Song, W.H.; Shen, Y.F.; Tang, W.; Zhang, Y.G.; Wang, X.; Yan, H.; Gao, R.F.; Ahmad, M.Q.; et al. Identification and functional characterization of a flavonol synthase gene from sweet potato [Ipomoea batatas (L.) Lam.]. Front. Plant Sci. 2023, 14, 1181173. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.T.; Sun, H.G.; Tian, C.; Su, W.W.; Garcia-Caparros, P.; Wang, J.M.; Zhou, Y.J.; Liu, T.X.; Gao, F. The AmMYB35-AmFLS module mediates the accumulation of flavonol induced by drought stress in Astragalus membranaceus. Food Biosci. 2025, 68, 106541. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, X.; Zhao, A.; Fu, L.; Wang, Y.; Ren, H.; Su, W.; Shi, M.; Liu, L.; Li, Y.; Li, D. Cloning and Functional Analysis of Flavonol Synthase Gene ZjFLS from Chinese Jujube (Ziziphus jujuba Mill.). Horticulturae 2025, 11, 729. https://doi.org/10.3390/horticulturae11070729
Xue X, Zhao A, Fu L, Wang Y, Ren H, Su W, Shi M, Liu L, Li Y, Li D. Cloning and Functional Analysis of Flavonol Synthase Gene ZjFLS from Chinese Jujube (Ziziphus jujuba Mill.). Horticulturae. 2025; 11(7):729. https://doi.org/10.3390/horticulturae11070729
Chicago/Turabian StyleXue, Xiaofang, Ailing Zhao, Le Fu, Yongkang Wang, Haiyan Ren, Wanlong Su, Meijuan Shi, Li Liu, Yi Li, and Dengke Li. 2025. "Cloning and Functional Analysis of Flavonol Synthase Gene ZjFLS from Chinese Jujube (Ziziphus jujuba Mill.)" Horticulturae 11, no. 7: 729. https://doi.org/10.3390/horticulturae11070729
APA StyleXue, X., Zhao, A., Fu, L., Wang, Y., Ren, H., Su, W., Shi, M., Liu, L., Li, Y., & Li, D. (2025). Cloning and Functional Analysis of Flavonol Synthase Gene ZjFLS from Chinese Jujube (Ziziphus jujuba Mill.). Horticulturae, 11(7), 729. https://doi.org/10.3390/horticulturae11070729





