Putative Upstream Regulators DoNF-YB3 and DoIDD12 Correlate with DoGSTF11 Expression and Anthocyanin Accumulation in Dendrobium officinale
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Gene Expression Analysis
2.2. Subcellular Localization of DoGSTF11 in Nicotiana tabacum
2.3. Agrobacterium Tumefaciens-Mediated Transformation in Solanum lycopersicum
2.4. Analysis of DoGSTF11 Transcriptional Autoactivation Activity
2.5. Yeast Two-Hybrid Assay
2.6. Bimolecular Fluorescence Complementation Experiment
2.7. Screening of Upstream Genes of DoGSTF11
3. Results
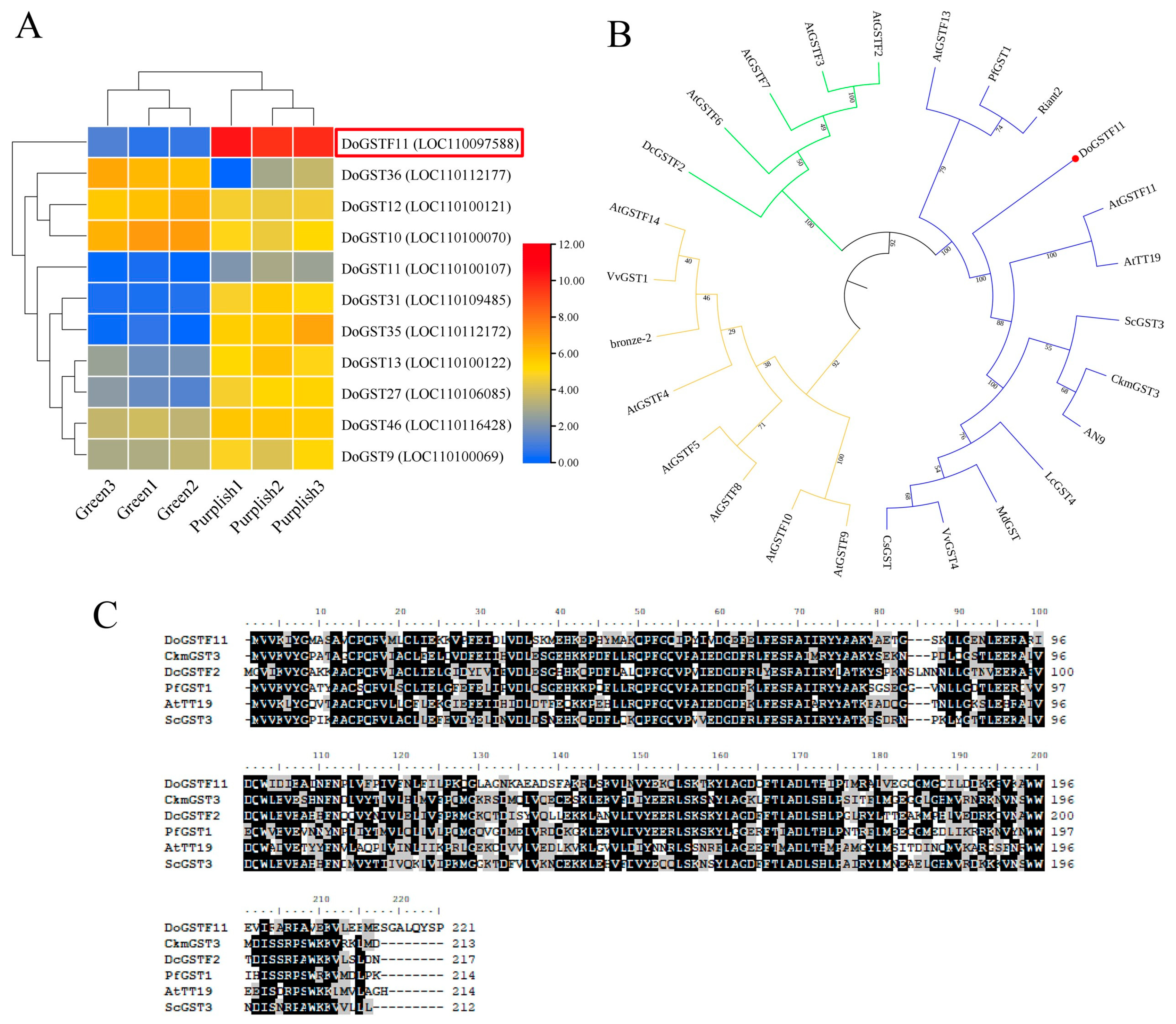
3.1. The Color Phenotype Analysis in D. officinale
3.2. Sequence Alignment and Phylogenetic Analysis
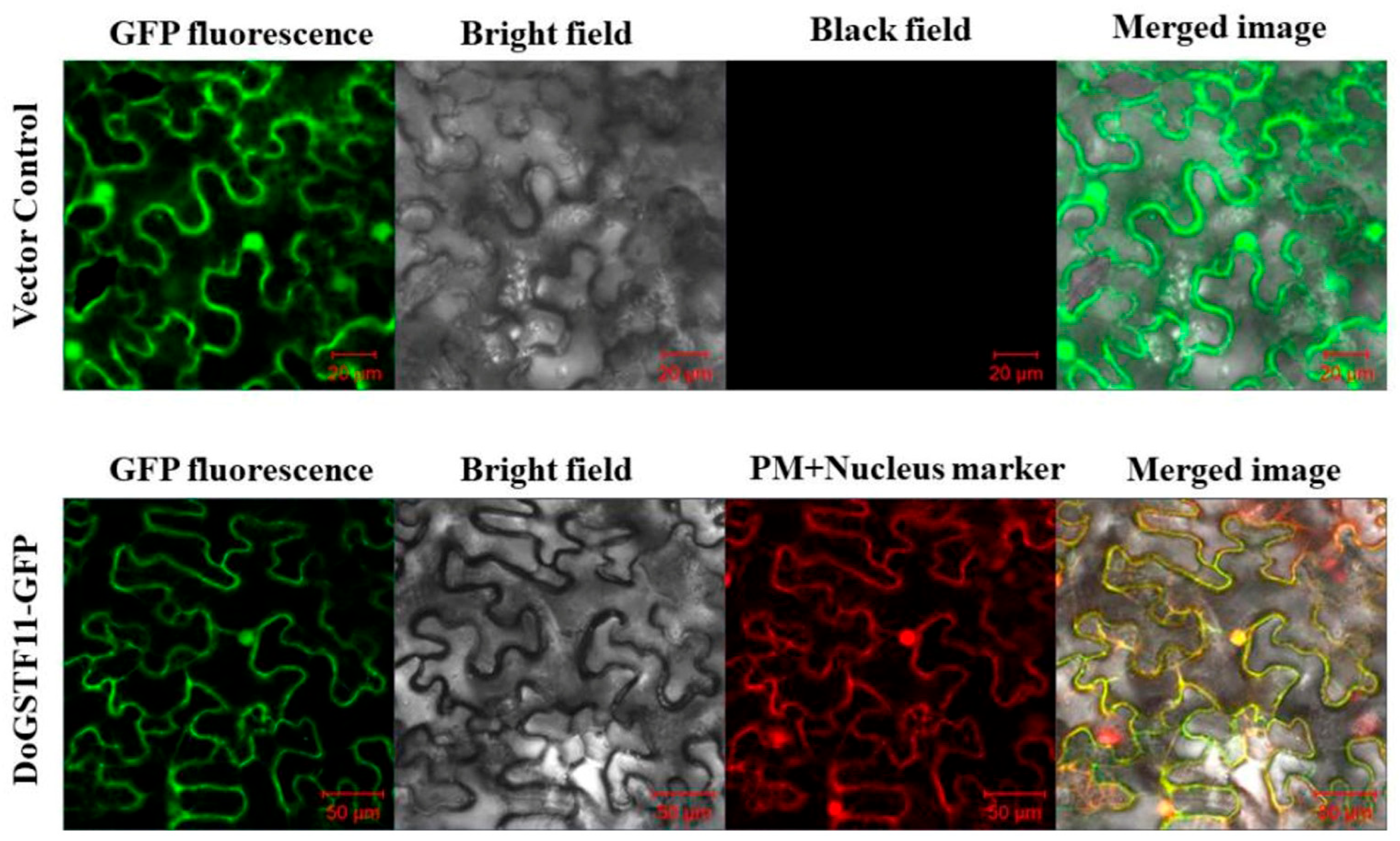
3.3. Subcellular Localization of DoGSTF11 in Nicotiana tabacum
3.4. Functional Validation in Transgenic Solanum lycopersicum
3.5. Interaction Between DoGSTF11 and DoGSTF31
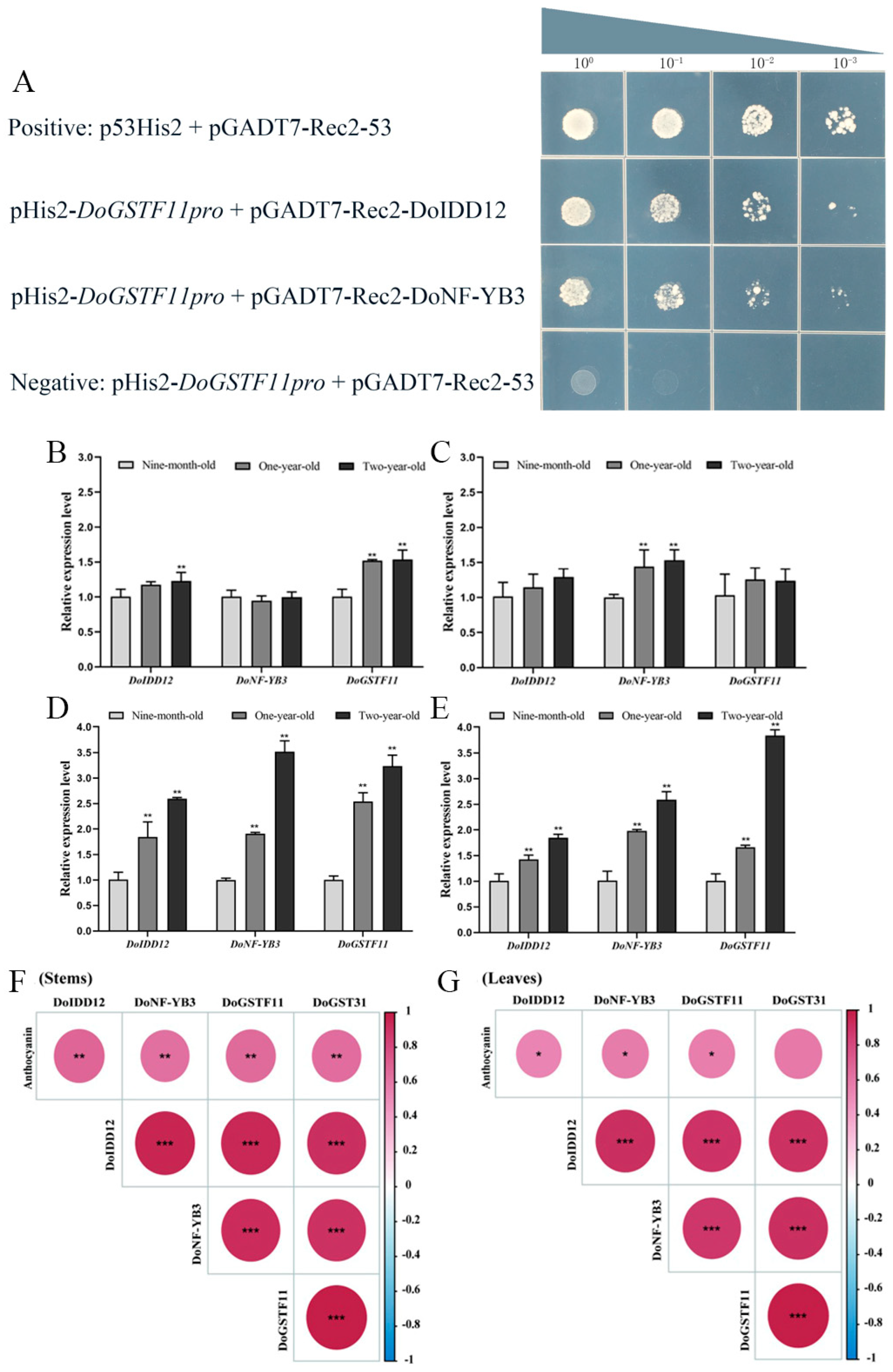
3.6. Identification of Upstream Regulatory Factors
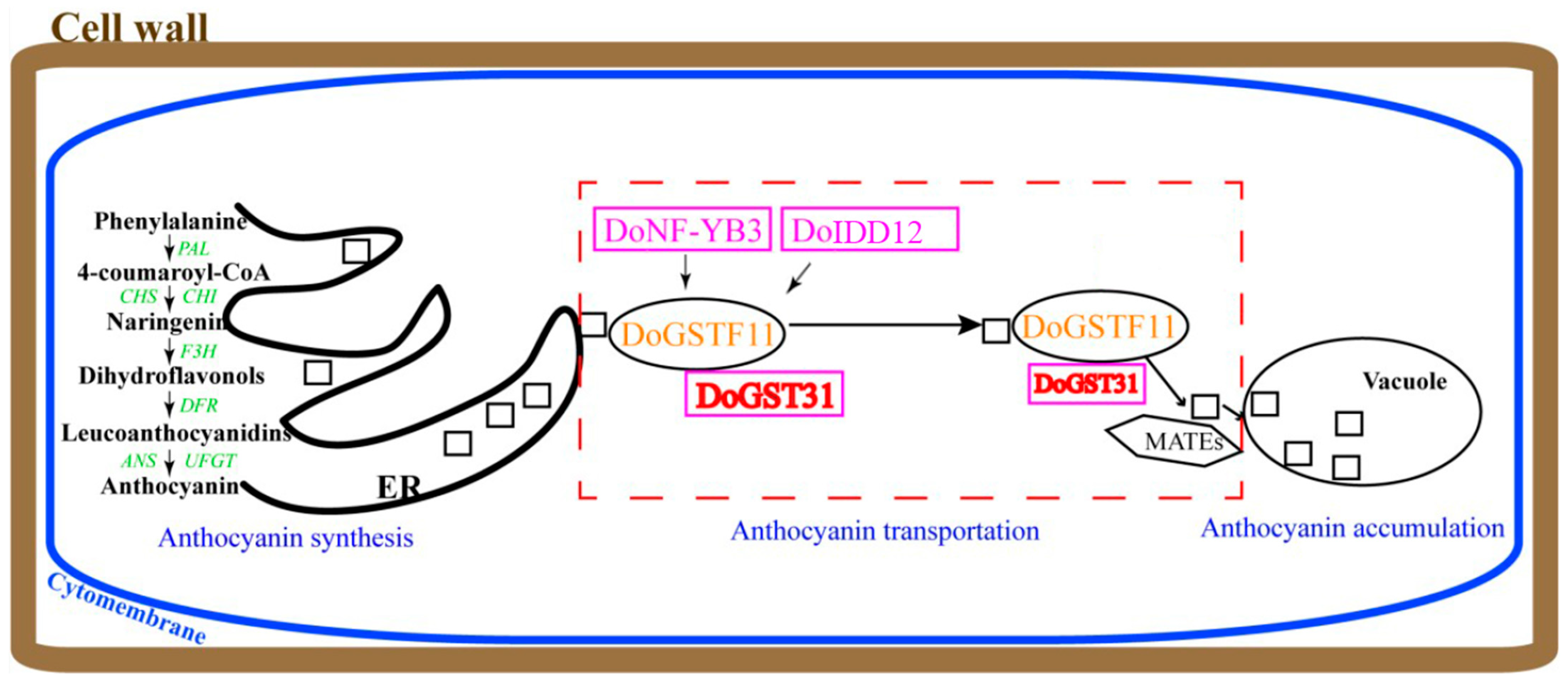
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Alvarez-Suarez, J.M.; Cuadrado, C.; Redondo, I.B.; Giampieri, F.; González-Paramás, A.M.; Santos-Buelga, C. Novel approaches in anthocyanin research-Plant fortification and bioavailability issues. Trends Food Sci. Technol. 2021, 117, 92–105. [Google Scholar] [CrossRef]
- He, F.; Mu, L.; Yan, G.-L.; Liang, N.-N.; Pan, Q.-H.; Wang, J.; Reeves, M.J.; Duan, C.-Q. Biosynthesis of anthocyanins and their regulation in colored grapes. Molecules 2010, 15, 9057–9091. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Brugliera, F.; Kalc, G.; Senior, M.; Dyson, B.; Nakamura, N.; Katsumoto, Y.; Chandler, S. Flower color modification by engineering of the flavonoid biosynthetic pathway: Practical perspectives. Biosci. Biotechnol. Biochem. 2010, 74, 1760–1769. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Lu, S. Biosynthesis and regulation of phenylpropanoids in plants. Crit. Rev. Plant Sci. 2017, 36, 257–290. [Google Scholar] [CrossRef]
- Jiang, N.; Gutierrez-Diaz, A.; Mukundi, E.; Lee, Y.S.; Meyers, B.C.; Otegui, M.S.; Grotewold, E. Synergy between the anthocyanin and RDR6/SGS3/DCL4 siRNA pathways expose hidden features of Arabidopsis carbon metabolism. Nat. Commun. 2020, 11, 2456. [Google Scholar] [CrossRef]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhang, X.; Zhao, M.; Chiang, V.L.; Sederoff, R.R.; Zhao, X. MYB-Mediated Regulation of Anthocyanin Biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef]
- Jiang, L.; Yue, M.; Liu, Y.; Zhang, N.; Lin, Y.; Zhang, Y.; Wang, Y.; Li, M.; Luo, Y.; Zhang, Y.; et al. A novel R2R3-MYB transcription factor FaMYB5 positively regulates anthocyanin and proanthocyanidin biosynthesis in cultivated strawberries (Fragaria × ananassa). Plant Biotechnol. J. 2023, 21, 1140–1158. [Google Scholar] [CrossRef]
- Qin, J.; Zhao, C.; Wang, S.; Gao, N.; Wang, X.; Na, X.; Wang, X.; Bi, Y. PIF4-PAP1 interaction affects MYB-bHLH-WD40 complex formation and anthocyanin accumulation in Arabidopsis. J. Plant Physiol. 2022, 268, 153558. [Google Scholar] [CrossRef]
- Lee, S.-H.; Kim, S.-H.; Park, T.-K.; Kim, Y.-P.; Lee, J.-W.; Kim, T.-W. Transcription factors BZR1 and PAP1 cooperate to promote anthocyanin biosynthesis in Arabidopsis shoots. Plant Cell 2024, 36, 3654–3673. [Google Scholar] [CrossRef]
- Cummins, I.; Dixon, D.P.; Freitag-Pohl, S.; Skipsey, M.; Edwards, R. Multiple roles for plant glutathione transferases in xenobiotic detoxification. Drug Metab. Rev. 2011, 43, 266–280. [Google Scholar] [CrossRef]
- Shi, H.; Wan, K.; Dou, B.; Ren, Y.; Huo, L.; Zhang, C.; Yue, S.; Li, Z.; Guo, H.; Dai, J. Genome-wide identification and expression analysis of the glutathione transferase gene family and its response to abiotic stress in rye (Secale cereale). BMC Genom. 2024, 25, 1142. [Google Scholar] [CrossRef]
- Jiang, W.; Wang, T.; Zhang, M.; Duan, X.; Chen, J.; Liu, Y.; Tao, Z.; Guo, Q. Genome-Wide Identification of Glutathione S-Transferase Family from Dendrobium officinale and the Functional Characterization of DoGST5 in Cadmium Tolerance. Int. J. Mol. Sci. 2024, 25, 8439. [Google Scholar] [CrossRef]
- Marrs, K.A.; Alfenito, M.R.; Lloyd, A.M.; Walbot, V. A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 1995, 375, 397–400. [Google Scholar] [CrossRef]
- Alfenito, M.R.; Souer, E.; Goodman, C.D.; Buell, R.; Mol, J.; Koes, R.; Walbot, V. Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell 1998, 10, 1135–1149. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, S.; Shikazono, N.; Tanaka, A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 2004, 37, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, H.; Huang, J.-R. Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol. Plant 2012, 5, 387–400. [Google Scholar] [CrossRef]
- Sasaki, N.; Nishizaki, Y.; Uchida, Y.; Wakamatsu, E.; Umemoto, N.; Momose, M.; Okamura, M.; Yoshida, H.; Yamaguchi, M.; Nakayama, M.; et al. Identification of the glutathione S-transferase gene responsible for flower color intensity in carnations. Plant Biotechnol. 2012, 29, 223–227. [Google Scholar] [CrossRef]
- Kitamura, S.; Akita, Y.; Ishizaka, H.; Narumi, I.; Tanaka, A. Molecular characterization of an anthocyanin-related glutathione S-transferase gene in cyclamen. J. Plant Physiol. 2012, 169, 636–642. [Google Scholar] [CrossRef]
- Jiang, S.; Chen, M.; He, N.; Chen, X.; Wang, N.; Sun, Q.; Zhang, T.; Xu, H.; Fang, H.; Wang, Y.; et al. MdGSTF6, activated by MdMYB1, plays an essential role in anthocyanin accumulation in apple. Hortic. Res. 2019, 6, 1–14. [Google Scholar] [CrossRef]
- Wei, K.; Wang, L.; Zhang, Y.; Ruan, L.; Li, H.; Wu, L.; Xu, L.; Zhang, C.; Zhou, X.; Cheng, H.; et al. A coupled role for CsMYB75 and CsGSTF1 in anthocyanin hyperaccumulation in purple tea. Plant J. 2019, 97, 825–840. [Google Scholar] [CrossRef]
- Tang, H.; Zhao, T.; Sheng, Y.; Zheng, T.; Fu, L.; Zhang, Y. Dendrobium officinale Kimura et Migo: A review on its ethnopharmacology, phytochemistry, pharmacology, and industrialization. Evid.-Based Complement. Altern. Med. 2017, 2017, 7436259. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Wu, Y.; Yuan, H.; Yang, Y.; Xiong, Q.; Liang, C.; Li, Z.; Li, C.; Zhang, G.; Lai, X.; et al. Dendrobium officinale polysaccharides attenuate learning and memory disabilities via anti-oxidant and anti-inflammatory actions. Int. J. Biol. Macromol. 2019, 126, 414–426. [Google Scholar] [CrossRef]
- Lei, Z.; Zhou, C.; Ji, X.; Wei, G.; Huang, Y.; Yu, W.; Luo, Y.; Qiu, Y. Transcriptome analysis reveals genes involved in flavonoid biosynthesis and accumulation in Dendrobium catenatum from different locations. Sci. Rep. 2018, 8, 6373. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Liao, Y.; da Silva, J.A.T.; Yang, Z.; Duan, J. Differential accumulation of anthocyanins in Dendrobium officinale stems with red and green peels. Int. J. Mol. Sci. 2018, 19, 2857. [Google Scholar] [CrossRef]
- Ng, T.B.; Liu, J.; Wong, J.H.; Ye, X.; Wing Sze, S.C.; Tong, Y.; Zhang, K.Y. Review of research on Dendrobium, a prized folk medicine. Appl. Microbiol. Biotechnol. 2012, 93, 1795–1803. [Google Scholar] [CrossRef]
- Zhang, Z.; Tian, C.; Zhang, Y.; Li, C.; Li, X.; Yu, Q.; Wang, S.; Wang, X.; Chen, X.; Feng, S. Transcriptomic and metabolomic analysis provides insights into anthocyanin and procyanidin accumulation in pear. BMC Plant Biol. 2020, 20, 129. [Google Scholar] [CrossRef]
- Shen, J.; Zou, Z.; Zhang, X.; Zhou, L.; Wang, Y.; Fang, W.; Zhu, X. Metabolic analyses reveal different mechanisms of leaf color change in two purple-leaf tea plant (Camellia sinensis L.) cultivars. Hortic. Res. 2018, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Maren, N.A.; Duduit, J.R.; Huang, D.; Zhao, F.; Ranney, T.G.; Liu, W. Stepwise Optimization of Real-Time RT-PCR Analysis. Methods Mol. Biol. 2023, 2653, 317–332. [Google Scholar]
- Zhong, C.; Tang, Y.; Pang, B.; Li, X.; Yang, Y.; Deng, J.; Feng, C.; Li, L.; Ren, G.; Wang, Y.; et al. The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida. Hortic. Res. 2020, 7, 78. [Google Scholar] [CrossRef]
- Zhao, D.; Tao, J. Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 2015, 6, 261. [Google Scholar] [CrossRef]
- Fu, M.; Yang, X.; Zheng, J.; Wang, L.; Yang, X.; Tu, Y.; Ye, J.; Zhang, W.; Liao, Y.; Cheng, S.; et al. Unraveling the regulatory mechanism of color diversity in Camellia japonica petals by integrative transcriptome and metabolome analysis. Front. Plant Sci. 2021, 12, 685136. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.Y.; Xie, Z.S.; Huang, Y.C.; Yuan, Y.U.A.N.; Zhou, C.J.; Wang, Y.W.; Wei, G. HPLC characteristic spectrum optimization of flavonoid glycosides on Dendrobium officinale and characteristics analysis of different provenances. Chin. J. Exp. Tradit. Med. Formulae 2019, 22–28. [Google Scholar]
- Lin, R.C.; Rausher, M.D. R2R3-MYB genes control petal pigmentation patterning in Clarkia gracilis ssp. sonomensis (Onagraceae). New Phytol. 2021, 229, 1147–1162. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.; Curtin, C.; Bézier, A.; Franco, C.; Zhang, W. Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J. Exp. Bot. 2008, 59, 3621–3634. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Ghanashyam, C.; Bhattacharjee, A. Comprehensive expression analysis suggests overlapping and specific roles of rice glutathione S-transferase genes during development and stress responses. BMC Genom. 2010, 11, 73. [Google Scholar] [CrossRef]
- Islam, S.; Rahman, I.A.; Islam, T.; Ghosh, A.; Chakrabarty, D. Genome-wide identification and expression analysis of glutathione S-transferase gene family in tomato: Gaining an insight to their physiological and stress-specific roles. PLoS ONE 2017, 12, e0187504. [Google Scholar] [CrossRef]
- Larsen, E.S.; Alfenito, M.R.; Briggs, W.R.; Walbot, V. A carnation anthocyanin mutant is complemented by the glutathione S-transferases encoded by maize Bz2 and petunia An9. Plant Cell Rep. 2003, 21, 900–904. [Google Scholar] [CrossRef]
- Mueller, L.A.; Goodman, C.D.; Silady, R.A.; Walbot, V. AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol. 2000, 123, 1561–1570. [Google Scholar] [CrossRef]
- Luo, H.; Dai, C.; Li, Y.; Feng, J.; Liu, Z.; Kang, C. Reduced Anthocyanins in Petioles codes for a GST anthocyanin transporter that is essential for the foliage and fruit coloration in strawberry. J. Exp. Bot. 2018, 69, 2595–2608. [Google Scholar] [CrossRef]
- Hu, B.; Zhao, J.; Lai, B.; Qin, Y.; Wang, H.; Hu, G. LcGST4 is an anthocyanin-related glutathione S-transferase gene in Litchi chinensis Sonn. Plant Cell Rep. 2016, 35, 831–843. [Google Scholar] [CrossRef]
- Cheng, J.; Liao, L.; Zhou, H.; Gu, C.; Wang, L.; Han, Y. A small indel mutation in an anthocyanin transporter causes variegated colouration of peach flowers. J. Exp. Bot. 2015, 66, 7227–7239. [Google Scholar] [CrossRef]
- Kaur, S.; Sharma, N.; Kapoor, P.; Chunduri, V.; Pandey, A.K.; Garg, M. Spotlight on the overlapping routes and partners for anthocyanin transport in plants. Physiol. Plant 2021, 171, 868–881. [Google Scholar] [CrossRef] [PubMed]
- Sylvia, C.; Sun, J.; Zhang, Y.; Ntini, C.; Ogutu, C.; Zhao, Y.; Han, Y. Genome-Wide Analysis of ATP Binding Cassette (ABC) Transporters in Peach (Prunus persica) and Identification of a Gene PpABCC1 Involved in Anthocyanin Accumulation. Int. J. Mol. Sci. 2023, 24, 1931. [Google Scholar] [CrossRef] [PubMed]
- Chaves-Silva, S.; dos Santos, A.L.; Chalfun-Júnior, A.; Zhao, J.; Peres, L.E.; Benedito, V.A. Understanding the genetic regulation of anthocyanin biosynthesis in plants—Tools for breeding purple varieties of fruits and vegetables. Phytochemistry 2018, 153, 11–27. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004, 39, 366–380. [Google Scholar] [CrossRef]
- Albert, N.W.; Butelli, E.; Moss, S.M.; Piazza, P.; Waite, C.N.; Schwinn, K.E.; Davies, K.M.; Martin, C. Discrete bHLH transcription factors play functionally overlapping roles in pigmentation patterning in flowers of Antirrhinum majus. New Phytol. 2021, 231, 849–863. [Google Scholar] [CrossRef]
- Matus, J.T.; Poupin, M.J.; Cañón, P.; Bordeu, E.; Alcalde, J.A.; Arce-Johnson, P. Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine (Vitis vinifera L.). Plant Mol. Biol. 2010, 72, 607–620. [Google Scholar] [CrossRef]
- Liu, B.; Wang, L.; Wang, S.; Li, W.; Liu, D.; Guo, X.; Qu, B. Transcriptomic analysis of bagging-treated ‘Pingguo’ pear shows that MYB4-like1, MYB4-like2, MYB1R1 and WDR involved in anthocyanin biosynthesis are up-regulated in fruit peels in response to light. Sci. Hortic. 2019, 244, 428–434. [Google Scholar] [CrossRef]
- Zhang, Q.; Feng, Y.-X.; Tian, P.; Lin, Y.-J.; Yu, X.-Z. Proline-mediated regulation on jasmonate signals repressed anthocyanin accumulation through the MYB-bHLH-WDR complex in rice under chromium exposure. Front. Plant Sci. 2022, 13, 953398. [Google Scholar] [CrossRef] [PubMed]
- Dou, F.; Phillip, F.O.; Liu, H. Combined Metabolome and Transcriptome Analysis Revealed the Accumulation of Anthocyanins in Grape Berry (Vitis vinifera L.) under High-Temperature Stress. Plants 2024, 13, 2394. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Risueno, M.A.; Sozzani, R.; Yardımcı, G.G.; Petricka, J.J.; Vernoux, T.; Blilou, I.; Alonso, J.; Winter, C.M.; Ohler, U.; Scheres, B.; et al. Transcriptional control of tissue formation throughout root development. Science 2015, 350, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Lan, C.; Li, X.; Xie, W.; Lin, F.; Liang, Y.; Tao, Z. A pair of nuclear factor Y transcription factors act as positive regulators in jasmonate signaling and disease resistance in Arabidopsis. J. Integr. Plant Biol. 2024, 66, 2042–2057. [Google Scholar] [CrossRef]
- Zhang, P.; Li, X.; Wang, Y.; Guo, W.; Chachar, S.; Riaz, A.; Geng, Y.; Gu, X.; Yang, L. PRMT6 physically associates with nuclear factor Y to regulate photoperiodic flowering in Arabidopsis. aBIOTECH 2021, 2, 403–414. [Google Scholar] [CrossRef]
- Sato, H.; Suzuki, T.; Takahashi, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NF-YB2 and NF-YB3 Have Functionally Diverged and Differentially Induce Drought and Heat Stress-Specific Genes. Plant Physiol. 2019, 180, 1677–1690. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Chen, J.; Duan, X.; Zhang, M.; Tao, Z.; Jiang, W. Putative Upstream Regulators DoNF-YB3 and DoIDD12 Correlate with DoGSTF11 Expression and Anthocyanin Accumulation in Dendrobium officinale. Horticulturae 2025, 11, 711. https://doi.org/10.3390/horticulturae11060711
Liu Y, Chen J, Duan X, Zhang M, Tao Z, Jiang W. Putative Upstream Regulators DoNF-YB3 and DoIDD12 Correlate with DoGSTF11 Expression and Anthocyanin Accumulation in Dendrobium officinale. Horticulturae. 2025; 11(6):711. https://doi.org/10.3390/horticulturae11060711
Chicago/Turabian StyleLiu, Yingying, Jiadong Chen, Xiaojing Duan, Man Zhang, Zhengming Tao, and Wu Jiang. 2025. "Putative Upstream Regulators DoNF-YB3 and DoIDD12 Correlate with DoGSTF11 Expression and Anthocyanin Accumulation in Dendrobium officinale" Horticulturae 11, no. 6: 711. https://doi.org/10.3390/horticulturae11060711
APA StyleLiu, Y., Chen, J., Duan, X., Zhang, M., Tao, Z., & Jiang, W. (2025). Putative Upstream Regulators DoNF-YB3 and DoIDD12 Correlate with DoGSTF11 Expression and Anthocyanin Accumulation in Dendrobium officinale. Horticulturae, 11(6), 711. https://doi.org/10.3390/horticulturae11060711





