Genome-Wide Identification of the Eceriferum Gene Family and Analysis of Gene Expression Patterns Under Different Treatments in Pepper (Capsicum annuum L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Retrieval and Identification of Eceriferum Genes in Pepper
2.2. Analysis of Physicochemical Properties and Subcellular Localization of CalCERs
2.3. Sequence Alignment and Construction of Phylogenetic Tree
2.4. Gene Structure and Motif Analysis of CalCERs
2.5. Cis-Regulatory Element Analysis of CalCER Promoters
2.6. RNA-Seq Data Analyses
3. Results
3.1. Identification and Physicochemical Properties of CER Genes in Pepper
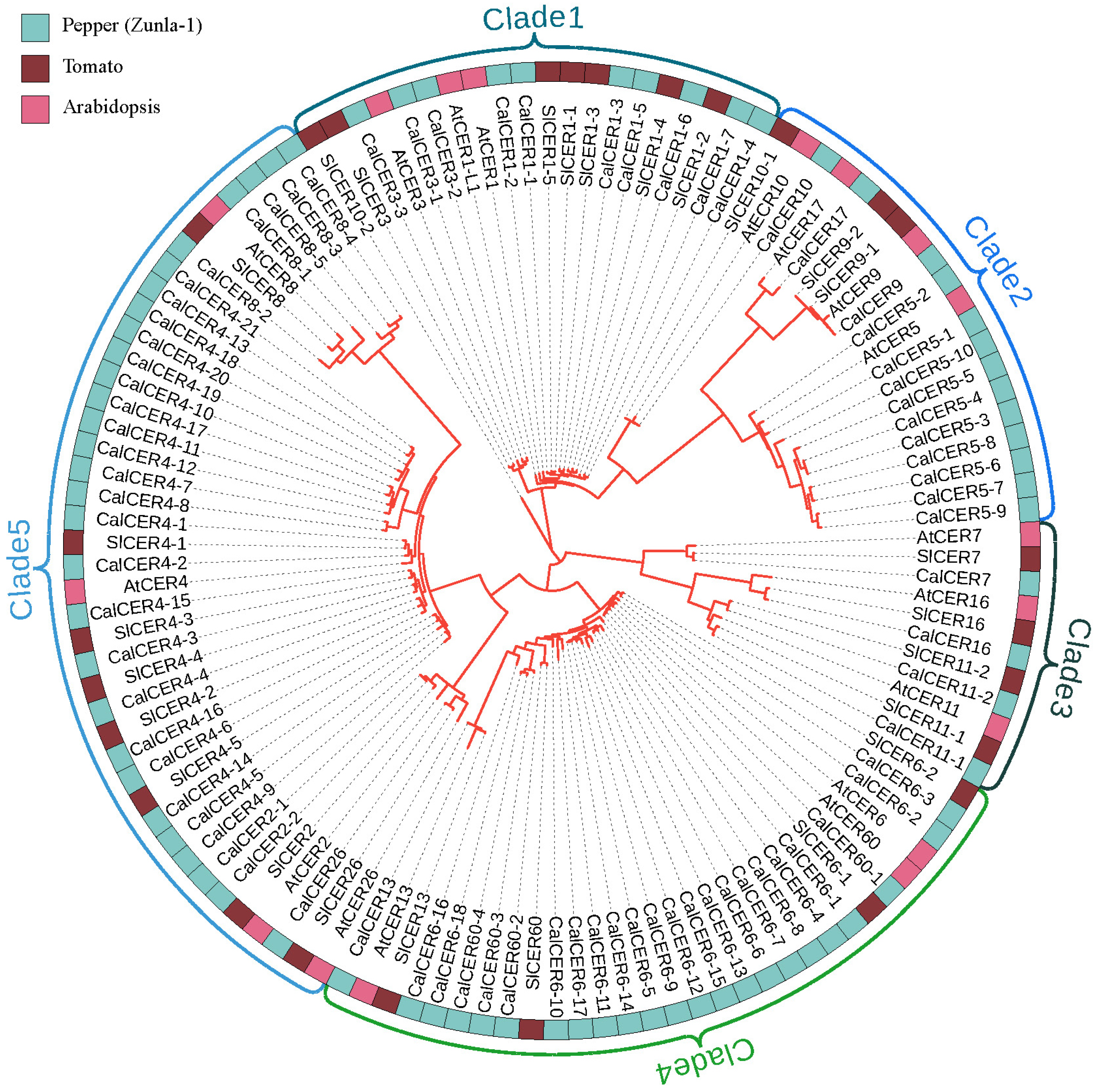
3.2. Phylogenetic Analysis of CalCER Proteins in Pepper
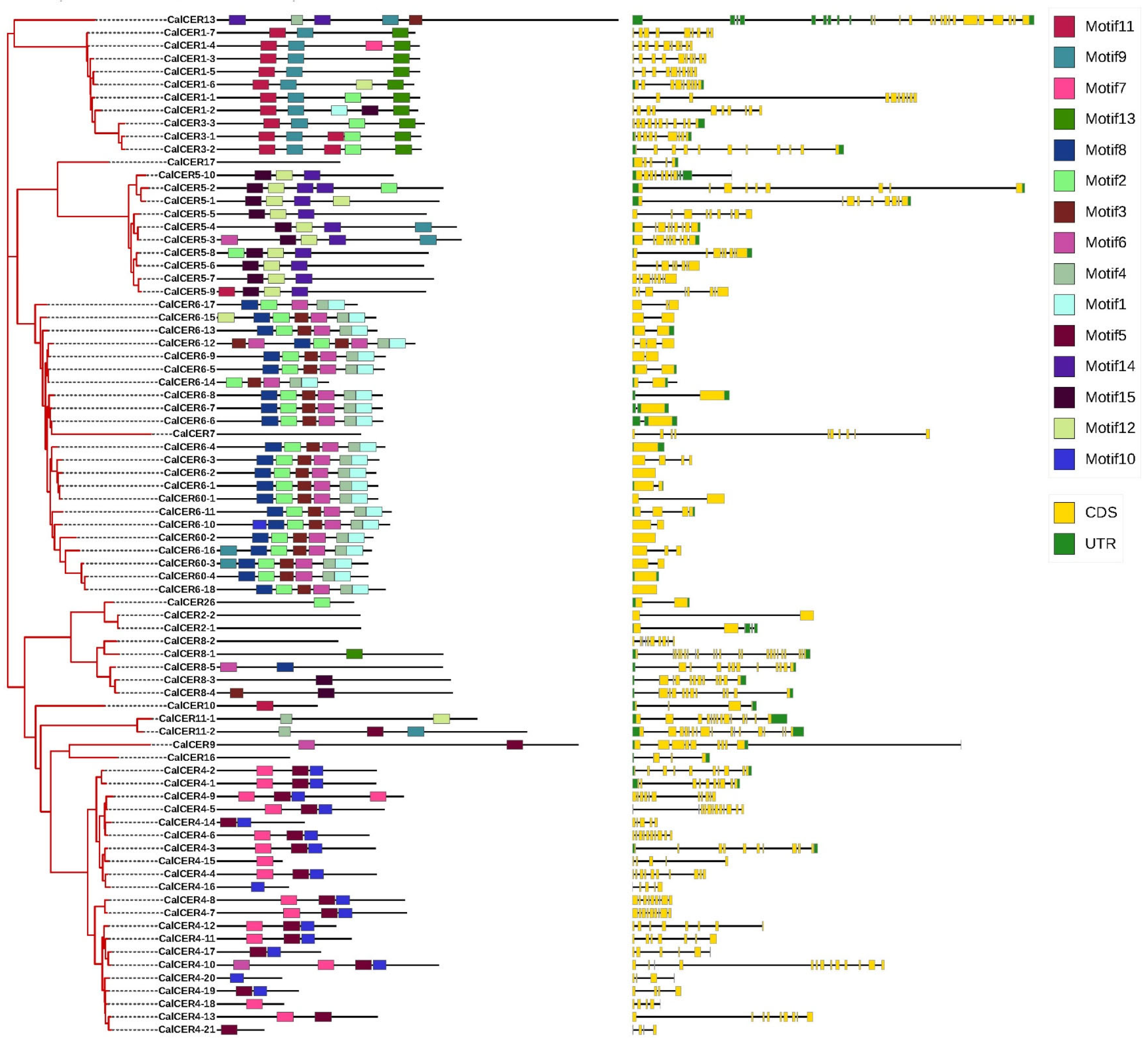
3.3. Motifs and Gene Structural Analysis of CalCERs
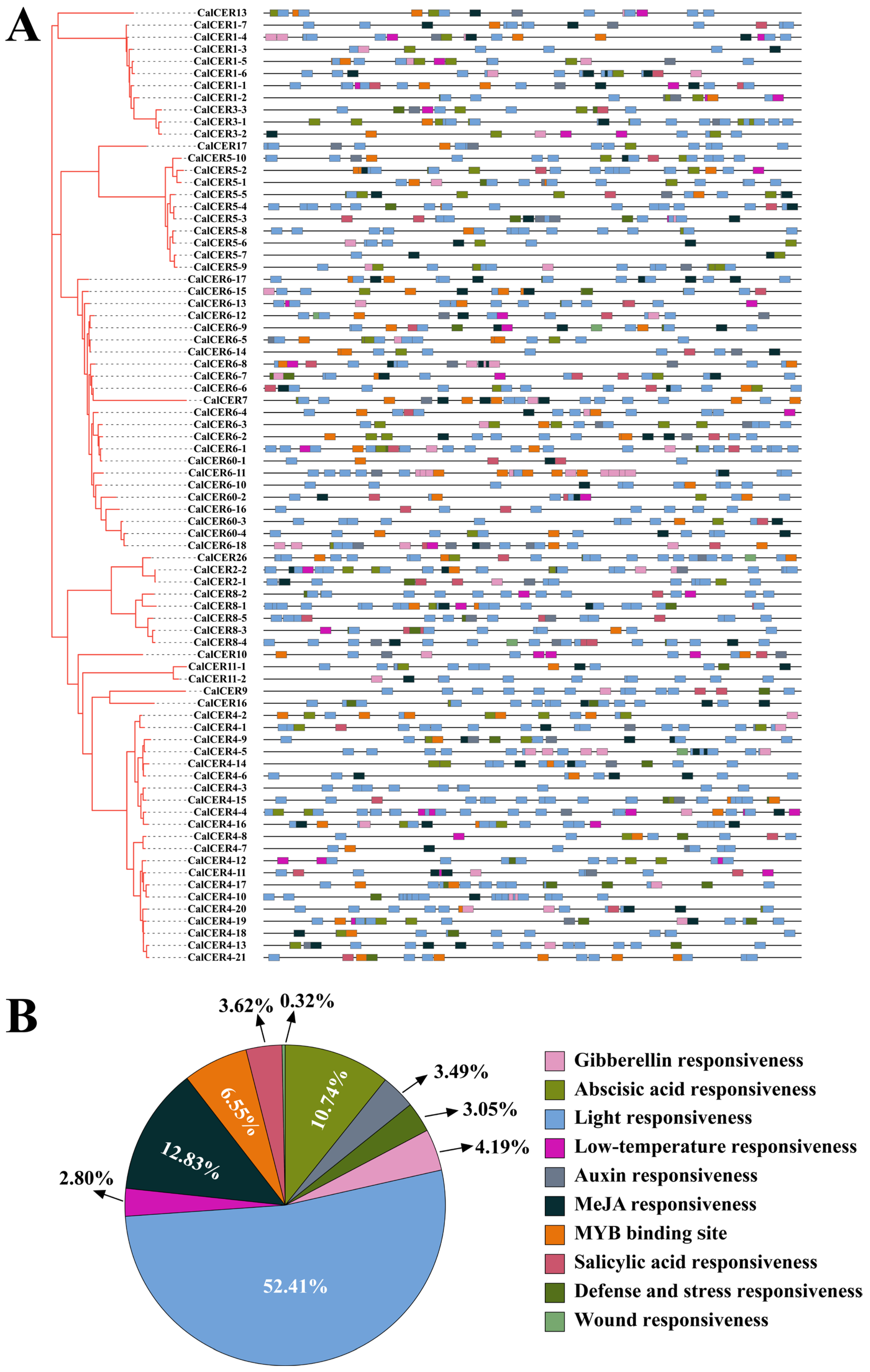
3.4. Cis-Regulatory Element Analysis of CalCERs
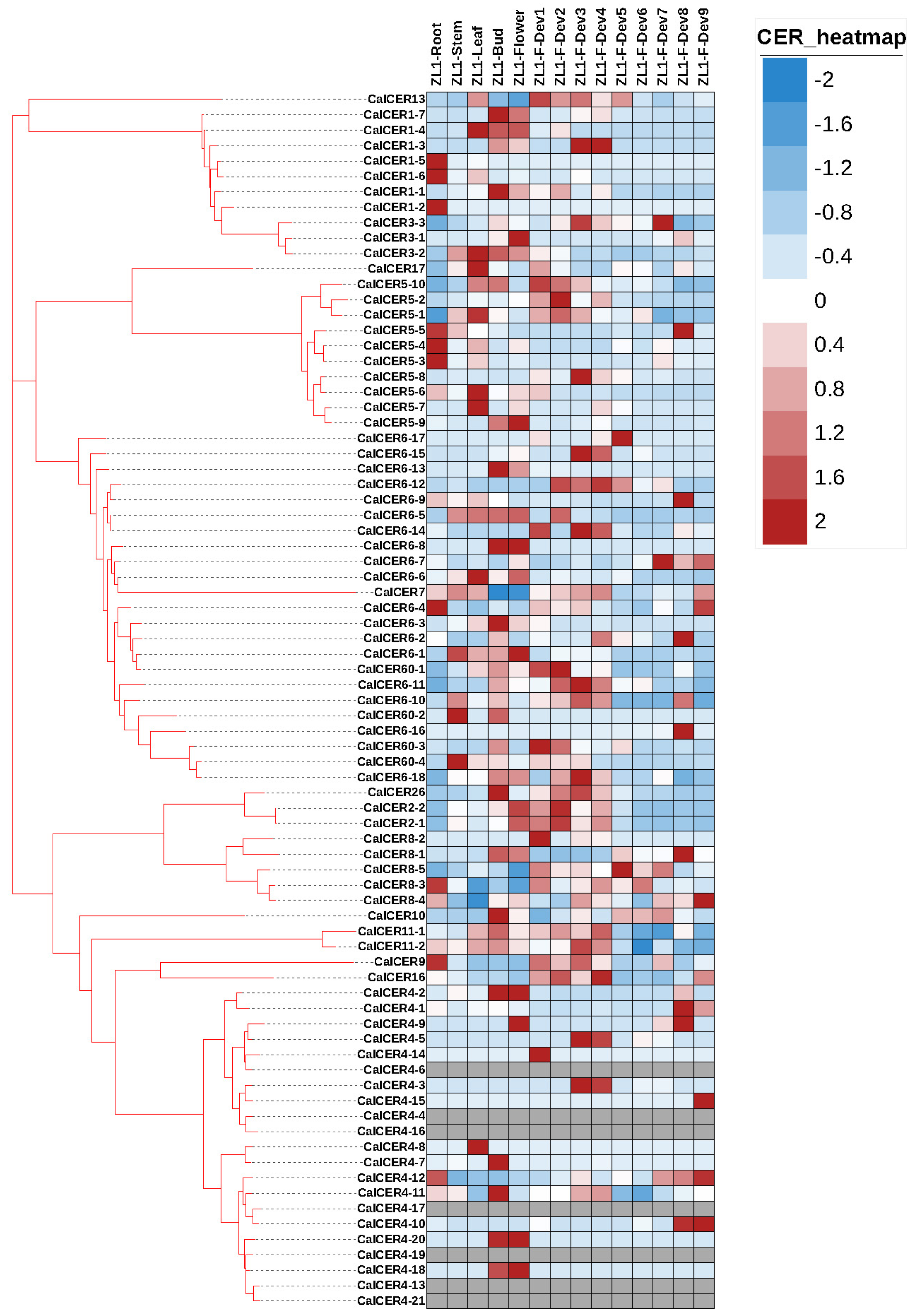
3.5. Expression Profiles of CalCER Genes During Different Development Stages
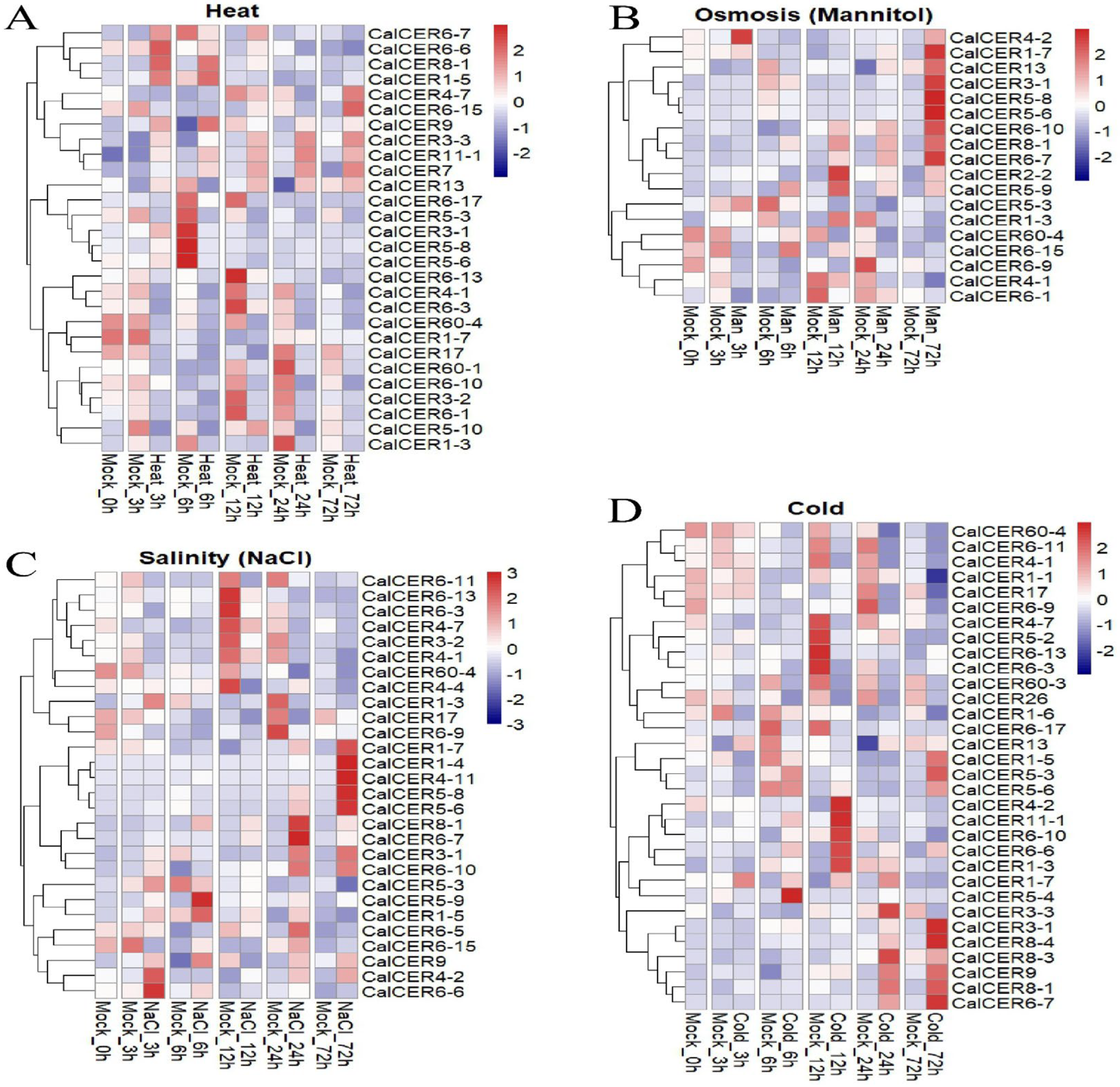
3.6. Expression Profiles of CalCERs Under Abiotic Stress
3.7. Expression Profiles of CalCERs Under Biotic Stress
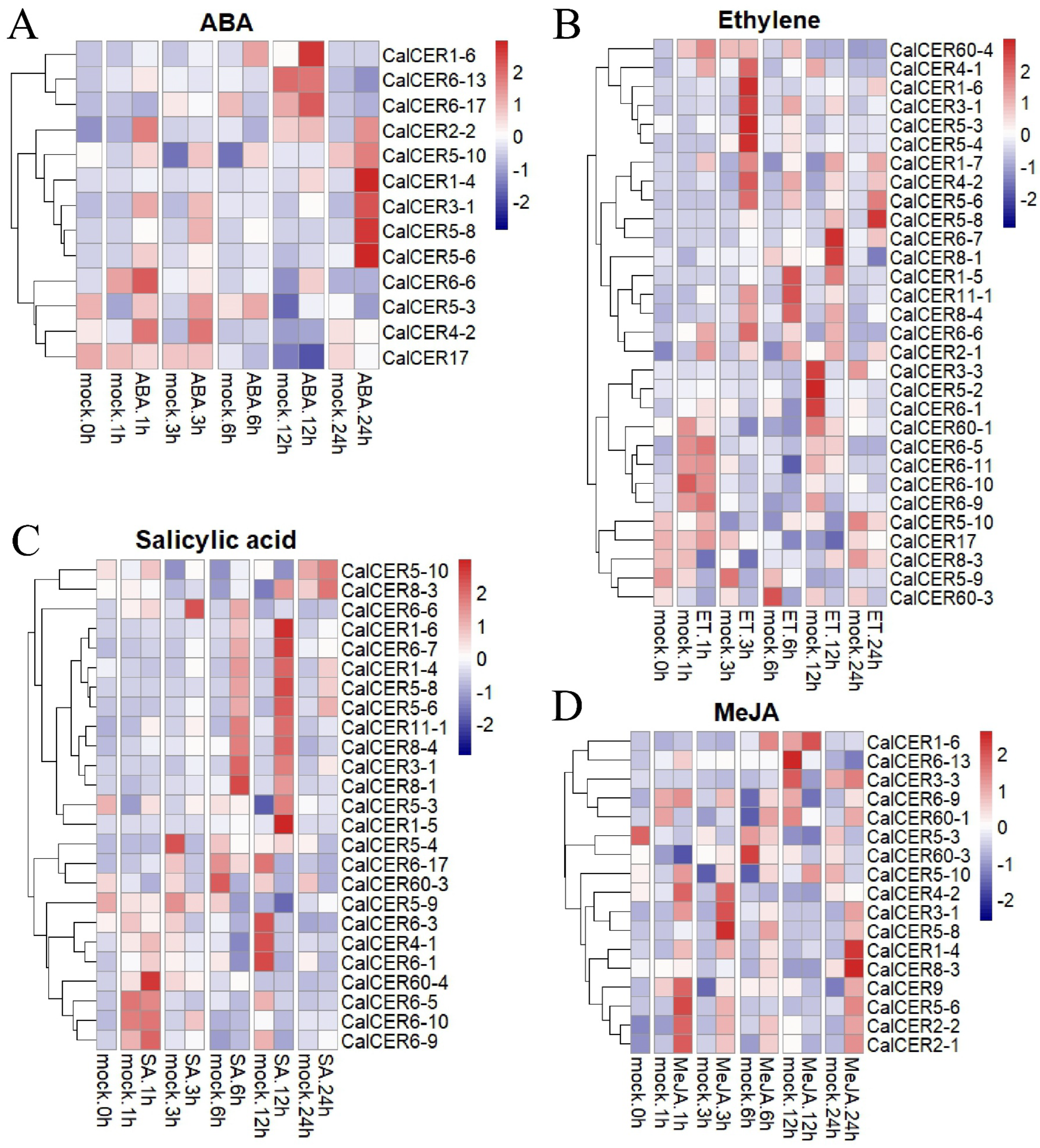
3.8. Expression Profiles of CalCERs Under Phytohormone Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Waters, E.R. Molecular adaptation and the origin of land plants. Mol. Phylogenetics Evol. 2003, 29, 456–463. [Google Scholar] [CrossRef]
- Borisjuk, N.; Hrmova, M.; Lopato, S. Transcriptional regulation of cuticle biosynthesis. Biotechnol. Adv. 2014, 32, 526–540. [Google Scholar] [CrossRef]
- Wang, X.; Guan, Y.; Zhang, D.; Dong, X.; Tian, L.; Qu, L.Q. A β-Ketoacyl-CoA synthase is involved in rice leaf cuticular wax synthesis and requires a CER2-LIKE protein as a cofactor. Plant Physiol. 2016, 173, 944–955. [Google Scholar] [CrossRef]
- Hannoufa, A.; McNevin, J.; Lemieux, B. Epicuticular waxes of eceriferum mutants of Arabidopsis thaliana. Phytochemistry 1993, 33, 851–855. [Google Scholar] [CrossRef]
- Samuels, L.; Kunst, L.; Jetter, R. Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 2008, 59, 683–707. [Google Scholar] [CrossRef]
- Bourdenx, B.; Bernard, A.; Domergue, F.; Pascal, S.; Léger, A.; Roby, D.; Pervent, M.; Vile, D.; Haslam, R.P.; Napier, J.A.; et al. Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 2011, 156, 29–45. [Google Scholar] [CrossRef]
- Aarts, M.G.; Keijzer, C.J.; Stiekema, W.J.; Pereira, A. Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 1995, 7, 2115–2127. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, Z.; Cheng, J.; Xian, X.; Li, C.; Wang, Y. Genome-wide identification of the CER1 gene family in apple and response of MdCER1-1 to drought stress. Funct. Integr. Genom. 2023, 23, 17. [Google Scholar] [CrossRef]
- Ni, E.; Zhou, L.; Li, J.; Jiang, D.; Wang, Z.; Zheng, S.; Qi, H.; Zhou, Y.; Wang, C.; Xiao, S.; et al. OsCER1 plays a pivotal role in very-long-chain alkane biosynthesis and affects plastid development and programmed cell death of tapetum in rice (Oryza sativa L.). Front. Plant Sci. 2018, 9, 1217. [Google Scholar] [CrossRef]
- Haslam, T.M.; Haslam, R.; Thoraval, D.; Pascal, S.; Delude, C.; Domergue, F.; Fernández, A.M.; Beaudoin, F.; Napier, J.A.; Kunst, L.; et al. ECERIFERUM2-LIKE proteins have unique biochemical and physiological functions in very-long-chain fatty acid elongation. Plant Physiol. 2015, 167, 682–692. [Google Scholar] [CrossRef]
- Zhong, M.S.; Jiang, H.; Cao, Y.; Wang, Y.X.; You, C.X.; Li, Y.Y.; Hao, Y.J. MdCER2 conferred to wax accumulation and increased drought tolerance in plants. Plant Physiol. Biochem. 2020, 149, 277–285. [Google Scholar] [CrossRef]
- McNevin, J.P.; Woodward, W.; Hannoufa, A.; Feldmann, K.A.; Lemieux, B. Isolation and characterization of eceriferum (cer) mutants induced by T-DNA insertions in Arabidopsis thaliana. Genome 1993, 36, 610–618. [Google Scholar] [CrossRef]
- Jenks, M.A.; Tuttle, H.A.; Eigenbrode, S.D.; Feldmann, K.A. Leaf epicuticular waxes of the eceriferum mutants in Arabidopsis. Plant Physiol. 1995, 108, 369–377. [Google Scholar] [CrossRef]
- Wang, Y.; Jin, S.; Xu, Y.; Li, S.; Zhang, S.; Yuan, Z.; Li, J.; Ni, Y. Overexpression of BnKCS1-1, BnKCS1-2, and BnCER1- 2 promotes cuticular wax production and increases drought tolerance in Brassica napus. Crop J. 2019, 8, 12. [Google Scholar] [CrossRef]
- Rowland, O.; Zheng, H.; Hepworth, S.R.; Lam, P.; Jetter, R.; Kunst, L. CER4 encodes an alcohol-forming fatty acyl-coenzyme a reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 2006, 142, 866–877. [Google Scholar] [CrossRef]
- Pighin, J.A.; Zheng, H.; Balakshin, L.J.; Goodman, I.P.; Western, T.L.; Jetter, R.; Kunst, L.; Samuels, A.L. Plant cuticular lipid export requires an ABC transporter. Science 2004, 306, 702–704. [Google Scholar] [CrossRef]
- Nawrath, C.; Schreiber, L.; Franke, R.B.; Geldner, N.; Reina-Pinto, J.J.; Kunst, L. Apoplastic diffusion barriers in Arabidopsis. Arab. Book 2013, 11, e0167. [Google Scholar] [CrossRef]
- Lü, S.; Song, T.; Kosma, D.K.; Parsons, E.P.; Rowland, O.; Jenks, M.A. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 2009, 59, 553–564. [Google Scholar] [CrossRef]
- Lee, S.B.; Suh, M.C. Regulatory mechanisms underlying cuticular wax biosynthesis. J. Exp. Bot. 2022, 73, 2799–2816. [Google Scholar] [CrossRef]
- Lü, S.; Zhao, H.; Des Marais, D.L.; Parsons, E.P.; Wen, X.; Xu, X.; Bangarusamy, D.K.; Wang, G.; Rowland, O.; Juenger, T.; et al. Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water status. Plant Physiol. 2012, 159, 930–944. [Google Scholar] [CrossRef]
- Zheng, H.; Rowland, O.; Kunst, L. Disruptions of the Arabidopsis Enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 2005, 17, 1467–1481. [Google Scholar] [CrossRef]
- Trenkamp, S.; Martin, W.; Tietjen, K. Specific and differential inhibition of very-long-chain fatty acid elongases from Arabidopsis thaliana by different herbicides. Proc. Natl. Acad. Sci. USA 2004, 101, 11903–11908. [Google Scholar] [CrossRef]
- Pascal, S.; Bernard, A.; Sorel, M.; Pervent, M.; Vile, D.; Haslam, R.P.; Napier, J.A.; Lessire, R.; Domergue, F.; Joubès, J. The Arabidopsis cer26 mutant, like the cer2 mutant, is specifically affected in the very long chain fatty acid elongation process. Plant J. 2013, 73, 733–746. [Google Scholar] [CrossRef]
- Zhukov, A.; Popov, V. Synthesis of C20-38 Fatty Acids in Plant Tissues. Int. J. Mol. Sci. 2022, 23, 4731. [Google Scholar] [CrossRef]
- Cameron, K.D.; Teece, M.A.; Smart, L.B. Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol. 2006, 140, 176–183. [Google Scholar] [CrossRef]
- Qi, C.; Jiang, H.; Zhao, X.; Mao, K.; Liu, H.; Li, Y.; Hao, Y. The Characterization, Authentication, and Gene Expression Pattern of the MdCER Family in Malus domestica. Hortic. Plant J. 2019, 5, 1–9. [Google Scholar] [CrossRef]
- Ahmad, H.M.; Wang, X.; Fiaz, S.; Nadeem, M.A.; Khan, S.A.; Ahmar, S.; Azeem, F.; Shaheen, T.; Mora-Poblete, F. Comprehensive genomics and expression analysis of eceriferum (CER) genes in sunflower (Helianthus annuus). Saudi J. Biol. Sci. 2021, 28, 6884–6896. [Google Scholar] [CrossRef]
- Li, N.; Li, X.; Song, Y.; Yang, S.; Li, L. Genome-wide identification, characterization, and expression profiling of the ECERIFERUM (CER) gene family in Ziziphus jujube. Russ. J. Plant Physiol. 2021, 68, 828–837. [Google Scholar] [CrossRef]
- Rizwan, H.M.; Waheed, A.; Ma, S.; Li, J.; Arshad, M.B.; Irshad, M.; Li, B.; Yang, X.; Ali, A.; Ahmed, M.A.A.; et al. Comprehensive Genome-Wide Identification and Expression Profiling of Eceriferum (CER) Gene Family in Passion Fruit (Passiflora edulis) Under Fusarium kyushuense and Drought Stress Conditions. Front. Plant Sci. 2022, 13, 898307. [Google Scholar] [CrossRef]
- Zhao, S.; Nie, X.; Liu, X.; Wang, B.; Liu, S.; Qin, L.; Xing, Y. Genome-wide identification of the CER Gene Family and significant features in Climate Adaptation of Castanea mollissima. Int. J. Mol. Sci. 2022, 23, 16202. [Google Scholar] [CrossRef]
- Pan, F.; Li, X.; Zhong, D.; Lu, X.; Pan, C.; Hu, J.; Su, W.; Zhang, H.; Zhang, C.; Shi, L.; et al. Eceriferum Genes in Tomato (Solanum lycopersicum): Genome-Wide Identification and Expression Analysis Reveal Their Potential Functions during Domestication. Horticulturae 2023, 9, 748. [Google Scholar] [CrossRef]
- Hamid, R.; Ghorbanzadeh, Z.; Jacob, F.; Nekouei, M.K.; Zeinalabedini, M.; Mardi, M.; Sadeghi, A.; Ghaffari, M.R. Decoding drought resilience: A comprehensive exploration of the cotton Eceriferum (CER) gene family and its role in stress adaptation. BMC Plant Biol. 2024, 24, 468. [Google Scholar] [CrossRef]
- Cao, W.; Sun, H.; Wang, C.; Yang, L.; Zhang, Y.; Zhuang, M.; Lv, H.; Wang, Y.; Liu, F.; Ji, J. Genome-wide identification of the ECERIFERUM (CER) gene family in cabbage and critical role of BoCER4.1 in wax biosynthesis. Plant Physiol. Biochem. 2025, 222, 109718. [Google Scholar] [CrossRef]
- Schreiber, L. Transport barriers made of cutin, suberin and associated waxes. Trends Plant Sci. 2010, 15, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Komatsuda, T.; Ma, J.F.; Li, C.; Yamaji, N.; Nevo, E. A functional cutin matrix is required for plant protection against water loss. Plant Signal. Behav. 2011, 6, 1297–1299. [Google Scholar] [CrossRef]
- Ahmad, H.M.; Azeem, F.; Shaheen, T.; Irshad, M.A. Genome-wide analysis of long chain Acyl-CoA synthetase (LACS) genes in sunflower (Helianthus annuus) suggests their role in drought stress. Int. J. Agric. Biol. 2020, 12, 6884–6896. [Google Scholar] [CrossRef]
- Duan, Y.; Jiang, Y.; Ye, S.; Karim, A.; Ling, Z.; He, Y.; Yang, S.; Luo, K. PtrWRKY73, a salicylic acid-inducible poplar WRKY transcription factor, is involved in disease resistance in Arabidopsis thaliana. Plant Cell Rep. 2015, 34, 831–841. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, X.; Chen, S.; Liu, Y.; Zhang, L.; Yang, X.; Yu, H.; Cao, Y.; Zhang, L.; Cai, C.; et al. The gap-free genome of pepper reveals the transposable element-driven expansion and rapid evolution of Pericentromeres. Plant Commun. 2024, 6, 101177. [Google Scholar] [CrossRef]
- Rhee, S.Y.; Beavis, W.; Berardini, T.Z.; Chen, G.; Dixon, D.; Doyle, A.; Garcia-Hernandez, M.; Huala, E.; Lander, G.; Montoya, M. The Arabidopsis Information Resource (TAIR): A model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003, 31, 224–228. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, A.G.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Yeom, S.I. Global co-expression network for key factor selection on environmental stress RNA-seq dataset in Capsicum annuum. Sci. Data 2023, 10, 692. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Yu, C.; Shen, Y.; Fang, X.; Chen, L.; Min, J.; Cheng, J.; Zhao, S.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef]
- Bernard, A.; Domergue, F.; Pascal, S.; Jetter, R.; Renne, C.; Faure, J.-D.; Haslam, R.P.; Napier, J.A.; Lessire, R.; Joubès, J. Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 2012, 24, 3106–3118. [Google Scholar] [CrossRef] [PubMed]
- Fiebig, A.; Mayfield, J.A.; Miley, N.L.; Chau, S.; Fischer, R.L.; Preuss, D. Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 2000, 12, 2001–2008. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, Y.; Xu, C.; Ren, J.; Liu, X.; Black, K.; Gai, X.; Wang, Q.; Ren, H. Cucumber ECERIFERUM1 (CsCER1), which influences the cuticle properties and drought tolerance of cucumber, plays a key role in VLC alkanes biosynthesis. Plant Mol. Biol. 2015, 87, 219–233. [Google Scholar] [CrossRef]
- Xia, Y.; Nikolau, B.J.; Schnable, P.S. Cloning and characterization of CER2, an Arabidopsis gene that affects cuticular wax accumulation. Plant Cell 1996, 8, 1291–1304. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Nikolau, B.J.; Schnable, P.S. Developmental and Hormonal Regulation of the Arabidopsis CER2 Gene That Codes for a Nuclear-Localized Protein Required for the Normal Accumulation of Cuticular Waxes. Plant Physiol. 1997, 115, 925–937. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, F.; Wei, K.; Zhang, Y.; Chang, X.; Yang, W.; Yao, Q.; Xiao, H. Genome-Wide Identification of the Eceriferum Gene Family and Analysis of Gene Expression Patterns Under Different Treatments in Pepper (Capsicum annuum L.). Horticulturae 2025, 11, 571. https://doi.org/10.3390/horticulturae11060571
Yang F, Wei K, Zhang Y, Chang X, Yang W, Yao Q, Xiao H. Genome-Wide Identification of the Eceriferum Gene Family and Analysis of Gene Expression Patterns Under Different Treatments in Pepper (Capsicum annuum L.). Horticulturae. 2025; 11(6):571. https://doi.org/10.3390/horticulturae11060571
Chicago/Turabian StyleYang, Fan, Kai Wei, Ying Zhang, Xiaoke Chang, Wenrui Yang, Qiuju Yao, and Huaijuan Xiao. 2025. "Genome-Wide Identification of the Eceriferum Gene Family and Analysis of Gene Expression Patterns Under Different Treatments in Pepper (Capsicum annuum L.)" Horticulturae 11, no. 6: 571. https://doi.org/10.3390/horticulturae11060571
APA StyleYang, F., Wei, K., Zhang, Y., Chang, X., Yang, W., Yao, Q., & Xiao, H. (2025). Genome-Wide Identification of the Eceriferum Gene Family and Analysis of Gene Expression Patterns Under Different Treatments in Pepper (Capsicum annuum L.). Horticulturae, 11(6), 571. https://doi.org/10.3390/horticulturae11060571



