Genome-Wide Analysis of the EIN3/EIL Transcription Factors in Osmanthus fragrans and Their Stress Response to Azacytidine (AZA) and Ethylene (ETH) Treatment
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Related Treatment Methods
2.2. Identification and Synteny Analysis of the O. fragrans OfEIL Gene Family
2.3. Physicochemical Information, Gene Family Structure, and Conserved Motif Analysis of OfEILs
2.4. Phylogenetic Tree and Ka/Ks Analysis
2.5. Expression Analysis of OfEILs at Different Flowering Stages and Under Demethylation and ETH Treatments
2.6. Statistical Analysis
3. Results
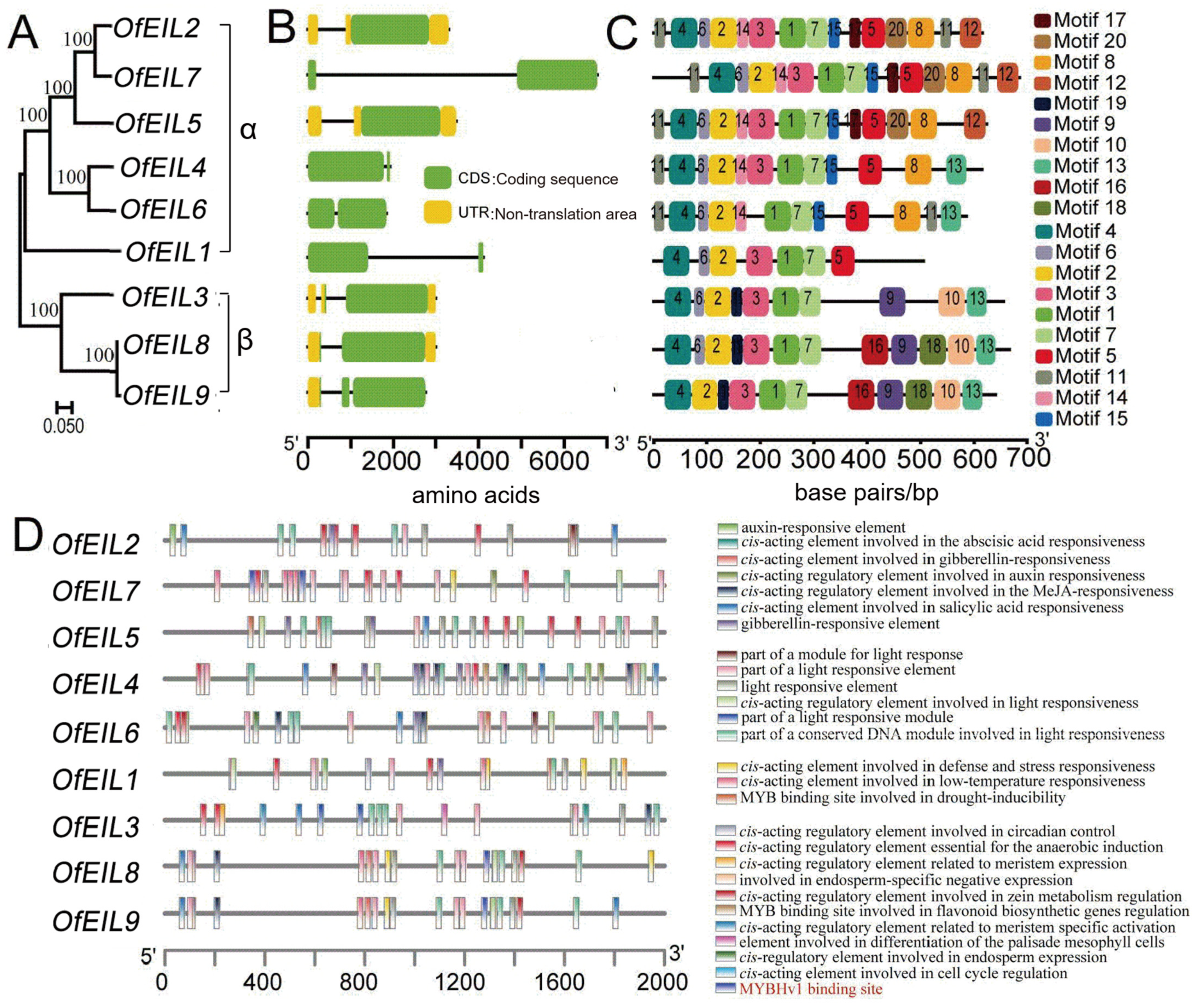
3.1. Identification and Synteny Analysis of O. fragrans OfEIL Genes
3.2. Physicochemical Properties of OfEILs in O. fragrans, Gene Family Structure, and Conserved Motif Analysis
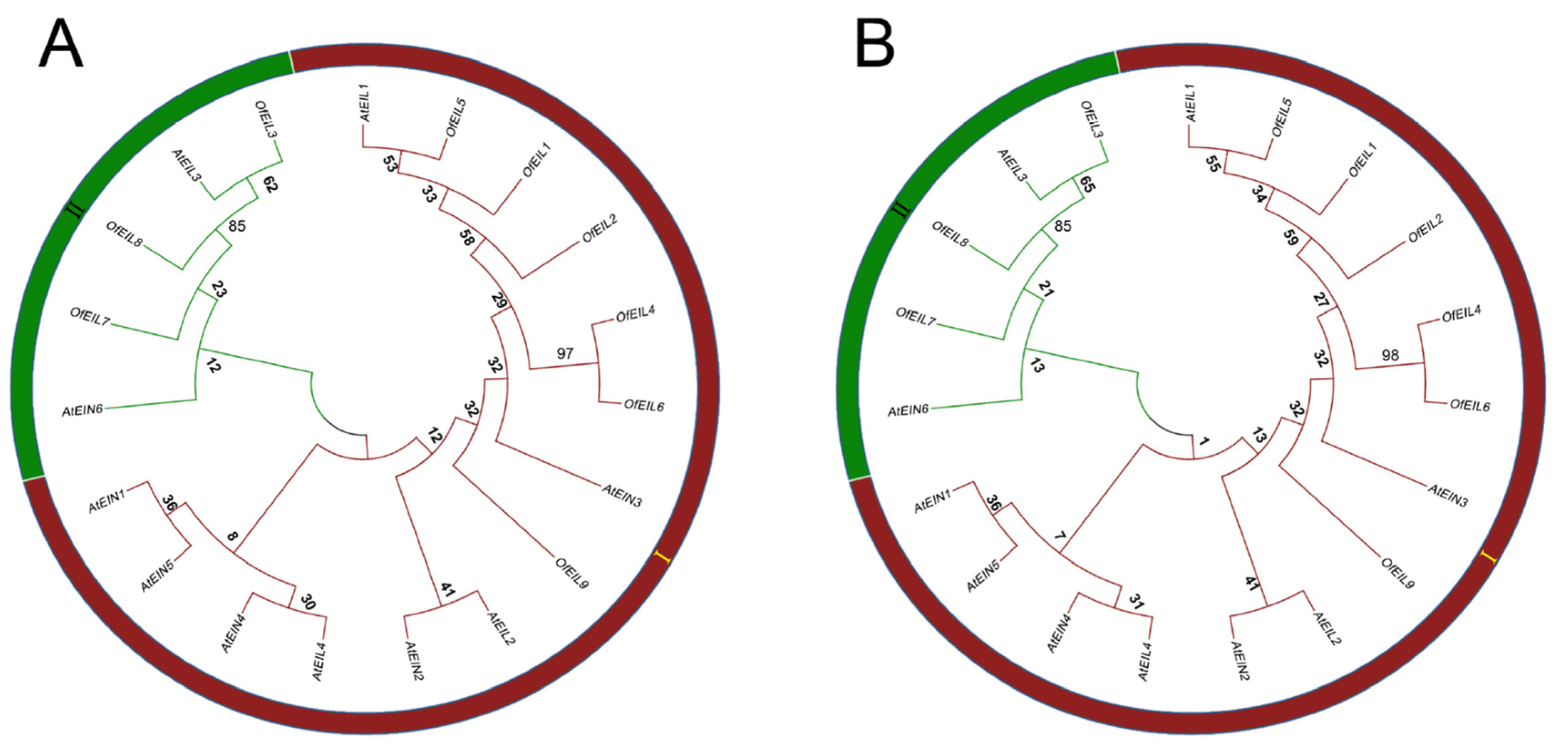
3.3. Phylogenetic and Selection Pressure Analysis of EIN3/EILs in O. fragrans and A. thaliana
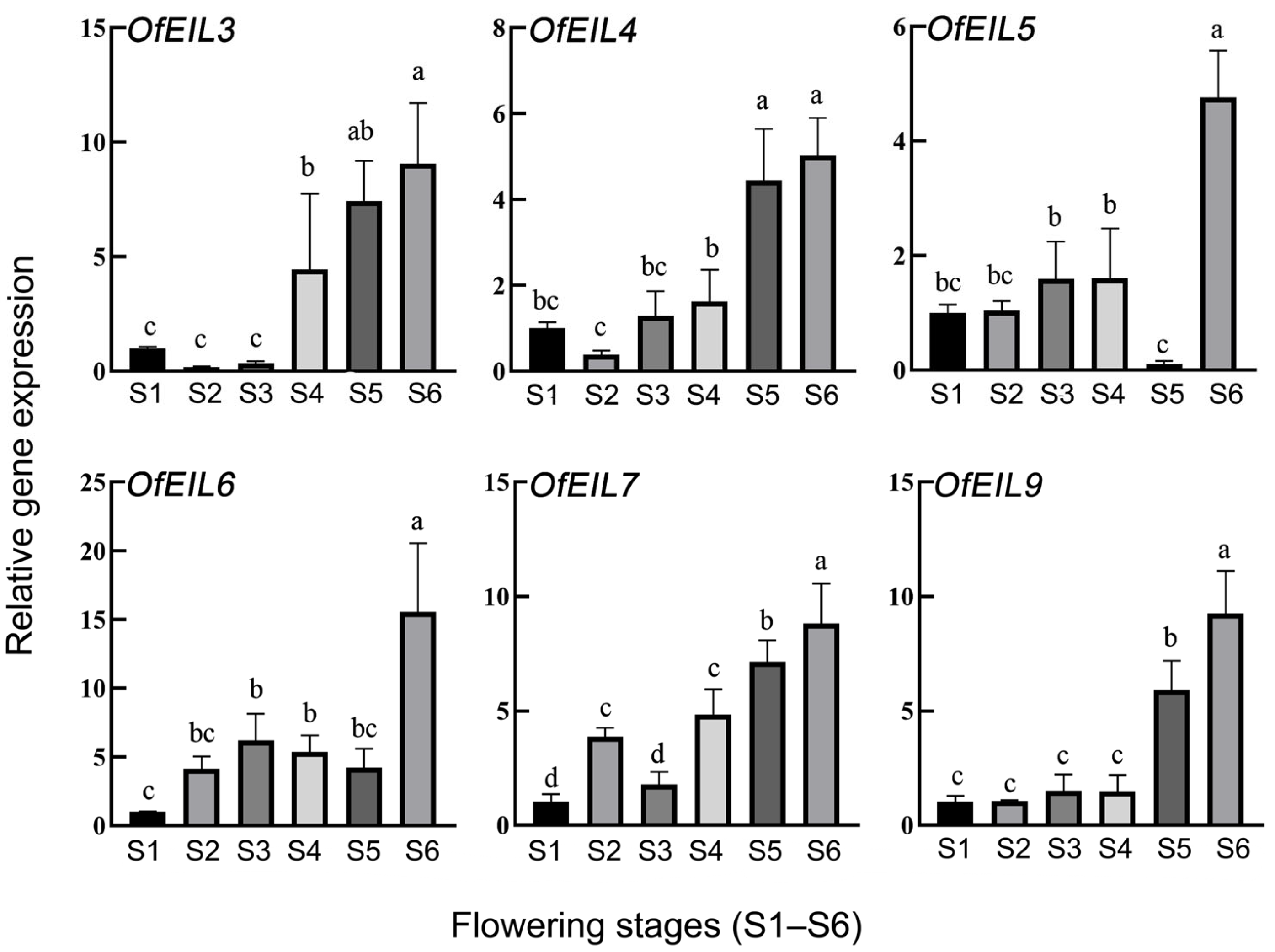
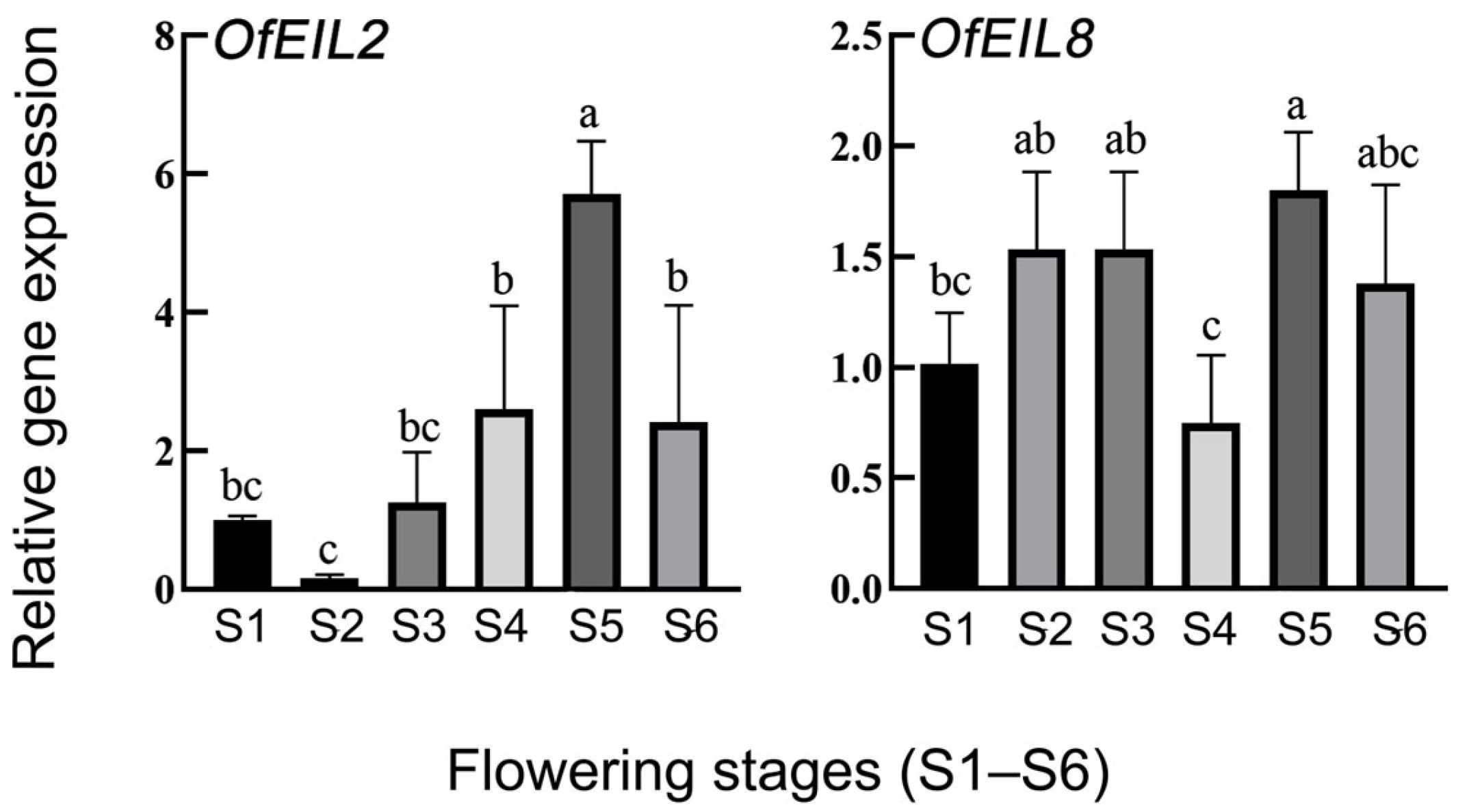
3.4. Expression Profiles and Real-Time Quantitative Fluorescence Analysis of O. fragrans During Natural Flowering Development
3.5. Expression Profiles and Real-Time Quantitative Fluorescence Analysis of O. fragrans Under Aza and ETH Treatments
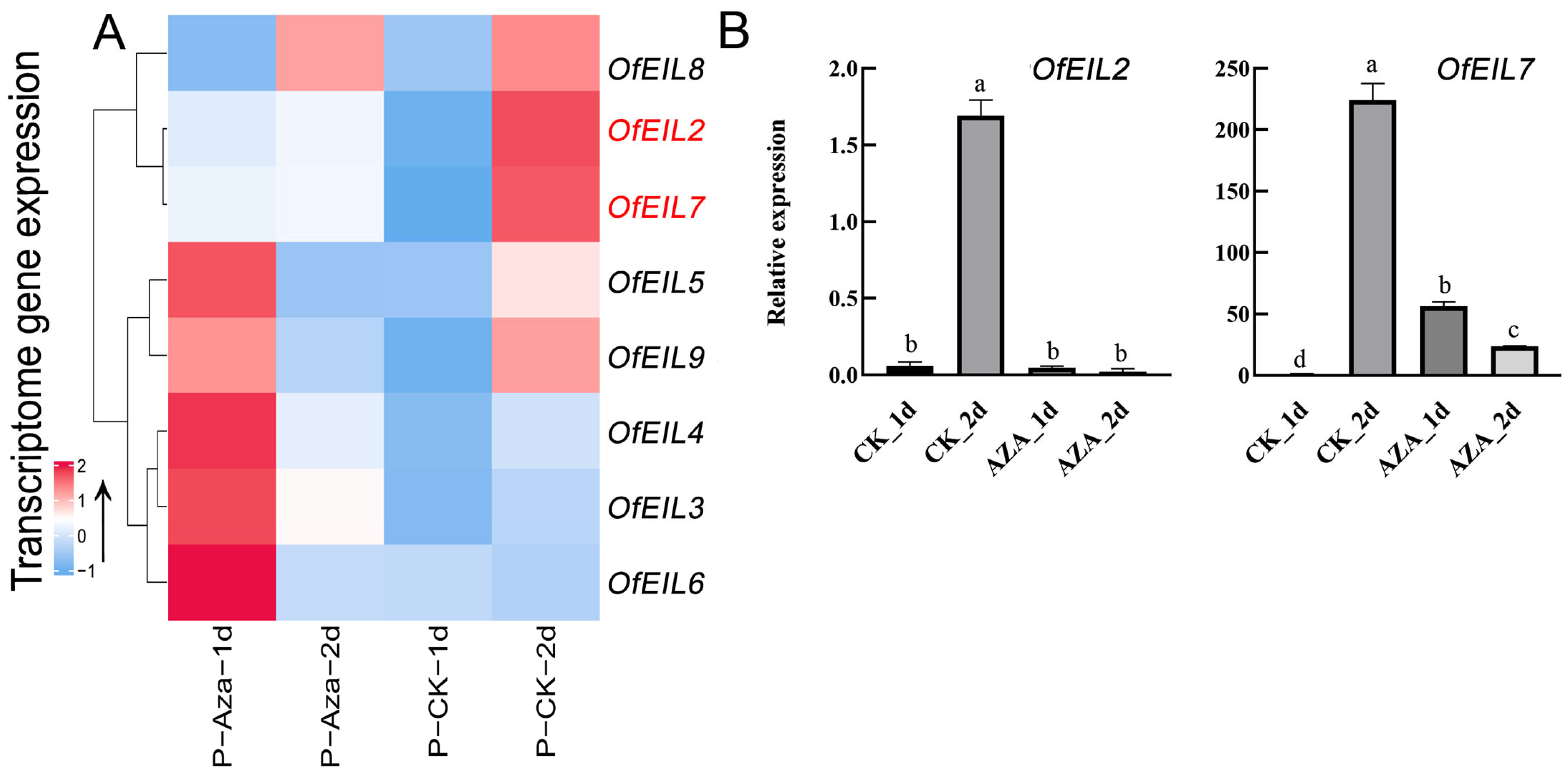
3.5.1. Expression Profile and Fluorescence Quantitative Expression Analysis of Disk-Inserted O. fragrans After Demethylation Treatment
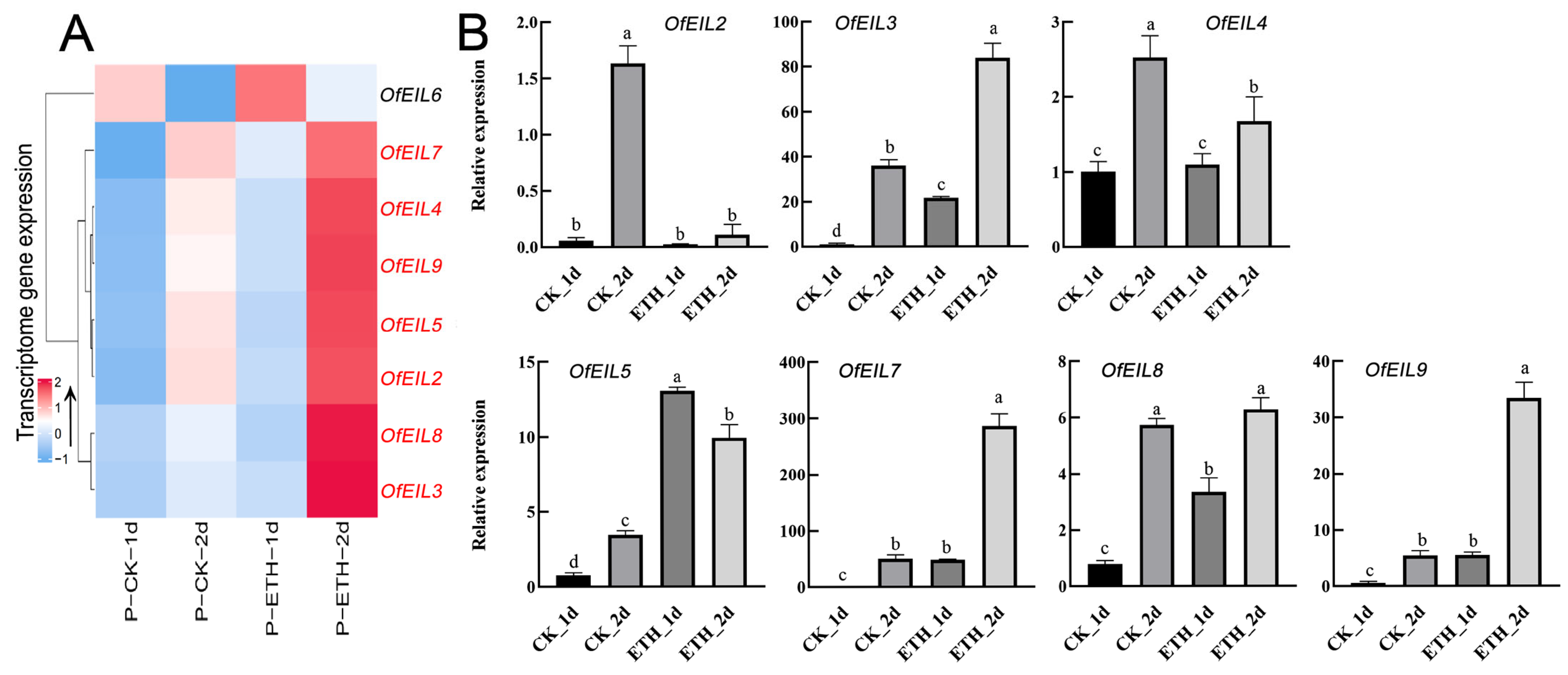
3.5.2. Expression Profile and Fluorescence Quantitative Expression Analysis of Disk-Inserted O. fragrans After Ethephon Treatment
4. Discussion
4.1. Evolutionary Conservation and Functional Divergence of OfEILs
4.2. Interspecies Synteny and Functional Homology
4.3. Functional Divergence Driven by Cis-Element Remodeling
4.4. Hormonal Cross-Talk and Regulatory Specificity in Flowering and Senescence Under Stress
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, M.; Wang, R.; Liang, Z.; Wu, X.; Wang, J. Genome-wide identification and analysis of the EIN3/EIL gene family in allotetraploid Brassica napus reveal its potential advantages during polyploidization. BMC Plant Biol. 2019, 19, 110. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Zhang, M.; Kong, Y.; Dai, S.; Wang, Y.; Meng, Q.; Ma, N.; Lv, W. Origin, expansion, and divergence of ETHYLENE-INSENSITIVE 3 (EIN3)/EIN3-LIKE transcription factors during streptophytes evolution. Front. Plant Sci. 2022, 13, 858477. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jin, Q.; Lin, Y.; Cai, Y. Genome-wide analysis suggests high level of microsynteny and purifying selection affect the evolution of EIN3/EIL family in Rosaceae. PeerJ 2017, 5, e3400. [Google Scholar] [CrossRef]
- Rieu, I.; Mariani, C.; Weterings, K. Expression analysis of five tobacco EIN3 family members in relation to tissue-specific ethylene responses. J. Exp. Bot. 2003, 54, 2239–2244. [Google Scholar] [CrossRef]
- Mao, C.; Wang, S.; Jia, Q.; Wu, P. OsEIL1, a rice homolog of the Arabidopsis EIN3 regulates the ethylene response as a positive component. Plant Mol. Biol. 2006, 61, 141–152. [Google Scholar] [CrossRef]
- Li, J.; Li, Z.; Tang, L.; Yang, Y.; Zouine, M.; Bouzayen, M. A conserved phosphorylation site regulates the transcriptional function of Ethylene-Insensitive3-like1 in tomato. J. Exp. Bot. 2012, 63, 427–439. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.; Zang, Y.; Chen, J.; Feng, S.; Zhang, Z.; Hu, Y.; Zhang, T. A truncated ethylene insensitive3-like protein, GhLYI, regulates senescence in cotton. Plant Physiol. 2023, 193, 1177–1196. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Z.; Quan, R.; Li, G.; Wang, R.; Huang, R. An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol. 2011, 157, 854–865. [Google Scholar] [CrossRef]
- Liu, C.; Li, J.; Zhu, P.; Yu, J.; Hou, J.; Wang, C.; Long, D.; Yu, M.; Zhao, A. Mulberry EIL3 confers salt and drought tolerances and modulates ethylene biosynthetic gene expression. PeerJ 2019, 7, e6391. [Google Scholar] [CrossRef]
- Shi, Y.; Tian, S.; Hou, L.; Huang, X.; Zhang, X.; Guo, H.; Yang, S. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 2012, 24, 2578–2595. [Google Scholar] [CrossRef]
- Chen, H.; Xue, L.; Chintamanani, S.; Germain, H.; Lin, H.; Cui, H.; Cai, R.; Zuo, J.; Tang, X.; Li, X.; et al. Ethylene insensitive3 and ethylene insensitive3-like1 repress salicylic acid induction deficient2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 2009, 21, 2527–2540. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, F.; Shi, X.; Liu, X.; Yang, H.; Zhang, Z. Identification and Characterization of the Cyclin-Dependent Kinases Gene Family in Silkworm, Bombyx mori. DNA Cell Biol. 2016, 35, 13–23. [Google Scholar] [CrossRef] [PubMed]
- An, F.; Zhang, X.; Zhu, Z.; Ji, Y.; He, W.; Jiang, Z.; Li, M.; Guo, H. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 2012, 22, 915–927. [Google Scholar] [CrossRef]
- Li, Z.; Peng, J.; Wen, X.; Guo, H. Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 2013, 25, 3311–3328. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, S.; Gao, Y.; Kan, C.; Wang, H.L.; Yang, Q.; Xia, X.; Ishida, T.; Sawa, S.; Guo, H.; et al. CLE42 delays leaf senescence by antagonizing ethylene pathway in Arabidopsis. New Phytol. 2022, 235, 550–562. [Google Scholar] [CrossRef]
- Zhu, C.; Huang, Z.; Sun, Z.; Feng, S.; Wang, S.; Wang, T.; Yuan, X.; Zhong, L.; Cheng, Y.; Bao, M.; et al. The mutual regulation between DcEBF1/2 and DcEIL3-1 is involved in ethylene induced petal senescence in carnation (Dianthus caryophyllus L.). Plant J. Cell Mol. Biol. 2023, 114, 636–650. [Google Scholar] [CrossRef]
- Shibuya, K.; Barry, K.G.; Ciardi, J.A.; Loucas, H.M.; Underwood, B.A.; Nourizadeh, S.; Ecker, J.R.; Klee, H.J.; Clark, D.G. The central role of PhEIN2 in ethylene responses throughout plant development in petunia. Plant Physiol. 2004, 136, 2900–2912. [Google Scholar] [CrossRef]
- Vaseva, I.I.; Simova-Stoilova, L.; Kirova, E.; Mishev, K.; Depaepe, T.; Van Der Straeten, D.; Vassileva, V. Ethylene signaling in salt-stressed Arabidopsis thaliana ein2-1 and ctr1-1 mutants–A dissection of molecular mechanisms involved in acclimation. Plant Physiol. Biochem. 2021, 167, 999–1010. [Google Scholar] [CrossRef]
- Lu, X.F.; Liu, Y.Z.; Xu, J.R.; Liu, X.J.; Chi, Y.Z.; Li, R.X.; Mo, L.J.; Shi, L.Y.; Liang, S.J.; Yu, W.J.; et al. Recent progress of molecular mechanisms of DNA methylation in plant response to abiotic stress. Environ. Exp. Bot. 2024, 218, 105599. [Google Scholar] [CrossRef]
- Gao, H.Y.; Xia, X.Y.; An, L.J. Critical roles of the activation of ethylene pathway genes mediated by DNA demethylation in Arabidopsis hyperhydricity. Plant Genome 2022, 15, e20202. [Google Scholar] [CrossRef]
- Wang, B.; Luan, F.; Bao, Y.; Peng, X.; Rao, Z.; Tang, Q.; Zeng, N. Traditional uses, phytochemical constituents and pharmacological properties of Osmanthus fragrans: A review. J. Ethnopharmacol. 2022, 293, 115273. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.J.; Zhou, Y.; Cai, X.; Wang, C.Y. Increases in DNA fragmentation and role of ethylene during the petal senescence of Osmanthus fragrans. Postharvest Biol. Technol. 2014, 93, 97–105. [Google Scholar] [CrossRef]
- Zou, J.J.; Cai, X.; Wang, C.Y. The spatial and temporal distribution of programmed cell death (PCD) duringpetal senescence of Osmanthus fragrans. Acta Hortic. 2017, 1185, 315–324. [Google Scholar] [CrossRef]
- Zou, J.J.; Cai, X.; Yang, J.; Zeng, X.; Liu, D.X.; Huang, S.; Chen, X.; Yang, Q.Y.; Wang, C.; Chen, H. DNA hypomEthylation mediates flower opening and senescence in sweet osmanthus through auxin and Ethylene responsive pathways. Postharvest Biol. Technol. 2023, 198, 112250. [Google Scholar] [CrossRef]
- Chen, H.G.; Zeng, X.L.; Yang, J.; Cai, X.; Shi, Y.M.; Zheng, R.R.; Wang, Z.Q.; Liu, J.Y.; Yi, X.X.; Xiao, S.W.; et al. Whole-genome resequencing of Osmanthus fragrans provides insights into flower color evolution. Hortic. Res. 2021, 8, 98. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Pei, J. Multiple protein sequence alignment. Curr. Opin. Struct. Biol. 2008, 18, 382–386. [Google Scholar] [CrossRef]
- Yang, X.; Scheffler, B.E.; Weston, L.A. Recent developments in primer design for DNA polymorphism and mRNA profiling in higher plants. Plant Methods 2006, 2, 4. [Google Scholar] [CrossRef]
- Zhang, C.; Fu, J.X.; Wang, Y.G.; Bao, Z.Y.; Zhao, H.B. Identification of Suitable Reference Genes for Gene Expression Normalization in the Quantitative Real-Time PCR Analysis of Sweet Osmanthus (Osmanthus fragrans Lour.). PLoS ONE 2015, 10, e0136355. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.Y.; Chen, J.Z.; Huang, W.J.; Huang, G.; Deng, M.Y.; Hong, S.M.; Ai, P.; Gao, C.; Zhou, H.K. OmicShare Tools: A Zero-Code Interactive Online Platform for Biological Data Analysis and Visualization. iMeta 2024, 3, e228. [Google Scholar] [CrossRef]
- Li, T.; Peng, Z.; Kangxi, D.; Inzé, D.; Dubois, M. ETHYLENE RESPONSE FACTOR6, A Central Regulator of Plant Growth in Response to Stress. Plant Cell Environ. 2025, 48, 882–892. [Google Scholar] [CrossRef]
- Kerk, D.; Bulgrien, J.; Smith, D.W.; Gribskov, M. Arabidopsis Proteins Containing Similarity to the Universal Stress Protein Domain of Bacteria. Plant Physiol. 2003, 131, 1209–1219. [Google Scholar] [CrossRef]
- Rakhra, G.; Dev Sharma, A.; Singh, J.P. Drought-Induced Changes in the Accumulation of Boiling-Soluble Proteins (p40, GST, HSP90) in the Grains of Drought-Tolerant and Drought-Sensitive Cultivars of Triticum aestivum. Crop and Pasture Science. 2015, 66, 904–911. [Google Scholar] [CrossRef]
- Meng, L.; Zhang, J.; Clarke, N. A Critical Review of Recent Advances in Maize Stress Molecular Biology. Int. J. Mol. Sci. 2024, 25, 12383. [Google Scholar] [CrossRef]
- Kulichikhin, K.Y.; Greenway, H.; Byrne, L.; Colmer, T.D. Regulation of intracellular pH during anoxia in rice coleoptiles in acidic and near neutral conditions. J. Exp. Bot. 2009, 60, 2119–2128. [Google Scholar] [CrossRef]
- Wang, C.Y.; Lin, T.T.; Wang, M.Q.; Qi, X.T. An AC-Rich Bean Element Serves as an Ethylene-Responsive Element in Arabidopsis. Plants 2020, 9, 1033. [Google Scholar] [CrossRef]
- Binder, B.M.; Walker, J.M.; Gagne, J.M.; Emborg, T.J.; Hemmann, G.; Bleecker, A.B.; Vierstra, R.D. The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 2007, 19, 509–523. [Google Scholar] [CrossRef]
- Chowdhury, Z.; Mohanty, D.; Giri, M.K.; Venables, B.J.; Chaturvedi, R.; Chao, A.; Petros, R.A.; Shah, J. Dehydroabietinal promotes flowering time and plant defense in Arabidopsis via the autonomous pathway genes flowering locus D, FVE, and relative of early flowering 6. J. Exp. Bot. 2020, 71, 4903–4913. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gao, J.; Gao, S.; Song, Y.; Yang, Z.; Kuai, B. The H3K27me3 demethylase REF6 promotes leaf senescence through directly activating major senescence regulatory and functional genes in Arabidopsis. PLoS Genet. 2019, 15, e1008068. [Google Scholar] [CrossRef]
- Antunez-Sanchez, J.; Naish, M.; Ramirez-Prado, J.S.; Ohno, S.; Huang, Y.; Dawson, A.; Opassathian, K.; Manza-Mianza, D.; Ariel, F.; Raynaud, C.; et al. A new role for histone demethylases in the maintenance of plant genome integrity. eLife 2020, 9, e58533. [Google Scholar] [CrossRef]
- Fernández-Milmanda, G.L. Touch me not! Jasmonic acid and ethylene converge on gibberellins breakdown to regulate touch-induced morphogenesis. Plant Physiol. 2024, 194, 601–603. [Google Scholar] [CrossRef]
- Zou, J.J.; Zhang, J.; Wang, X.Q.; Xia, H.; Zeng, X.L.; Cai, X.; Yang, J.; Zeng, J.; Li, Z.Q.; Zhang, G.F.; et al. Comprehensive transcriptome analysis of AP2/ERFs in Osmanthus fragrans reveals the role of OfERF017-mediated organic acid metabolism pathway in flower senescence. Front. Plant Sci. 2024, 15, 1467232. [Google Scholar] [CrossRef]


| Gene Number | Primer | Primer Sequences (5′→3′) |
|---|---|---|
| OfEIL2 | LYG006439-F | ATTTCTTCTCGTCTGCTCCTCCA |
| LYG006439-R | TCTTTTTCCTCCTCGCTTGTTCT | |
| OfEIL3 | LYG010377-F | GGAAGAGTTGGAGAGACGAATGTG |
| LYG010377-R | ATTGGCAGAAGAAAAAGTGGATAA | |
| OfEIL4 | LYG016518-F | CGGTAATCTTGATTTCCTCTCTACT |
| LYG016518-R | ACTGACTCCTTGTTCTTCTTCTGCT | |
| OfEIL5 | LYG018604-F | TGGTGATTTTGATTTCTTTTCCTCT |
| LYG018604-R | TCATTTCTTTCAGTCTTTTGTGTCG | |
| OfEIL6 | LYG028598-F | AATGGGTTTTTGTGGTAATCTTGA |
| LYG028598-R | TGGACTCCTTGTTCTTTTTCTGCT | |
| OfEIL7 | LYG035007-F | ATTTCTTTTCGTCTGCTCCTCTCA |
| LYG035007-R | TCAACACCGTCCTTACCTTTATTC | |
| OfEIL8 | LYG035276-F | CCAAAAGTTAGCCGTAGTAACAAACA |
| LYG035276-R | GCACAAAAAAAGTCACAATCAGAATC | |
| OfEIL9 | LYG038346-F | AGCCGGTTGTGGATGATGATTA |
| LYG038346-R | TGAGATTGCCGTTGTTTGGAA | |
| / | OfACT-F | AGTCCTCTTCCAGCCTTCTTT |
| OfACT-R | ATTTCCTTGCTCATACGGTCA |
| Gene Name | Total Number of Atoms | Molecular mass/ku | Theoretical Isoelectric Point | Hydrophilicity Average | Instability Index | Protein Properties | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| OfEIL1 | 8116 | 57.92 | 5.51 | −0.766 | 54.3 | Unstable hydrophilic proteins | Nucleus |
| OfEIL2 | 9626 | 69.69 | 5.63 | −0.757 | 56.67 | Unstable hydrophilic proteins | Nucleus |
| OfEIL3 | 10,216 | 73.53 | 5.88 | −0.827 | 52.52 | Unstable hydrophilic proteins | Nucleus |
| OfEIL4 | 9624 | 69.70 | 5.02 | −0.718 | 48.6 | Unstable hydrophilic proteins | Nucleus |
| OfEIL5 | 9669 | 70.21 | 5.84 | −0.823 | 52.66 | Unstable hydrophilic proteins | Nucleus |
| OfEIL6 | 9158 | 66.07 | 5.38 | −0.673 | 54.22 | Unstable hydrophilic proteins | Nucleus |
| OfEIL7 | 10,829 | 77.88 | 5.66 | −0.777 | 59.9 | Unstable hydrophilic proteins | Nucleus |
| OfEIL8 | 10,316 | 74.17 | 5.8 | −0.775 | 59.43 | Unstable hydrophilic proteins | Nucleus |
| OfEIL9 | 3927 | 67.91 | 8.64 | −0.904 | 48.66 | Unstable hydrophilic proteins | Nucleus |
| Gene Pairs | Non-Synonymous, Ka | Synonymous, Ks | Ka/Ks | Evolutionary Direction |
|---|---|---|---|---|
| OfEIL2&OfEIL7 | 0.05 | 0.31 | 0.16 | Purify selection |
| OfEIL5&OfEIL7 | 0.13 | 0.71 | 0.18 | Purify selection |
| OfEIL5&OfEIL9 | 0.11 | 0.35 | 0.31 | Purify selection |
| OfEIL4&OfEIL6 | 0.07 | 0.36 | 0.2 | Purify selection |
| OfEIL7&OfEIL9 | 0.11 | 0.92 | 0.12 | Purify selection |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, D.; Xu, C.; Ma, W.; Zhu, X.; Yu, Q.; Zhang, Y.; Yang, J.; Zeng, X.; Cai, X.; Zou, J. Genome-Wide Analysis of the EIN3/EIL Transcription Factors in Osmanthus fragrans and Their Stress Response to Azacytidine (AZA) and Ethylene (ETH) Treatment. Horticulturae 2025, 11, 572. https://doi.org/10.3390/horticulturae11060572
Pan D, Xu C, Ma W, Zhu X, Yu Q, Zhang Y, Yang J, Zeng X, Cai X, Zou J. Genome-Wide Analysis of the EIN3/EIL Transcription Factors in Osmanthus fragrans and Their Stress Response to Azacytidine (AZA) and Ethylene (ETH) Treatment. Horticulturae. 2025; 11(6):572. https://doi.org/10.3390/horticulturae11060572
Chicago/Turabian StylePan, Dou, Chun Xu, Wanlu Ma, Xinyi Zhu, Qiangjun Yu, Yingting Zhang, Jie Yang, Xiangling Zeng, Xuan Cai, and Jingjing Zou. 2025. "Genome-Wide Analysis of the EIN3/EIL Transcription Factors in Osmanthus fragrans and Their Stress Response to Azacytidine (AZA) and Ethylene (ETH) Treatment" Horticulturae 11, no. 6: 572. https://doi.org/10.3390/horticulturae11060572
APA StylePan, D., Xu, C., Ma, W., Zhu, X., Yu, Q., Zhang, Y., Yang, J., Zeng, X., Cai, X., & Zou, J. (2025). Genome-Wide Analysis of the EIN3/EIL Transcription Factors in Osmanthus fragrans and Their Stress Response to Azacytidine (AZA) and Ethylene (ETH) Treatment. Horticulturae, 11(6), 572. https://doi.org/10.3390/horticulturae11060572






