The Identification of Regulatory Genes Involved in Light-Induced Anthocyanin Accumulation in Aft Tomato Developing Fruits
Abstract
:1. Introduction
2. Materials and Methods
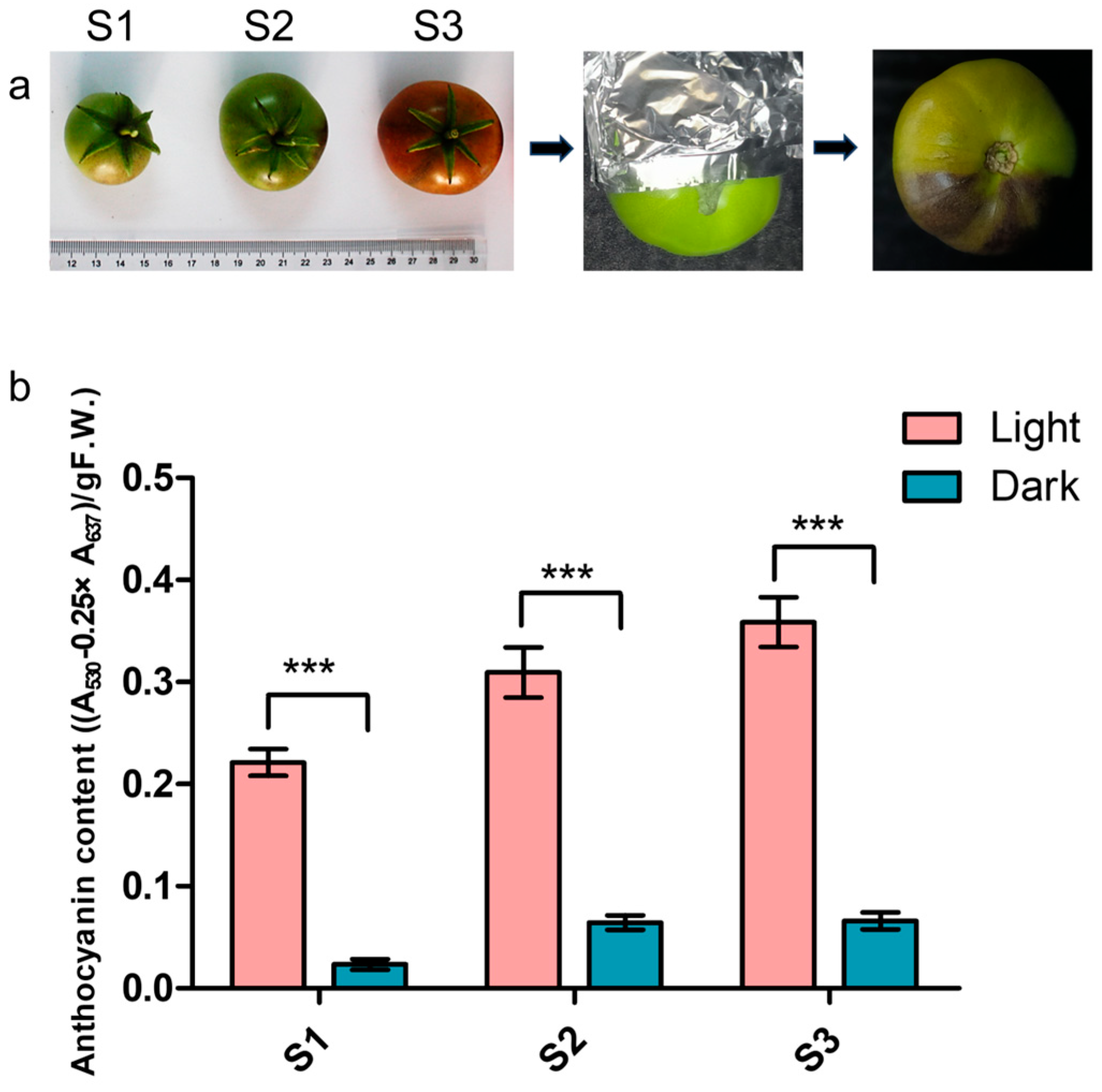
2.1. Plant Material and Light Treatment
2.2. Anthocyanin Content Measurement
2.3. RNA Extraction, Library Construction and Illumina High-Throughput Sequencing
2.4. RNA-Seq and DEGs Annotation
2.5. qRT-PCR Analysis
2.6. Co-Expression Network Analysis
3. Results
3.1. Light-Induced Anthocyanin Accumulation in Fruits of Aft Tomato
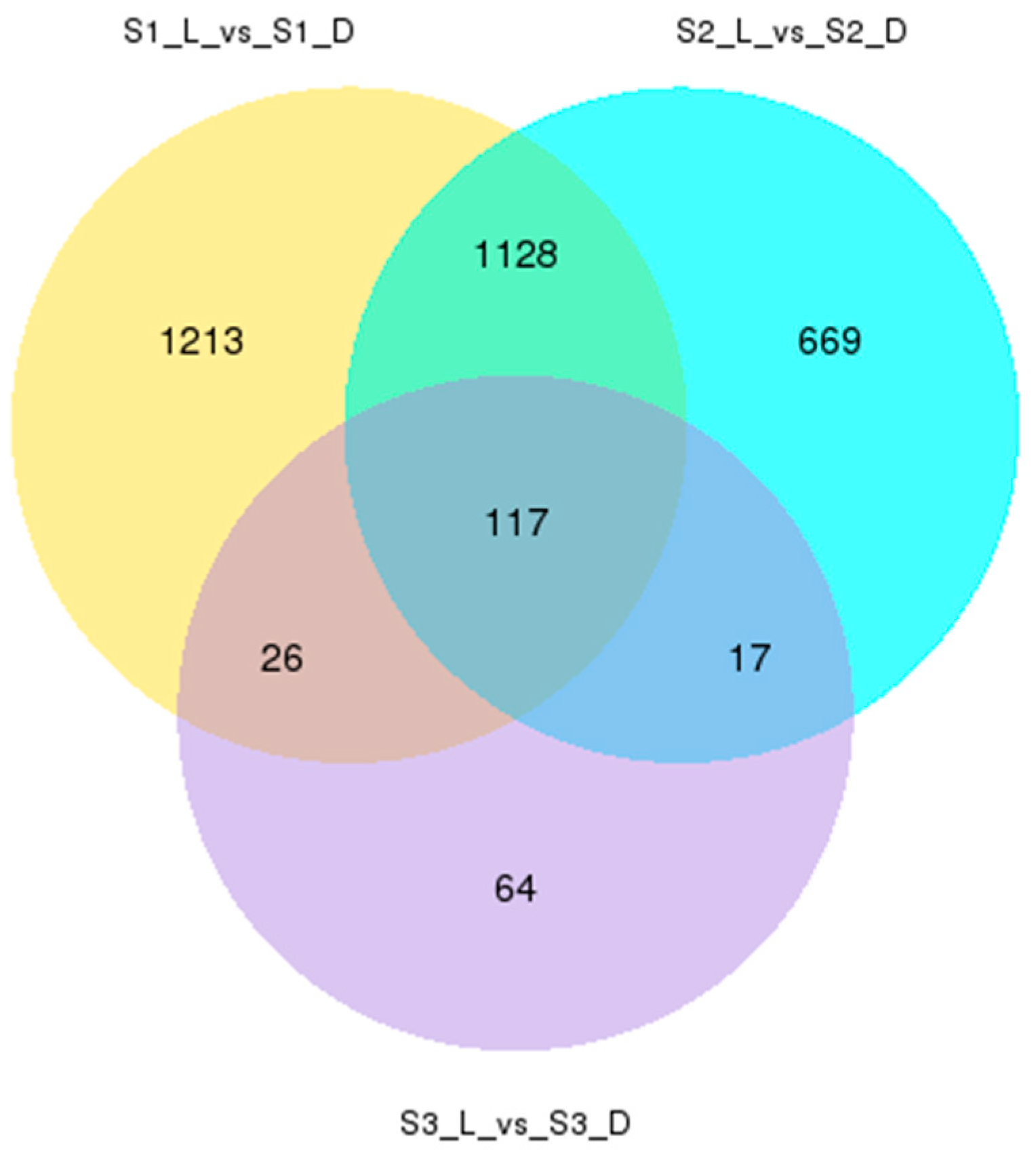
3.2. Analysis of Transcriptome Sequencing and Differential Gene Expression
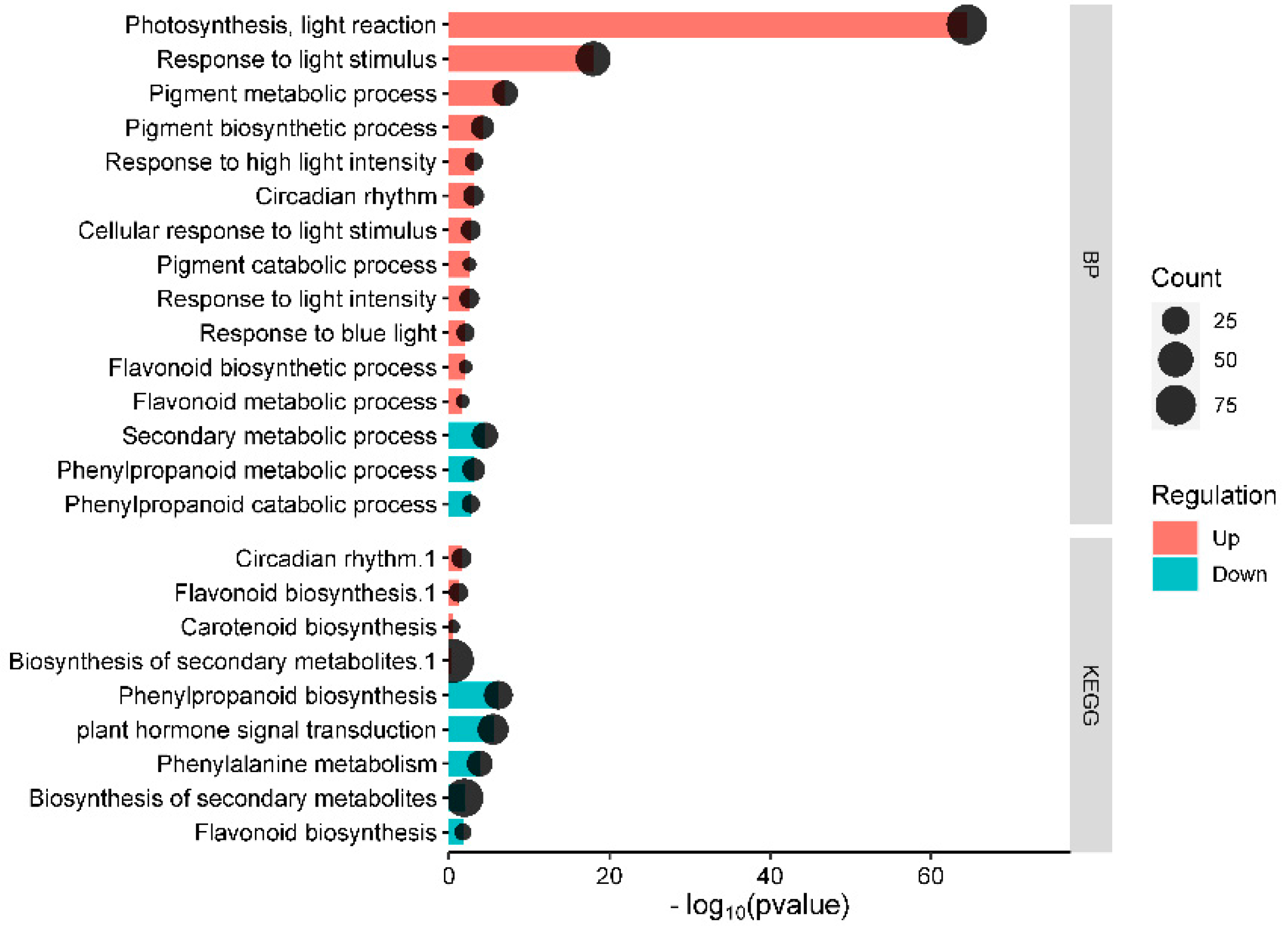
3.3. GO and KEGG Analysis of DEGs
3.4. Identification of Candidate Genes
3.4.1. Genes Related to the Light Response and Signal Transduction
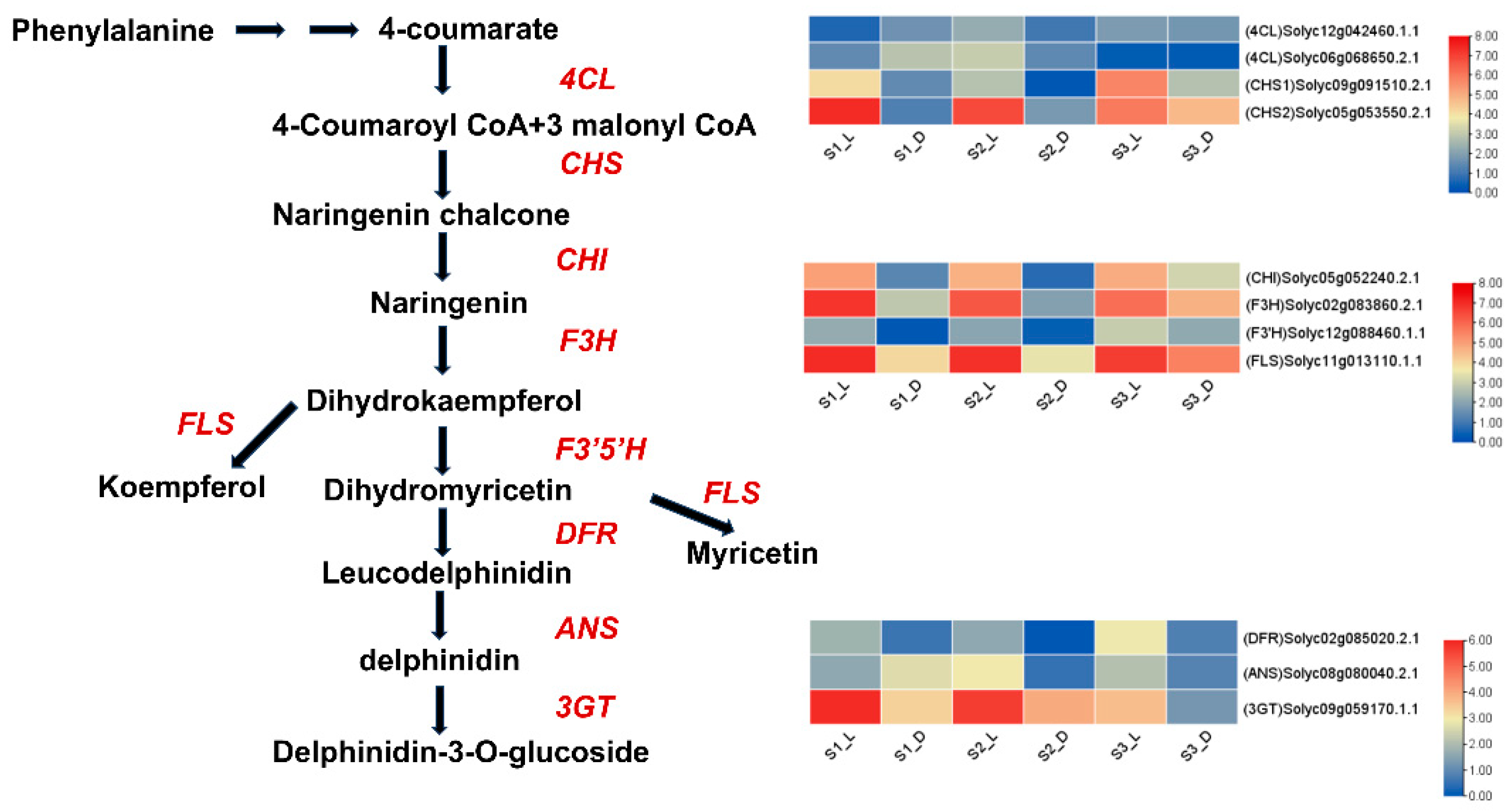
3.4.2. Genes Involved in Anthocyanin Biosynthesis
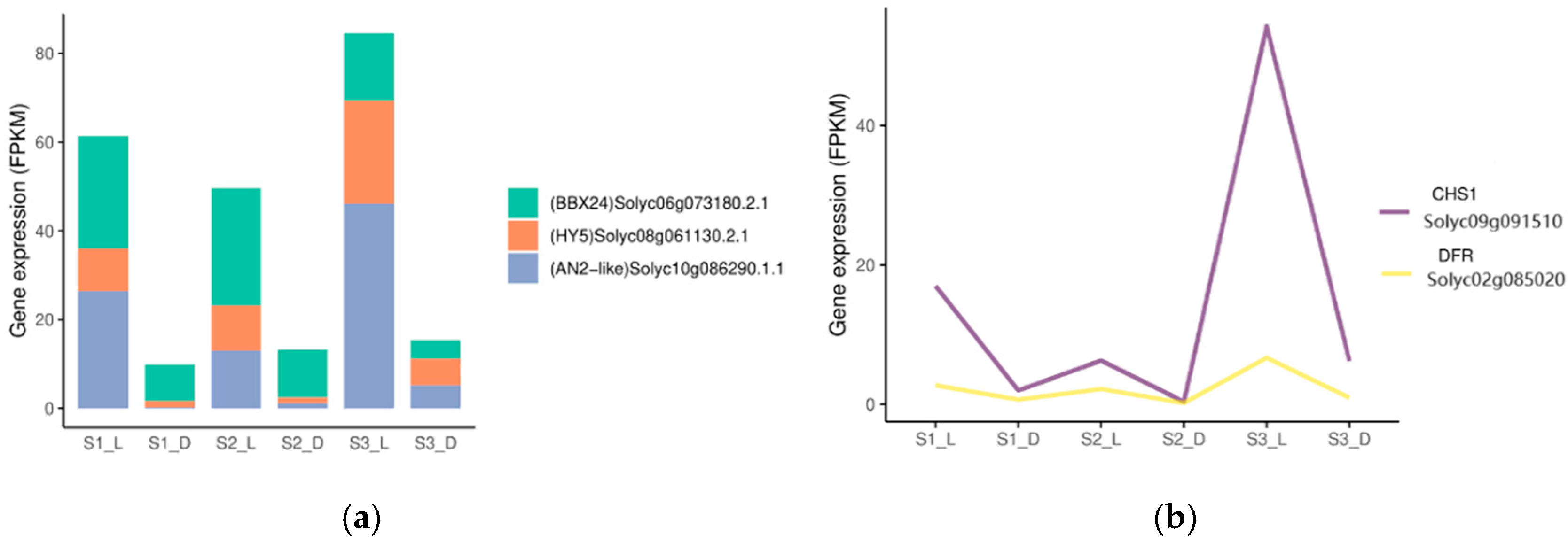
3.4.3. Gene Expression Associated with Anthocyanin Biosynthesis During Various Fruit Development Stages
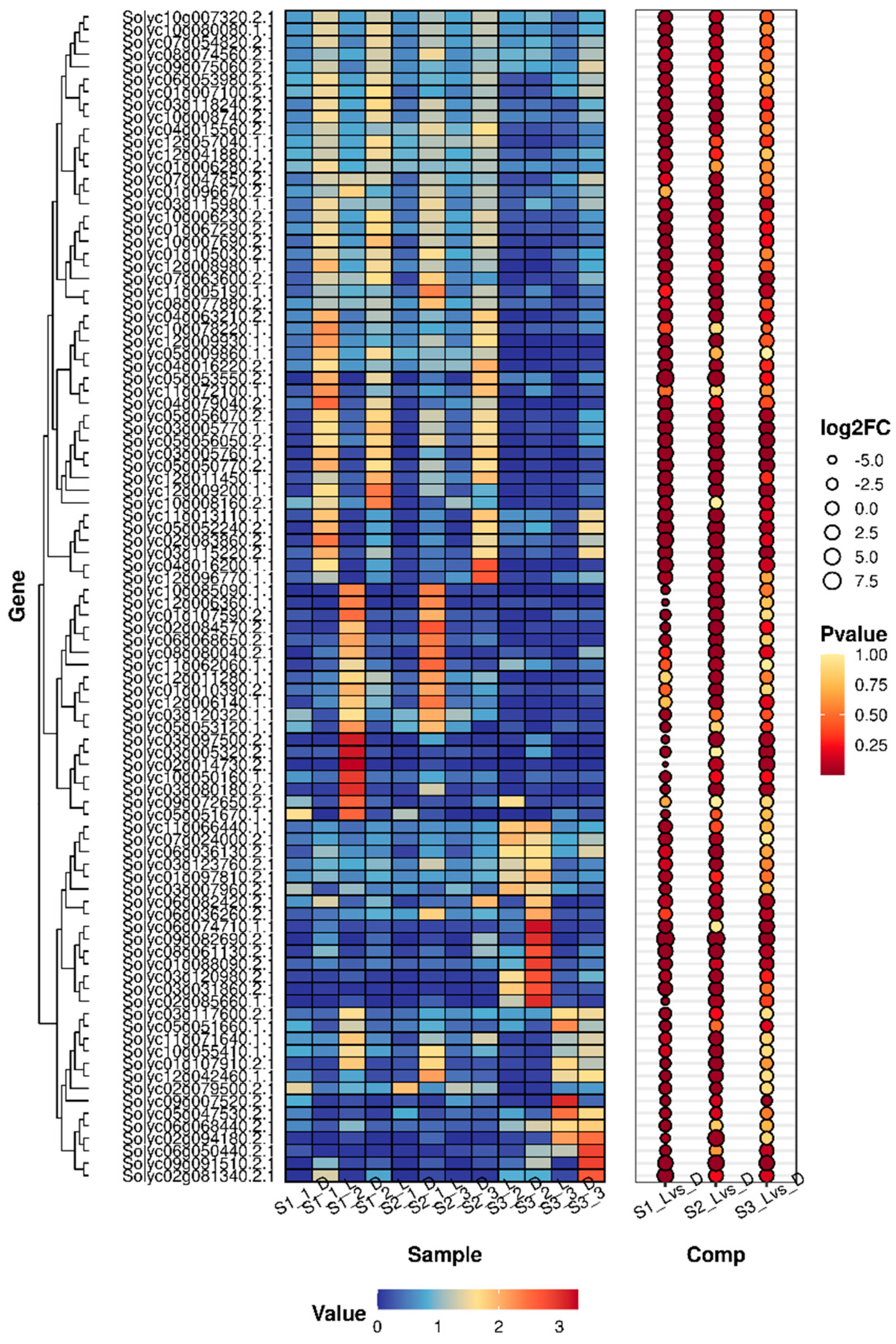
3.4.4. Analysis of Differentially Expressed Genes (DEGs) Through Co-Expression Networks
3.4.5. Verification of Related Genes in Tomato Fruits Under Light Treatment Using qRT-PCR
4. Discussion
4.1. Factors Involved in Light Signal Transduction That Activate Anthocyanin Biosynthesis
4.2. Expression of Regulatory and Structural Genes in the Anthocyanin Metabolic Pathway Is Influenced by Light
4.3. Developmental Signals Also Regulate Anthocyanin Accumulation in Fruits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Belwal, T.; Singh, G.; Jeandet, P.; Pandey, A.; Giri, L.; Ramola, S.; Bhatt, I.D.; Venskutonis, P.R.; Georgiev, M.I.; Clement, C.; et al. Anthocyanins, multi-functional natural products of industrial relevance: Recent biotechnological advances. Biotechnol. Adv. 2020, 43, 107600. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Han, T.; Lyu, L.; Li, W.; Wu, W. Research progress in understanding the biosynthesis and regulation of plant anthocyanins. Sci. Hortic. 2023, 321, 112374. [Google Scholar] [CrossRef]
- Jones, C.M.; Mes, P.; Myers, J.R. Characterization and inheritance of the Anthocyanin fruit (Aft) tomato. J. Hered. 2003, 94, 449–456. [Google Scholar] [CrossRef]
- Mes, P.J.; Boches, P.; Myers, J.R.; Durst, R. Characterization of tomatoes expressing anthocyanin in the fruit. J. Am. Soc. Hortic. Sci. 2008, 133, 262–269. [Google Scholar] [CrossRef]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, X.W.; Liu, X.; Zheng, P.F.; Su, L.; Wang, G.L.; Wang, X.F.; Li, Y.Y.; You, C.X.; An, J.P. Phytochrome interacting factor MdPIF7 modulates anthocyanin biosynthesis and hypocotyl growth in apple. Plant Physiol. 2022, 188, 2342–2363. [Google Scholar] [CrossRef]
- Tao, R.; Bai, S.; Ni, J.; Yang, Q.; Zhao, Y.; Teng, Y. The blue light signal transduction pathway is involved in anthocyanin accumulation in ‘Red Zaosu’ pear. Planta 2018, 248, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Kadomura-Ishikawa, Y.; Miyawaki, K.; Noji, S.; Takahashi, A. Phototropin 2 is involved in blue light-induced anthocyanin accumulation in Fragaria × ananassa fruits. J. Plant Res. 2013, 126, 847–857. [Google Scholar] [CrossRef]
- Li, T.; Yamane, H.; Tao, R. Preharvest long-term exposure to UV-B radiation promotes fruit ripening and modifies stage-specific anthocyanin metabolism in highbush blueberry. Hortic. Res. 2021, 8, 67. [Google Scholar] [CrossRef]
- Ma, Y.; Ma, X.; Gao, X.; Wu, W.; Zhou, B. Light Induced Regulation Pathway of Anthocyanin Biosynthesis in Plants. Int. J. Mol. Sci. 2021, 22, 11116. [Google Scholar] [CrossRef]
- Pan, C.; Liao, Y.; Shi, B.; Zhang, M.; Zhou, Y.; Wu, J.; Wu, H.; Qian, M.; Bai, S.; Teng, Y.; et al. Blue light-induced MiBBX24 and MiBBX27 simultaneously promote peel anthocyanin and flesh carotenoid biosynthesis in mango. Plant Physiol. Biochem. 2025, 219, 109315. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Wang, X.; Wu, H.; Yao, Y. The nuclear and cytoplasmic colocalization of MdGST12 regulated by MdWRKY26 and MdHY5 promotes anthocyanin accumulation by forming homodimers and interact with MdUFGT and MdDFR under light conditions in Malus. Int. J. Biol. Macromol. 2025, 289, 138666. [Google Scholar] [CrossRef]
- Sun, Y.; Shou, J.; Dai, Z.; Yang, X.; Jiang, G.; Shi, Y.; Chen, K. The HD-Zip transcription factor FaANL2 represses FaMYB10 to inhibit anthocyanin biosynthesis in strawberry fruit. Plant Growth Regul. 2025, 105, 515–527. [Google Scholar] [CrossRef]
- Chen, D.; Xu, G.; Tang, W.; Jing, Y.; Ji, Q.; Fei, Z.; Lin, R. Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 2013, 25, 1657–1673. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.H.; Liu, Y.H.; Zhao, H.B.; Xu, J.M.; Zheng, P.; Liu, S.Q.; Sun, B.M. CsHY5 Regulates Light-Induced Anthocyanin Accumulation in. Int. J. Mol. Sci. 2025, 26, 3253. [Google Scholar] [CrossRef]
- Wang, W.; Wang, P.; Li, X.; Wang, Y.; Tian, S.; Qin, G. The transcription factor SlHY5 regulates the ripening of tomato fruit at both the transcriptional and translational levels. Hortic. Res. 2021, 8, 83. [Google Scholar] [CrossRef]
- Xu, Y.; You, C.; Xu, C.; Zhang, C.; Hu, X.; Li, X.; Ma, H.; Gong, J.; Sun, X. Red and blue light promote tomato fruit coloration through modulation of hormone homeostasis and pigment accumulation. Postharvest Biol. Technol. 2024, 207, 112588. [Google Scholar] [CrossRef]
- Sun, C.; Deng, L.; Du, M.; Zhao, J.; Chen, Q.; Huang, T.; Jiang, H.; Li, C.B.; Li, C. A Transcriptional Network Promotes Anthocyanin Biosynthesis in Tomato Flesh. Mol. Plant 2020, 13, 42–58. [Google Scholar] [CrossRef]
- Li, Y.; Shan, X.; Gao, R.; Han, T.; Zhang, J.; Wang, Y.; Kimani, S.; Wang, L.; Gao, X. MYB repressors and MBW activation complex collaborate to fine-tune flower coloration in Freesia hybrida. Commun. Biol. 2020, 3, 396. [Google Scholar] [CrossRef]
- Yang, S.Y.; Wang, Y.; Wang, W.X.; Wang, N.; Yan, R.; Li, S.Y.; Zhang, T.L.; Liu, J.L.; Zeng, X.F.; Zhao, S.N.; et al. Analysis of WD40 genes in kiwifruit reveals the key role of the light-induced AcTTG1-AcMYB75-AcbHLH2 complex in anthocyanin accumulation. Int. J. Biol. Macromol. 2025, 297, 139758. [Google Scholar] [CrossRef]
- Xu, X.W.; Zhu, Y.; Yuan, Y.; Sohail, H.; He, S.Y.; Ye, Y.; Wang, M.X.; Lv, M.; Qi, X.H.; Yang, X.D.; et al. R2R3-MYB transcription factor controls mature fruit skin color by regulating flavonoid accumulation in cucumber. Plant J. 2024, 119, 796–813. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Xu, H.; Yang, G.; Wang, N.; Zhang, J.; Wang, Y.; Jiang, S.; Fang, H.; Zhang, Z.; Chen, X. Molecular mechanism of MYB111 and WRKY40 involved in anthocyanin biosynthesis in red-fleshed apple callus. Plant Cell Tissue Organ Cult. (PCTOC) 2019, 139, 467–478. [Google Scholar] [CrossRef]
- Liu, W.; Mei, Z.; Yu, L.; Gu, T.; Li, Z.; Zou, Q.; Zhang, S.; Fang, H.; Wang, Y.; Zhang, Z.; et al. The ABA-induced NAC transcription factor MdNAC1 interacts with a bZIP-type transcription factor to promote anthocyanin synthesis in red-fleshed apples. Hortic. Res. 2023, 10, uhad049. [Google Scholar] [CrossRef]
- Li, D.L.; Zhu, M.L.; Li, S.H.; Wang, P.Q.; Li, J.Y.; Ge, H.Y.; Liu, Y.; Chen, H.Y. Eggplant transcription factor SmERF118 mediates light signaling regulation of anthocyanin synthesis. Plant Physiol. Biochem. 2025, 221, 109584. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.X.; Lei, Y.Y.; Jiao, H.B.; Zhang, Z.; Yao, J.X.; Li, H.; Dai, H.Y.; Zhang, Z.H.; Zhang, J.X. Ubiquitin-mediated degradation of the inhibitor FvMYB1 and the activator FvBBX20 by FvCSN5 balances anthocyanin biosynthesis in strawberry fruit. Plant J. 2025, 121, e70021. [Google Scholar] [CrossRef]
- Hosmani, P.S.; Flores-Gonzalez, M.; van de Geest, H.; Maumus, F.; Bakker, L.V.; Schijlen, E.; van Haarst, J.; Cordewener, J.; Sanchez-Perez, G.; Peters, S.; et al. An improved de novo assembly and annotation of the tomato reference genome using single-molecule sequencing, Hi-C proximity ligation and optical maps. bioRxiv 2019, 767764. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Chen, T.; Liu, Y.X.; Huang, L. ImageGP: An easy-to-use data visualization web server for scientific researchers. Imeta 2022, 1, e5. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.Y.; Chao, F.; Zhuo, Z.; Ma, Z.; Li, W.; Chen, G. Identification of hub genes in prostate cancer using robust rank aggregation and weighted gene co-expression network analysis. Aging 2019, 11, 4736–4756. [Google Scholar] [CrossRef]
- Yang, T.; Ma, H.; Li, Y.; Zhang, Y.; Zhang, J.; Wu, T.; Song, T.; Yao, Y.; Tian, J. Apple MPK4 mediates phosphorylation of MYB1 to enhance light-induced anthocyanin accumulation. Plant J. 2021, 106, 1728–1745. [Google Scholar] [CrossRef]
- Xing, Y.; Sun, W.; Sun, Y.; Li, J.; Zhang, J.; Wu, T.; Song, T.; Yao, Y.; Tian, J. MPK6-mediated HY5 phosphorylation regulates light-induced anthocyanin accumulation in apple fruit. Plant Biotechnol. J. 2023, 21, 283–301. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Chen, N.; Huang, Z.; Li, D.; Zhi, J.; Yu, B.; Liu, X.; Cao, B.; Qiu, Z. Anthocyanin Fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 2020, 225, 2048–2063. [Google Scholar] [CrossRef]
- Colanero, S.; Tagliani, A.; Perata, P.; Gonzali, S. Alternative Splicing in the Anthocyanin Fruit Gene Encoding an R2R3 MYB Transcription Factor Affects Anthocyanin Biosynthesis in Tomato Fruits. Plant Commun. 2019, 1, 100006. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.A.; Rahman, M.; Sakhi, S.; Nawaz, G.; Khan, A.; Ahmad, T.; Adnan, M.; Khan, S. PpMYB39 Activates PpDFR to Modulate Anthocyanin Biosynthesis during Peach Fruit Maturation. Horticulturae 2022, 8, 332. [Google Scholar] [CrossRef]
- Xu, P.B.; Lian, H.L.; Wang, W.X.; Xu, F.; Yang, H.Q. Pivotal Roles of the Phytochrome-Interacting Factors in Cryptochrome Signaling. Mol. Plant 2016, 9, 496–497. [Google Scholar] [CrossRef]
- Pierik, R.; Ballare, C.L. Control of Plant Growth and Defense by Photoreceptors: From Mechanisms to Opportunities in Agriculture. Mol. Plant 2022, 15, 1825. [Google Scholar] [CrossRef]
- Tissot, N.; Ulm, R. Cryptochrome-mediated blue-light signalling modulates UVR8 photoreceptor activity and contributes to UV-B tolerance in Arabidopsis. Nat. Commun. 2020, 11, 1323. [Google Scholar] [CrossRef]
- Legris, M.; Ince, Y.C.; Fankhauser, C. Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat. Commun. 2019, 10, 5219. [Google Scholar] [CrossRef] [PubMed]
- Schrager-Lavelle, A.; Herrera, L.A.; Maloof, J.N. Tomato phyE Is Required for Shade Avoidance in the Absence of phyB1 and phyB2. Front. Plant Sci. 2016, 7, 1275. [Google Scholar] [CrossRef]
- Velez-Ramirez, A.I.; Vreugdenhil, D.; Millenaar, F.F.; van Ieperen, W. Phytochrome A Protects Tomato Plants from Injuries Induced by Continuous Light. Front. Plant Sci. 2019, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Fantini, E.; Sulli, M.; Zhang, L.; Aprea, G.; Jimenez-Gomez, J.M.; Bendahmane, A.; Perrotta, G.; Giuliano, G.; Facella, P. Pivotal Roles of Cryptochromes 1a and 2 in Tomato Development and Physiology. Plant Physiol. 2019, 179, 732–748. [Google Scholar] [CrossRef]
- Liu, H.; Yu, X.; Li, K.; Klejnot, J.; Yang, H.; Lisiero, D.; Lin, C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef]
- Liu, H.; Liu, B.; Zhao, C.; Pepper, M.; Lin, C. The action mechanisms of plant cryptochromes. Trends Plant Sci. 2011, 16, 684–691. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Y.; Chang, H.; Ren, H.; Wu, X.; Wen, J.; Guan, Z.; Ma, L.; Qiu, L.; Yan, J.; et al. RUP2 facilitates UVR8 redimerization via two interfaces. Plant Commun. 2023, 4, 100428. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Q.; Guo, H.; Yu, X.; Liang, W.; Chen, Y.; Yin, R.; Lin, L. Tomato SlRUP is a negative regulator of UV-B photomorphogenesis. Mol. Hortic. 2021, 1, 8. [Google Scholar] [CrossRef]
- Liu, W.; Giuriani, G.; Havlikova, A.; Li, D.; Lamont, D.J.; Neugart, S.; Velanis, C.N.; Petersen, J.; Hoecker, U.; Christie, J.M.; et al. Phosphorylation of Arabidopsis UVR8 photoreceptor modulates protein interactions and responses to UV-B radiation. Nat. Commun. 2024, 15, 1221. [Google Scholar] [CrossRef]
- Bhatnagar, A.; Singh, S.; Khurana, J.P.; Burman, N. HY5-COP1: The central module of light signaling pathway. J. Plant Biochem. Biotechnol. 2020, 29, 590–610. [Google Scholar] [CrossRef]
- Qiu, Z.; Wang, H.; Li, D.; Yu, B.; Hui, Q.; Yan, S.; Huang, Z.; Cui, X.; Cao, B. Identification of Candidate HY5-Dependent and -Independent Regulators of Anthocyanin Biosynthesis in Tomato. Plant Cell Physiol. 2019, 60, 643–656. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.Q.; Chen, Y.Z.; Chen, Y.S.; Wang, X.; Phyon, J.M.; Zhou, S.P.; Cao, Z.Q.; Wang, Y.; Yang, J.F. Light regulates SlCOP1-mediated degradation of SlJAF13, a transcription factor essential for anthocyanin biosynthesis in tomato fruit. Plant Physiol. Biochem. 2025, 221, 109572. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Bu, Q.; Xu, X.; Paik, I.; Huang, X.; Hoecker, U.; Deng, X.W.; Huq, E. CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat. Commun. 2015, 6, 7245. [Google Scholar] [CrossRef]
- Paik, I.; Chen, F.; Ngoc Pham, V.; Zhu, L.; Kim, J.I.; Huq, E. A phyB-PIF1-SPA1 kinase regulatory complex promotes photomorphogenesis in Arabidopsis. Nat. Commun. 2019, 10, 4216. [Google Scholar] [CrossRef] [PubMed]
- Bursch, K.; Toledo-Ortiz, G.; Pireyre, M.; Lohr, M.; Braatz, C.; Johansson, H. Identification of BBX proteins as rate-limiting cofactors of HY5. Nat. Plants 2020, 6, 921–928. [Google Scholar] [CrossRef]
- Xu, D. COP1 and BBXs-HY5-mediated light signal transduction in plants. New Phytol. 2020, 228, 1748–1753. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB-bHLH-WD Repeat (MBW) Pigment Regulatory Model: Addition of a WRKY Factor and Co-option of an Anthocyanin MYB for Betalain Regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef]
- Sun, C.; Wang, C.; Zhang, W.; Liu, S.; Wang, W.; Yu, X.; Song, T.; Yu, M.; Yu, W.; Qu, S. The R2R3-type MYB transcription factor MdMYB90-like is responsible for the enhanced skin color of an apple bud sport mutant. Hortic. Res. 2021, 8, 156. [Google Scholar] [CrossRef]
- Zhong, C.; Tang, Y.; Pang, B.; Li, X.; Yang, Y.; Deng, J.; Feng, C.; Li, L.; Ren, G.; Wang, Y.; et al. The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida. Hortic. Res. 2020, 7, 78. [Google Scholar] [CrossRef]
- Jian, W.; Cao, H.; Yuan, S.; Liu, Y.; Lu, J.; Lu, W.; Li, N.; Wang, J.; Zou, J.; Tang, N.; et al. SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res. 2019, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Menconi, J.; Perata, P.; Gonzali, S. Novel R2R3 MYB transcription factors regulate anthocyanin synthesis in Aubergine tomato plants. BMC Plant Biol. 2023, 23, 148. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, Y.; Xiao, Z.; Shen, H.; Li, Y.; Wang, Y. Multilevel regulation of anthocyanin-promoting R2R3-MYB transcription factors in plants. Front. Plant Sci. 2022, 13, 1008829. [Google Scholar] [CrossRef]
- Li, H.; Bai, Y.; Yang, Y.; Zheng, H.; Xu, X.; Li, H.; Wang, W.; Tao, J. Transcriptomic analyses reveal light-regulated anthocyanin accumulation in ‘ZhongShan-HongYu’ grape berries. Sci. Hortic. 2023, 309, 111669. [Google Scholar] [CrossRef]
- Sun, L.; Li, S.; Tang, X.; Fan, X.; Zhang, Y.; Jiang, J.; Liu, J.; Liu, C. Transcriptome analysis reveal the putative genes involved in light-induced anthocyanin accumulation in grape ‘Red Globe’ (V. vinifera L.). Gene 2020, 728, 144284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, C.; Cui, Y.; Du, Q.; Tang, W.; Yang, W.; Kou, G.; Tang, W.; Chen, H.; Gong, R. Potential regulatory genes of light induced anthocyanin accumulation in sweet cherry identified by combining transcriptome and metabolome analysis. Front. Plant Sci. 2023, 14, 1238624. [Google Scholar] [CrossRef]
- Liu, H.; Su, J.; Zhu, Y.; Yao, G.; Allan, A.C.; Ampomah-Dwamena, C.; Shu, Q.; Lin-Wang, K.; Zhang, S.; Wu, J. The involvement of PybZIPa in light-induced anthocyanin accumulation via the activation of PyUFGT through binding to tandem G-boxes in its promoter. Hortic. Res. 2019, 6, 134. [Google Scholar] [CrossRef]
- Liu, H.; Shu, Q.; Lin-Wang, K.; Allan, A.C.; Espley, R.V.; Su, J.; Pei, M.; Wu, J. The PyPIF5-PymiR156a-PySPL9-PyMYB114/MYB10 module regulates light-induced anthocyanin biosynthesis in red pear. Mol. Hortic. 2021, 1, 14. [Google Scholar] [CrossRef]
- Zhao, L.; Sun, J.; Cai, Y.; Yang, Q.; Zhang, Y.; Ogutu, C.O.; Liu, J.; Zhao, Y.; Wang, F.; He, H.; et al. PpHYH is responsible for light-induced anthocyanin accumulation in fruit peel of Prunus persica. Tree Physiol. 2022, 42, 1662–1677. [Google Scholar] [CrossRef]
- Liu, Y.; Schouten, R.E.; Tikunov, Y.; Liu, X.; Visser, R.G.F.; Tan, F.; Bovy, A.; Marcelis, L.F.M. Blue light increases anthocyanin content and delays fruit ripening in purple pepper fruit. Postharvest Biol. Technol. 2022, 192, 112024. [Google Scholar] [CrossRef]
- Guo, X.L.; Hu, J.B.; Wang, D.L. Effect of Light Intensity on Blueberry Fruit Coloration, Anthocyanin Synthesis Pathway Enzyme Activity, and Gene Expression. Russ. J. Plant Physiol. 2023, 70, 136. [Google Scholar] [CrossRef]
- Zhang, H.N.; Li, W.C.; Wang, H.C.; Shi, S.Y.; Shu, B.; Liu, L.Q.; Wei, Y.Z.; Xie, J.H. Transcriptome Profiling of Light-Regulated Anthocyanin Biosynthesis in the Pericarp of Litchi. Front. Plant Sci. 2016, 7, 963. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.C.; Zhuo, M.G.; Abbas, F.; Hu, G.B.; Wang, H.C.; Huang, X.M. Transcription factor LcNAC002 coregulates chlorophyll degradation and anthocyanin biosynthesis in litchi. Plant Physiol. 2023, 192, 1913–1927. [Google Scholar] [CrossRef] [PubMed]
- Honda, C.; Kotoda, N.; Wada, M.; Kondo, S.; Kobayashi, S.; Soejima, J.; Zhang, Z.; Tsuda, T.; Moriguchi, T. Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiol. Biochem. 2002, 40, 955–962. [Google Scholar] [CrossRef]
- Do, V.G.; Kim, S.; Lee, Y.; Yang, S.; Kim, J.-H.; Win, N.M.; Kwon, Y.-S.; Park, J.; Park, J.-T. Differential Coloration, Pigment Biosynthesis-Related Gene Expression, and Accumulation According to Developmental Stage in the ‘Enbu’ Apple. Horticulturae 2023, 9, 1072. [Google Scholar] [CrossRef]
- Luan, A.; Zhang, W.; Yang, M.; Zhong, Z.; Wu, J.; He, Y.; He, J. Unveiling the molecular mechanism involving anthocyanins in pineapple peel discoloration during fruit maturation. Food Chem. 2023, 412, 135482. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, Y.; Zhang, G.; Wang, Y.; Huang, J.; Feng, Y.; Li, Y.; Jiang, J.; Zhang, Y. Overexpression of a BR inactivating enzyme gene GhPAG1 impacts eggplant fruit development and anthocyanin accumulation mainly by altering hormone homeostasis. Plant Sci. 2024, 341, 112014. [Google Scholar] [CrossRef]
- Jia, H.F.; Chai, Y.M.; Li, C.L.; Lu, D.; Luo, J.J.; Qin, L.; Shen, Y.Y. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 2011, 157, 188–199. [Google Scholar] [CrossRef]
- Rudell, D.R.; Fellman, J.K.; Mattheis, J.P. Preharvest Application of Methyl Jasmonate to ‘Fuji’ Apples Enhances Red Coloration and Affects Fruit Size, Splitting, and Bitter Pit Incidence. HortScience 2005, 40, 1760–1762. [Google Scholar] [CrossRef]
- Jeong, S.T.; Goto-Yamamoto, N.; Kobayashi, S.; Esaka, M. Effects of plant hormones and shading on the accumulation of anthocyanins and the expression of anthocyanin biosynthetic genes in grape berry skins. Plant Sci. 2004, 167, 247–252. [Google Scholar] [CrossRef]
- Awad, M.A.; de Jager, A. Formation of flavonoids, especially anthocyanin and chlorogenic acid in ‘Jonagold’ apple skin: Influences of growth regulators and fruit maturity. Sci. Hortic. 2002, 93, 257–266. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Li, Y.Y.; Song, L.Q.; Zhao, L.L.; You, C.X.; Hao, Y.J. EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 Act in a Regulatory Loop That Synergistically Modulates Ethylene Biosynthesis and Anthocyanin Accumulation. Plant Physiol. 2018, 178, 808–823. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Liu, L.; Yuan, C.; Guan, J. Molecular Characterization of Ethylene-Regulated Anthocyanin Biosynthesis in Plums During Fruit Ripening. Plant Mol. Biol. Report. 2016, 34, 777–785. [Google Scholar] [CrossRef]
- Ni, J.B.; Bai, S.L.; Zhao, Y.; Tao, R.Y.; Teng, Y.W. Ethylene Suppresses Anthocyanin Accumulation in ‘Red Zaosu’ Pear; International Society for Horticultural Science (ISHS): Leuven, Belgium, 2020; pp. 513–520. [Google Scholar]
- Figueroa, N.E.; Gatica-Meléndez, C.; Figueroa, C.R. Ethylene application at the immature stage of Fragaria chiloensis fruit represses the anthocyanin biosynthesis with a concomitant accumulation of lignin. Food Chem. 2021, 358, 129913. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Li, J.; Su, R.; Yan, H.; Zhao, F.; Xu, Q.; Zhou, B. The Identification of Regulatory Genes Involved in Light-Induced Anthocyanin Accumulation in Aft Tomato Developing Fruits. Horticulturae 2025, 11, 535. https://doi.org/10.3390/horticulturae11050535
Li J, Li J, Su R, Yan H, Zhao F, Xu Q, Zhou B. The Identification of Regulatory Genes Involved in Light-Induced Anthocyanin Accumulation in Aft Tomato Developing Fruits. Horticulturae. 2025; 11(5):535. https://doi.org/10.3390/horticulturae11050535
Chicago/Turabian StyleLi, Jiazhen, Ji Li, Rui Su, Haifang Yan, Fei Zhao, Qijiang Xu, and Bo Zhou. 2025. "The Identification of Regulatory Genes Involved in Light-Induced Anthocyanin Accumulation in Aft Tomato Developing Fruits" Horticulturae 11, no. 5: 535. https://doi.org/10.3390/horticulturae11050535
APA StyleLi, J., Li, J., Su, R., Yan, H., Zhao, F., Xu, Q., & Zhou, B. (2025). The Identification of Regulatory Genes Involved in Light-Induced Anthocyanin Accumulation in Aft Tomato Developing Fruits. Horticulturae, 11(5), 535. https://doi.org/10.3390/horticulturae11050535





