Abstract
Tomato fruit ripening is a complex process that determines the formation of fruit quality. Transcription factors (TFs) play key roles in regulating fruit ripening and quality formation. MADS-box genes, a crucial class of genes involved in virtually all aspects of plant development, are regarded as important candidate members among them. In this study, we present a detailed overview of the phylogeny and expression of 32 tomato MIKC-type MADS-box genes. Moreover, 20 genes contained many phytohormone-related elements. In combination with higher expression in fruit, eight genes are suggested to be involved in plant hormone pathways that regulate fruit ripening. A virus-induced gene silencing (VIGS) experiment revealed that TM4, TAGL11, SlMADS6, SlMADS99, TAGL1, SlMADS1, RIN, and MC may positively regulate fruit ripening. Measurements of the endogenous phytohormones in silenced TM4, TAGL11, SlMADS6, SlMADS99, TAGL1, SlMADS1, RIN, or MC fruit suggest that eight MIKC-type MADS-box genes, as well as medicated abscisic acid (ABA), salicylic acid (SA), gibberellin (GA3), indole-3-acetic acid (IAA), and/or methyl jasmonate (MeJA) pathways, positively regulate fruit ripening in tomatoes.
1. Introduction
The ripening of fleshy fruit involves a broad suite of metabolic pathway changes in texture, pigmentation, sugar and organic acid content, aroma, flavor, and nutritional qualities. Coordinate activation of the changes contributes to the appearance of high fruit quality, which not only makes fruit assist in seed dispersal but also provides essential nutrition for human and animal diets. Notably, the plant hormone ethylene, an important phytohormone, drives the coordinated activation of the above metabolic pathways during the fruit ripening stage, which is even more important in climacteric fruits [1]. Tomato (Solanum lycopersicum L.), a climacteric species, acts as a model plant for studying fruit ripening. To date, RNAi inhibition of SlACS2 or SlACO1 delayed fruit ripening [1,2]. Besides the functional plant hormone ethylene, additional phytohormones are also necessary for fruit ripening, such as abscisic acid (ABA) [3], salicylic acid (SA) [4], gibberellin (GA3) [5], indole-3-acetic acid (IAA) [6], and methyl jasmonate (MeJA) [7]. To further study fruit ripening, a series of tomato fruit ripening-deficient mutants, such as ripening inhibitor (rin), never ripe (Nr), nonripening (nor), and color nonripening (cnr), were obtained, suggesting that transcription factors (TFs) regulate fruit ripening, such as the MADS-box transcription factor (TF) [8,9,10].
The name MADS originates from the first letters of four MADS-box gene classes: MCM1, AGAMOUS, DEFICIENS, and SRF [11]. Members of MADS-box TF families are widely distributed in animals and plants. In plants, MADS-box genes were first found in Arabidopsis [12,13]. After that, MADS-box genes were also found to exist in tomatoes [11]. MADS-box genes function in nearly every aspect of plant growth and development, including seed germination [14], root development [15], floral development and evolution [16], and so on. In addition, a previous study indicated that TOMATOAGAMOUS1 (TAG1), TOMATO AGAMOUS-LIKE1 (TAGL1), TOMATO MADS BOX4 (TM4), and TOMATO MADS BOX6 (TM6) have been identified to be associated with fruit development and ripening. RNAi suppression of the TAG1 gene in tomatoes leads to misshapen fruits and homeotic conversion of stamens into petaloid organs [17,18], while TAGL1 plays an important role in regulating fruit ripening. To be more specific, the antisense suppression of TAGL1 results in ripening inhibition and pericarp thickness reduction. Furthermore, overexpression of TAGL1 leads to ripening-like sepals and lycopene-enhanced fruits [19,20,21]. TM4 is a homolog of the Arabidopsis FRUITFULL (FUL) gene and has also been reported to be related to fruit ripening [22]. The expression of TM4 is repressed in the rin, cnr, and nor mutants [23,24]. Additionally, TM6 transcripts mainly accumulate in carpel, primordial, and young tomato fruits and have been considered to be involved in fruit ripening [17,22].
MADS-box genes constitute one of the largest families of plant TFs [25]. They cluster into two phylogenetically distinct groups: type I and type II [12]. The function of most type I MADS-box genes remains to be elucidated, while several type II genes are key domestication genes in different eudicot and monocot crops [26]. Plant type II MADS-domain proteins possess a typical domain structure, which is composed of the MADS, I, K, and C-terminal domains [27]. Furthermore, because of this characteristic domain structure, type II genes are also referred to as MIKC-type MADS-box genes [27]. The MADS domain enables the DNA binding, nuclear localization, and dimerization of the TF [28,29], while the I and K domains facilitate dimerization and the higher-order complex formation of two or more MADS-domain proteins [30,31,32]. The C-terminal domain allows for the transcriptional activation of some MADS-domain proteins [33]. Overall, MADS-domain proteins bind as dimers to DNA sequences, with variable I, K, and C domains controlling dimer selectivity, helix formation, and transcriptional functions [28]. MIKC-type MADS-box genes are involved in virtually all aspects of plant development, including root, flower, seed, and embryo development [34]. They have also been reported to be involved in different stress responses [35,36,37]. Moreover, MIKC-type MADS-box genes, such as SlMADS1 [38], FaMADS1a [39], MaMADS1/MaMADS2 [40], etc., also affect fruit ripening. The mechanism of MIKC-type MADS-box-mediated phytohormone-regulating fruit ripening was also further confirmed. For instance, PaMADS7 regulates sweet cherry fruit ripening and softening by mediating the ABA pathway [41]. SlMBP15 positively regulated fruit ripening through GA biosynthesis in tomatoes [42]. Although these results suggest that some MIKC-type MADS-box genes regulate fruit ripening through phytohormone pathways, the regulatory network diagram between MIKC-type MADS-box genes and phytohormones is still incomplete. Here, we comprehensively analyzed the biological characteristics and expression patterns of the MIKC-type MADS-box genes in tomatoes and further explored the relationship between them and phytohormones, which provides a theoretical reference for research on the interaction of MIKC-type MADS-box genes with phytohormones during fruit ripening in plants.
2. Materials and Methods
2.1. Identification of the MIKC-Type MADS-box Genes in Tomatoes
The protein sequences of Arabidopsis thaliana AGL1 (At3g58780) and Oryza sativa OsMADS20 (Os12g31748) were downloaded from the TAIR database (https://www.arabidopsis.org, accessed on 1 October 2023) and PlantTFDB (http://planttfdb.gao-lab.org, accessed on 1 October 2023). The whole-genome data (GFF, FASTA, PEP, and CDS) of the tomato (SL4.0) were obtained from the Solanaceae Genomics Network (https://solgenomics.net/, accessed on 1 October 2023). The characteristic domains of MIKC-type MADS-box proteins (PF00319 and PF01486) were obtained from the PFAM database (https://www.ebi.ac.uk/interpro/, accessed on 1 October 2023). The physiochemical features of MIKC-type MADS-box proteins in tomatoes were examined using Expasy (https://web.expasy.org/protparam, accessed on 1 October 2023), including chromosome position, amino acid length, molecular weight (MW), isoelectric point (pI), instability index (II), and grand average of hydropathicity (GRAVY). The chromosome positions were determined using TBtools (v2.154) software, as previously described by us [43].
2.2. Phylogenetic Relationship
All MIKC-type MADS-box protein sequences of Arabidopsis thaliana (TAIR 10) and Oryza sativa subsp. japonica were downloaded from the TAIR database (https://www.arabidopsis.org, accessed on 2 October 2023) and PlantTFDB (http://planttfdb.gao-lab.org, accessed on 2 October 2023). A total of 105 protein sequences were aligned using MEGA7 software (megasoftware, https://www.megasoftware.net, accessed on 2 October 2023). The method of maximum likelihood (ML) was adopted to construct the evolutionary tree (the bootstrap parameter was set to 1000). Additionally, the phylogenetic tree was beautified using the iTOL online web tool (https://itol.embl.de, accessed on 2 October 2023) and Adobe Illustrator 2023 (Adobe Systems Incorporated, San Jose, CA, USA).
2.3. Analysis of Exon–Intron Structures and Conserved Motifs
MEGA7 was employed to sequence multiple alignments using the MUSCLE method with default parameters, and Espript 3.0 (https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi, accessed on 18 April 2025) was used for the presentation of the results [44]. We used TBtools software to analyze the exon–intron structure distribution of MIKC-type MADS-box genes in tomatoes. The conserved motifs were analyzed using the MEME online server (https://meme-suite.org, accessed on 3 October 2023). We utilized the TBtools software to visualize the distribution of exon–intron structures [44].
2.4. Analysis of Promoter Cis-Acting Elements
We choose the upstream region of the promoter sequence (2000 bp) of MIKC-type MADS-box genes to analyze. Then, we submitted the sequences of the promoter to PlantCar (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 4 October 2023). The cis-acting elements were screened and integrated using Excel software (Microsoft Inc., Seattle, WA, USA), and TBtools was used for further analysis.
2.5. Expression Patterns of MIKC-Type MADS-box Genes at Different Growth Stages
The members of MIKC-type MADS-box genes were searched for in the Tomato eFP Browser (https://bar.utoronto.ca/efp_tomato/cgi-bin/efpWeb.cgi, accessed on 4 October 2023). We sorted out output data and drew the expression patterns of MIKC-type MADS-box genes at different growth stages using Rstudio (Posit Inc., Boston, MA, USA). We applied STRING (https://string-db.org, accessed on 18 April 2025) to perform predictions of interacting proteins and co-expression in 32 candidate proteins.
2.6. Plant Materials and Growth Conditions
The tomatoes (Solanum. lycopersicum cv. Micro-Tom) were cultivated for further study. Firstly, the seeds were disinfected with 1% NaClO solution for 5 min. Second, the disinfected seeds were germinated in a conical flask containing pure water and placed in a shaking incubator (180 rpm, 28 °C, darkness) for 3 days. Lastly, the germinated seeds were planted in the pot and then transplanted into a vegetable base (Guangxi University). The breaker fruits were picked for further study.
2.7. Virus-Induced Gene Silencing (VIGS)
To generate the necessary constructs for VIGS, we employed the VIGS tool in the Sol Genomics Network (http://solgenomics.net, accessed on 18 April 2025) to clone a specific 300 bp segment of TM4/TAGL11/SlMADS6/SlMADS99/TAGL1/SlMADS1/RIN/MC CDS into the pTRV2 vector [45]. The plasmids pTRV1, pTRV2, and pTRV2-TM4/TAGL11/SlMADS6/SlMADS99/TAGL1/SlMADS1/RIN/MC were transformed with Agrobacterium tumefaciens GV3101. The mixture of A. tumefaciens pTRV2-TM4/TAGL11/SlMADS6/SlMADS99/TAGL1/SlMADS1/RIN/MC (pTRV1:pTRV2-TM4/TAGL11/SlMADS6/SlMADS99/TAGL1/SlMADS1/RIN/MC = 1:1, v/v) and a control mixture (pTRV1:pTRV2 = 1:1, v/v) was subsequently injected into intact and similarly sized tomato fruits at the Br stages using a needleless syringe with a capacity of 1 mL. The infiltration was conducted on ten fruits for each VIGS construct. Subsequently, the tomato fruits were placed in a growth chamber, subjected to darkness, and maintained at a temperature of 25 °C for a duration of 3 days. After 3 days of infiltration, the tomato fruits were photographed and pericarp tissues showing obvious inhibition of ripening were collected. The collected tissue samples were promptly flash-frozen in liquid nitrogen and stored at −80 °C until they were required for subsequent utilization.
2.8. Carotenoid (Car) Content Measurement
The quantification of total Car content was extracted with 90% ethanol, and the OD value was obtained using an ultraviolet spectrophotometer [46].
2.9. Extraction and Determination of Phytohormones
The fruit sample (0.2 g) was pulverized in liquid nitrogen and then the powder was transferred to tubes. Subsequently, 2 mL of a mixture containing 2-propanol, H2O, and HCl in a ratio of 2:1:0.002 (V/V/V) was added. After being shaken at 4 °C for an hour, 1 mL of dichloromethane was added swiftly, and the tube was further agitated for an additional hour. The sample was centrifuged at 4 °C for 5 min at a speed of 12,000× g. The liquid from the lower phase was subsequently transferred into a new tube. The nitrogen evaporator was utilized for the purpose of desiccating the sample. The samples were redissolved in 0.5 mL of 80% methanol and then the solution was filtered through a 0.22 μm organic filter and transferred to a 2 mL brown bottle. All samples were stored in a refrigerator at −20 °C until they were analyzed.
The concentrations of ABA, auxin (IAA), gibberellic acid 3 (GA3), and SA were measured using ultra-performance convergence chromatography UPC2 (Waters Inc., Milford, MA, USA) equipped with a HSS C18 SB column. The elution mode was set to isocratic elution with a mobile phase consisting of 90% CO2, 5% methanol, and 5% acetonitrile. The flow rate was maintained at 0.5 mL/min, while the back pressure was set to 1600 psi. The column temperature was kept at 30 °C. The detection wavelengths for ABA, GA3, and SA were set at 210 nm, while IAA was detected at 254 nm.
The Agilent 7890A/5975C gas chromatography-mass spectrometry system (Agilent Inc., Santa Clara, CA, USA) was utilized for the determination of jasmonic acid methyl ester (MeJA) content. The carrier gas used was helium with a purity of ≥99.999%, and it was delivered at a flow rate of 1.0 mL/min. The injector temperature was maintained at 250 °C. The temperature program was as follows: initially held at 150 °C for 1 min, then increased to 190 °C at a rate of 20 °C per minute, and finally raised to 215 °C at a rate of 5 °C per minute for a duration of 2 min.
2.10. Statistical Analysis
The experiment was conducted with three replications, and the results were reported as mean ± SE. Analysis of variance (ANOVA) was performed using SPSS 22.0 (SPSS Institute Inc., Armonk, NY, USA), and the treatment means were compared using Tukey’s test at a 0.05 level of probability. All figures were generated with OriginPro 2017 (OriginLab Institute Inc., Northampton, MA, USA).
3. Results
3.1. Genome-Wide Identification and Phylogenetic Analyses of MIKC-Type MADS-box Genes
The members of the MIKC-type MADS-box gene family were identified through homologous alignments and a simple HMM search (PF00319 and PF01486) using TBtools software. We identified a total of 32 MIKC-type MADS-box genes from the tomato genome, which were designated based on previous studies and their locations on the chromosomes [47] (Figure 1A; Table S1). Meanwhile, the gene locus is Solyc10g044965, which was renamed SlMADS99 by us (Table S1). Interestingly, the members were located unevenly on 11 chromosomes, excluding chromosome 9 (Figure 1A). The number of amino acids of 32 MIKC-type MADS-box genes varied from 179 to 337, molecular weight (MW) ranged from 20.7 to 38.6 Da, and the isoelectric point (pI) was distributed between 5.69 and 9.72 (Table S1). In order to better understand the interactions among tomato MIKC-type MADS-box proteins, we constructed a gene network using the MIKC-type MADS-box protein sequences. It indicated that SlMADS1 and TM5/TDR5/LeSEP3 interact with TAGL1. Moreover, TM5/TDR5/LeSEP3 also interact with TAGL11 (Figure S2). To understand evolutionary relationships, a phylogenetic tree was constructed, including the MIKC-type MADS-box proteins of tomatoes, rice, and Arabidopsis. The results demonstrated that based on the branching structure of the phylogenetic tree, the members of MIKC-type MADS-box proteins were divided into four phylogenetic subgroups (I to IV; Figure 1B). Subgroup IV had 44 members, making it the largest group. The smallest, subgroup I, had four MIKC-type MADS-box family members. Furthermore, subgroups II and III included 37 and 20 members, respectively (Figure 1B). These results suggest that these MIKC-type MADS-box genes may play important roles in the evolution of the tomato genome.
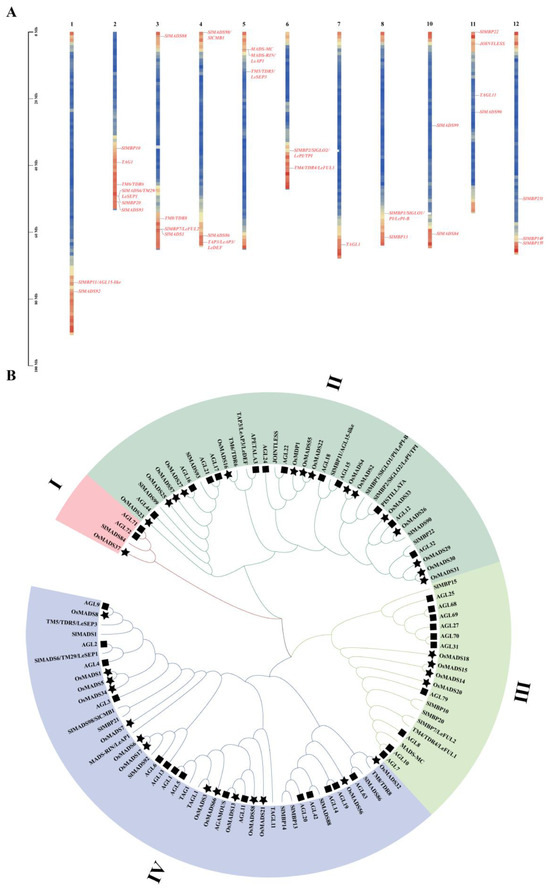
Figure 1.
Chromosomal locations (A) and phylogenetic analysis (B) of tomato (Solanum lycopersicum) MIKC-type MADS-box genes. Chromosomal names are indicated at the top of each bar, and MIKC-type MADS-box genes are distributed unevenly across chromosomes 1–12, with the exception of 9. Clustal W was used to align the protein sequences of tomato, Arabidopsis, and rice using the complete protein sequences, and phylogenetic trees were generated using the neighbor-joining (NJ) method in MEGA 7.0 software. A total of 1000 bootstrap replicates were used to assess tree reliability, and different shapes were used to distinguish different species. Simply put, the absence of markers represents tomatoes, the squares represent Arabidopsis, and the pentagram represents rice.
3.2. Exon–Intron Structure, Conserved Motifs, and Cis-Acting Elements of MIKC-Type MADS-box Genes
As shown in Figure S1A, 32 MIKC-type MADS-box genes displayed similar exon–intron structures. Furthermore, through the analysis of protein motifs using NCBI, we also discovered that MIKC-type MADS-box proteins all contain MADS domains, and most of them contain K-box domains. However, the conservation of I and C domains among MADS-box protein sequences is relatively low, and there are no common motifs (Figure S3). Additionally, we also analyzed the putative conserved motifs using the MEME website and seven divergent motifs were identified in MIKC-type MADS-box genes, which were named as motifs 1–8 (Figure S1B). Interestingly, all of the MIKC-type MADS-box genes contain one, two, four, and five motifs, which implies that they are highly conserved (Figure S1B). To predict their biological functions, the cis-acting elements of 32 MIKC-type MADS-box genes were analyzed. Figure 2A shows 45 types of elements in the promoter region (2000 bp). Among them, twenty-seven elements are related to light response, thirteen elements are hormone-responsive elements, and five elements are related to stress responses (Figure 2A), suggesting that MADS-box genes might be involved in the regulation of light, hormones, and stresses. Additionally, 20 candidate genes were chosen on the basis of an expectation greater than 1.5 (Figure 2B; Table 1). These genes contained many phytohormone-related elements, such as ABRE (related to ABA), a TCA element (related to SA), a TATC box (related to GA3), a TGA element and an O2 site (related to auxin), and a CGTCA motif (related to MeJA) (Figure 2B). The results indicated that 20 MADS-box genes play crucial roles in plant phytohormone pathways.
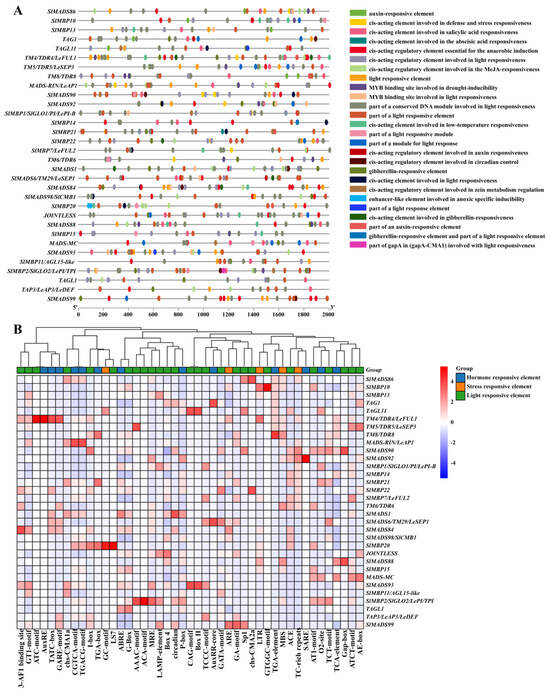
Figure 2.
The schematic diagram (A) and hierarchical clustering (B) of cis-elements in the promoters of MIKC-type MADS-box genes in tomatoes (Solanum lycopersicum).

Table 1.
Prediction of phytohormone-related cis-acting elements in 20 genes of tomato MIKC-type MADS-box family members.
3.3. Expression Patterns of MIKC-Type MADS-box Genes During Growth and Development
To investigate the expression patterns of MIKC-type MADS-box genes during tomato growth and development, the levels of 32 genes at various growth stages, including unopened flower bud (UFB), fully opened flower (FOF), leaves, root, 1 cm fruit (1F), 2 cm fruit (2F), 3 cm fruit (3F), mature green fruit (MGF), breaker fruit (BF), breaker fruit + 10 (BF10), Pimpinellifolium immature green fruit (PIGF), Pimpinellifolium breaker fruit (PBF), Pimpinellifolium breaker + 5 fruit (PB5F), and Pimpinellifolium leave (PL), were analyzed (Figure 3). Of twenty candidate genes, eight genes were selected based on their high expression of MADS-box genes at the fruit and breaker stages. The eight genes included TM4/TDR4/LeFUL1, TAGL11, SlMADS6/TM29/LeSEP1, TAGL1, SlMADS1, MADS-RIN/LeAP1, and MADS-MC, (Figure 3). Although the expression data of SlMADS99 were not found in the database, it was selected to explore its function.
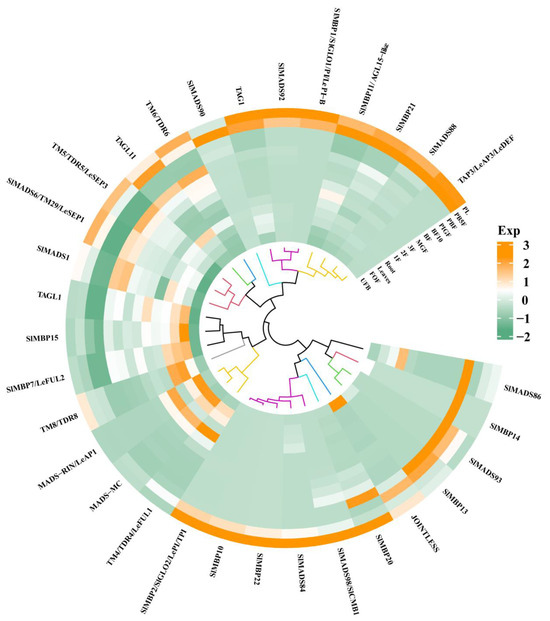
Figure 3.
The expression patterns of MIKC-type MADS-box genes during the growth and development of tomatoes (Solanum lycopersicum). UFB represents flower bud, FOF represents fully opened flower, 1F represents 1 cm fruit, 2F represents 2 cm fruit, 3F represents 3 cm fruit, MGF represents mature green fruit, BF represents breaker fruit, BF10 represents breaker fruit + 10, PIGF represents Pimpinellifolium immature green fruit, PBF represents Pimpinellifolium breaker fruit, PB5F represents Pimpinellifolium breaker + 5 fruit, and PL represents Pimpinellifolium. The values of gene expression levels are standardized.
3.4. Silencing of MADS-box Genes Affects Tomato Fruit Ripening
To ascertain whether there is a potential relationship between MADS-box genes and the process of tomato fruit ripening, we treated 30 ‘Micro-Tom’ tomatoes at the breaker stage using pTRV2-TM4/TAGL11/SlMADS6/SlMADS99/TAGL1/SlMADS1/RIN/MC constructs (Figure 4). The infected fruits showcased distinct mottled green and orange areas that were clearly demarcated by a distinct border (Figure 4), and the Car content was decreased, suggesting that TM4, TAGL11, SlMADS6, SlMADS99, TAGL1, SlMADS1, RIN, and MC may positively regulate fruit ripening.
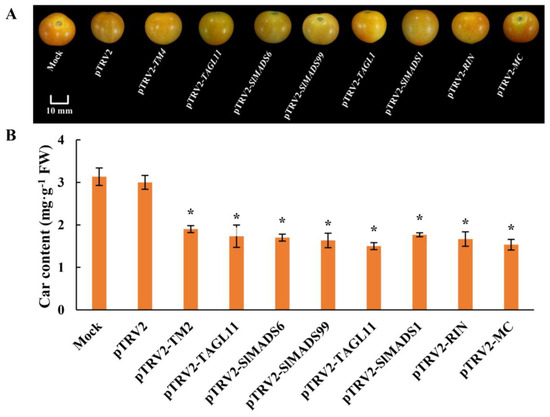
Figure 4.
S3 fruit phenotype (A) and the Car content (B) of pTRV2 and pTRV2-TM4/TAGL11/SlMADS6/SlMADS99/TAGL1/SlMADS1/RIN/MC. Car: carotenoids (* p < 0.05).
3.5. Silencing of MADS-box Genes Regulated Endogenous Plant Hormone Contents
To explore the relationship between TM4/TAGL11/SlMADS6/SlMADS99/TAGL1/SlMADS1/RIN/MC and ABA/SA/GA3/IAA/MeJA, the endogenous plant hormones of tomato fruits infected with A. tumefaciens cultures were measured using chromatography. As Figure 5A shows, the silenced MC significantly enhanced ABA content, and silenced TM4 or TAGL11 significantly suppressed ABA content. Silencing of SlMADS99, TAGL1, RIN, or MC resulted in an increase in SA content, and silencing of TM4 resulted in a decrease in SA content (Figure 5B). Endogenous GA3 was significantly increased when TM4/TAGL11/SlMADS6/SlMADS99/TAGL1/SlMADS1/RIN/MC was silenced (Figure 5C). Figure 5D shows that the silenced MC significantly increased IAA content, and silenced TM4 or SlMADS1 significantly decreased IAA content. Silencing of TM4/TAGL11/SlMADS6/SlMADS99/TAGL1/SlMADS1/MC significantly suppressed MeJA content (Figure 5E). The results showed that TM4/TAGL11/SlMADS6/SlMADS99/TAGL1/SlMADS1/RIN/MC may positively regulate fruit ripening by modulating ABA, SA, GA3, IAA, and/or MeJA pathway(s).
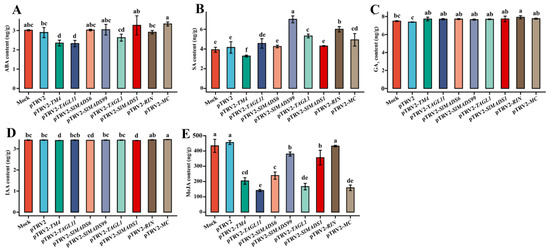
Figure 5.
ABA (A), SA (B), GA3 (C), IAA (D), and MeJA (E) contents of pTRV2 and pTRV2- TM4/TAGL11/SlMADS6/SlMADS99/TAGL1/SlMADS1/RIN/MC in fruit. ABA represents abscisic acid, SA represents salicylic acid, GA3 represents gibberellin, IAA represents indole-3-acetic acid, and MeJA represents methyl jasmonate. Bars denoted by the same letter presented no signiffcant difference at p < 0.05 according to Duncan’s test.
4. Discussion
MIKC-type MADS-box genes are of great significance for development and fruit ripening in plants. A total of 108, 72, 168, and 160 MADS-box genes were identified in Arabidopsis [48], wheat [49], rape [46], and wheel wingnut [50]. In this study, a genome-wide identification of the MIKC-type MADS-box gene family was carried out in tomatoes (Table S1). The identified 32 MIKC-type MADS-box genes were located unevenly on 11 chromosomes (Figure 1A). Phylogenetic tree analyses showed that their family members had high homology with Arabidopsis and rice (Figure 1B). Additionally, the gene structures and conserved motifs of each subclass were similar (Figure S1), and all of the MIKC-type MADS-box genes contain one, two, four, and five motifs, which implies that they are highly conserved (Figure S1B).
By analyzing the promoter region, we found that most of the components of cis-elements in MIKC-type MADS-box genes were associated with light, phytohormones, and stresses (Figure 2A). It is indicated that MIKC-type MADS-box genes in tomatoes respond to both hormonal and environmental factors. This has also been studied in other plants in related research. There is a relationship between MADS-box TF AaSEPALLATA1 (AaSEP1) and CONSTITUTIVE PHOTOMORPHOGENIC 1 (AaCOP1), a major repressor of light signaling that regulates glandular secretory trichome initiation in Artemisia annua [51]. SAUR50-like genes, which are MADS-box genes, are involved in light signal-mediated pedicel or stem development in Phoebe bournei [52]. A light cis-element was identified in the MIKC-type MADS-box genes of Arabidopsis [48], wheat [49], rape [46], etc. The regulatory mechanism of interaction between MIKC-type MADS-box genes and light signals is lacking and should be further investigated in the future. In peanuts, 166 MIKC-type MADS-box genes were identified, and five selected AhMADS genes, including AhMADS9, AhMADS21, AhMADS34, AhMADS50, and AhMADS64, were induced by cold, hot, drought, and salt stresses [53]. SiMADS51, a MIKC-type MADS-box gene, was reported to be a negative regulator of drought stress tolerance. These results suggested that MIKC-type MADS-box genes play an important role in response to stresses. The results of our study provide a theoretical basis for further research on the mechanisms of action of MIKC-type MADS-box genes with respect to stresses.
The expression patterns of MIKC-type MADS-box genes showed that six genes, including TM4/TDR4/LeFUL1, TAGL11, SlMADS6/TM29/LeSEP1, TAGL1, SlMADS1, and MADS-RIN/LeAP1, were highly expressed at the fruit and breaker stages, revealing that MIKC-type MADS-box genes play a regulatory role in the fruit ripening process (Figure 3). Tomato fruit ripening was delayed when TM4, TAGL11, SlMADS6, SlMADS99, TAGL1, SlMADS1, RIN, or MC was silenced (Figure 4), suggesting that TM4, TAGL11, SlMADS6, SlMADS99, TAGL1, SlMADS1, RIN, and MC may positively regulate fruit ripening. The rin mutation led to extended shelf lives in tomatoes [10], demonstrating that RIN is an essential regulator of tomato fruit ripening. RIN interacted with the promoters of genes involved in ripening regulation, ethylene metabolism, cell wall metabolism, and carotenoid biosynthesis, thereby regulating fruit ripening [54]. The single FUL1 (previously called TDR4) and FUL2 knockdown lines only showed very mild changes in fruit pigmentation, whereas the double-silenced lines significantly decreased lycopene levels, thereby exhibiting an orange, ripe fruit phenotype, revealing that FUL1 and FUL2 play redundant roles in fruit ripening [55]. Additionally, TDR4 is involved in ripening and nutrient synthesis in tomato fruits and is therefore an important regulator of fruit quality [56]. TAG1 controls placenta and seed formation, while TAGL1 participates in cuticle development and lignin biosynthesis inhibition [57]. Silencing of TAGL1 resulted in altered fruit pigmentation, while overexpression of TAGL1 reduces carotenoid and ethylene levels, inhibits chlorophyll breakdown, and downregulates genes related to fruit ripening to delay fruit ripening [19]. Double RNAi lines showed cell abnormalities in their stamens and carpels and produced extremely small fruit-like organs displaying some sepaloid features, suggesting that redundant and divergent functions of TAG1 and TAGL1 genes regulated the reproductive development of tomatoes [57]. Tomatoes with SlMADS1 silenced by RNA interference (RNAi) had shortened ripening times in comparison with the control, suggesting that SlMADS1 is a repressive modulator of fruit ripening [38]. The MC TF may play key roles in pedicel abscission zones (AZ) and the development of abscission zones [58]. These results indicated that MIKC-type MADS-box genes have crucial roles in fruit ripening. However, some MIKC-type MADS-box genes, as mentioned above, have not been reported in the regulation of fruit ripening. Therefore, their functions and regulatory mechanisms need further study.
The relationship between phytohormones and tomato fruit ripening has been extensively examined in recent scientific investigations [47]. To investigate whether MIKC-type MADS-box transcription factors mediate phytohormone-regulated fruit ripening, the cis-elements of MIKC-type MADS-box genes were analyzed. The results indicated that many phytohormone-related elements, such as ABR, TCA elements, TATC boxes, TGA elements, O2 sites, and the CGTCA motif, exist in promoter regions (Figure 2B), suggesting that MADS-box genes play crucial roles in fruit ripening through the phytohormone pathway. As Figure 5 shows, TM4/MC-mediated ABA, SA, GA3, IAA, and MeJA, TAGL11-mediated ABA, GA3, and MeJA, SlMADS6/SlMADS9-mediated SA, GA3, and MeJA, TAGL1-mediated SA, GA3, IAA, and MeJA, SlMADS1-mediated GA3 and MeJA, and RIN-mediated SA, GA3, and MeJA regulated fruit ripening. Yin et al. [42]. reported that when a MIKC-type MADS-box gene SlMBP15 was silenced, a lower GA3 content was observed than in the WT. Meanwhile, they also found that expression levels of the GA biosynthesis-related genes GA3ox1, GA3ox2, GA20ox1, GA20ox2, GAST1, and GID2 were greatly repressed in SlMBP15-silenced tomato fruits, demonstrating that SlMBP15-mediated GA positively regulated tomato fruit ripening [42]. In sweet cherry fruit, ABA regulated PaMADS7 expression. Silencing of PaMADS7 influenced ABA content and suppressed fruit ripening, which was rescued by exogenous ABA. These results indicated that PaMADS7 is a positive regulator of sweet cherry fruit ripening, mediated by the ABA pathway [41]. Two MADS-box genes, VCM1 and VCM2, regulated the proliferation activity of the vascular cambium and secondary growth in Populus by modulating auxin homeostasis [59]. MADS2 positively participated in the regulation of defense in postharvest peach through a combination of SA-dependent NPR1 activation and ABA signaling-induced callose accumulation [60]. There results suggested that a MIKC-type MADS-box TF regulated fruit ripening through a mechanism that is dependent on the phytohormone pathway. Additionally, increasing studies show that fruit ripening is regulated by the combined actions of TFs and phytohormones. For instance, the NAC TF SlNAC4 influences the process of fruit softening, which operates through an ABA-dependent pathway [61]. MpSNAC67, a NAC TF from banana, was reported to induce the senescence of fruit, which is dependent on the SA pathway [62]. Therefore, more research is needed on the relationship between MIKC-type MADS-box TFs and phytohormones during fruit ripening and softening in the future.
5. Conclusions
In this study, we first identified 32 MIKC-type MADS-box genes in the tomato genome. Subsequently, comprehensive bioinformatics analyses, including chromosomal locations, phylogenetics, exon–intron structures, and conserved motifs, were performed. Analysis of cis-acting elements reveals that 20 genes contained many phytohormone-related elements. Gene expression patterns indicate that TM4, TAGL11, SlMADS6, SlMADS99, TAGL1, SlMADS1, RIN, and MC may positively regulate fruit ripening. This conclusion was further confirmed by VIGS experiments. Measuring the endogenous phytohormones in infected fruits revealed that eight MIKC-type MADS-box genes mediate the ABA, SA, GA3, IAA, and/or MeJA pathways and positively regulate fruit ripening in tomatoes. Taken together, the function analysis of the MIKC-type MADS-box genes in tomatoes will provide an effective and practical way to study the functions of the MADS-box genes and to further explore the regulation mechanism of the MADS-box genes in tomatoes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae11050487/s1, Figure S1: Analysis of the conserved motifs and gene structures of the MIKC-type MADS-box gene family; Figure S2: Alignment of the amino acid sequences of the MIKC-type MADS-box; Figure S3: Analysis of the interaction relationship of MIKC-type MADS-box proteins; Table S1: Overview of the physiochemical features of MIKC-type MADS-box proteins in tomatoes.
Author Contributions
Conceptualization, formal analysis, and writing—original draft, C.L.; visualization—Y.L. (Yushi Lu); investigation—J.X.; investigation—J.C.; data curation—Y.L. (Yunzhi Liu); conceptualization—W.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Nature Science Foundation of China (31660568), Guangxi Science and Technology Major Project (GuikeAA22068088), the Science and Technology Major Project of Guangxi (AA22068088-2), Start-up Funding for Introduced Talents in Guangxi University (to C.L.), the Guangxi Colleges and Universities Young and Middle-aged Teachers’ Basic Scientific Research Ability Improvement Project (2024KY0010), the Guangxi Graduate Education Innovation Program (YCSW2024093), and the Guangxi University Student Innovation and Entrepreneurship Training Program Funding Project (202310593704; 202310593714; 202410953044S).
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cara, B.; Giovannoni, J.J. Molecular biology of ethylene during tomato fruit development and maturation. Plant Sci. 2008, 175, 106–113. [Google Scholar] [CrossRef]
- Yokotani, N.; Nakano, R.; Imanishi, S.; Nagata, M.; Inaba, A.; Kubo, Y. Ripening-associated ethylene biosynthesis in tomato fruit is autocatalytically and developmentally regulated. J. Exp. Bot. 2009, 60, 3433–3442. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Yang, S.; Chai, L.; Wu, C.; Zhou, J.; Liu, Y.; Xue, Z. Abscisic acid and fruit ripening: Multifaceted analysis of the effect of abscisic acid on fleshy fruit ripening. Sci. Hortic. 2021, 281, 109999. [Google Scholar] [CrossRef]
- Shi, H.Y.; Cao, L.W.; Xu, Y.; Yang, X.; Liu, S.L.; Liang, Z.S.; Li, G.C.; Yang, Y.P.; Zhang, Y.X.; Chen, L. Transcriptional profiles underlying the effects of salicylic acid on fruit ripening and senescence in pear (Pyrus pyrifolia Nakai). J. Integr. Agric. 2021, 20, 2424–2437. [Google Scholar] [CrossRef]
- Wu, M.; Liu, K.; Li, H.; Li, Y.; Zhu, Y.; Su, D.; Zhang, Y.; Deng, H.; Wang, Y.; Liu, M. Gibberellins involved in fruit ripening and softening by mediating multiple hormonal signals in tomato. Hortic. Res. 2023, 11, uhad275. [Google Scholar] [CrossRef]
- Böttcher, C.; Keyzers, R.A.; Boss, P.K.; Davies, C. Sequestration of auxin by the indole-3-acetic acid-amido synthetase GH3-1 in grape berry (Vitis vinifera L.) and the proposed role of auxin conjugation during ripening. J. Exp. Bot. 2010, 61, 3615–3625. [Google Scholar] [CrossRef] [PubMed]
- Zuñiga, P.E.; Castañeda, Y.; Arrey-Salas, O.; Fuentes, L.; Aburto, F.; Figueroa, C.R. Methyl jasmonate applications from flowering to ripe fruit stages of strawberry (Fragaria × ananassa ‘Camarosa’) reinforce the fruit antioxidant response at post-harvest. Front. Plant Sci. 2020, 11, 538. [Google Scholar] [CrossRef]
- Tigchelaar, E.; Tomes, M.; Kerr, E.; Barman, R. A new fruit ripening mutant, non-ripening (nor). Rep. Tomato Genet. Coop. 1973, 23, 33. [Google Scholar]
- Wilkinson, J.Q.; Lanahan, M.B.; Yen, H.C.; Giovannoni, J.J.; Klee, H.J. An ethylene-inducible component of signal transduction encoded by never-ripe. Science 1995, 270, 1807–1809. [Google Scholar] [CrossRef]
- Vrebalov, J.; Ruezinsky, D.; Padmanabhan, V.; White, R.; Medrano, D.; Drake, R.; Schuch, W.; Giovannoni, J. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 2002, 296, 343–346. [Google Scholar] [CrossRef]
- Messenguy, F.; Dubois, E. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 2003, 316, 1–21. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; de Pouplana, L.R.; Martínez-Castilla, L.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef]
- Becker, A.; Theissen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenetics Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef]
- Yu, L.H.; Wu, J.; Zhang, Z.S.; Miao, Z.Q.; Zhao, P.X.; Wang, Z.; Xiang, C.B. Arabidopsis MADS-box transcription factor AGL21 acts as environmental surveillance of seed germination by regulating ABI5 expression. Mol. Plant 2017, 10, 834–845. [Google Scholar] [CrossRef]
- Sun, C.H.; Yu, J.Q.; Duan, X.; Wang, J.H.; Zhang, Q.Y.; Gu, K.D.; Hu, D.G.; Zheng, C.S. The MADS transcription factor CmANR1 positively modulates root system development by directly regulating CmPIN2 in chrysanthemum. Hortic. Res. 2018, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Saedler, H.; Becker, A.; Winter, K.U.; Kirchner, C.; Theissen, G. MADS-box genes are involved in floral development and evolution. Acta Biochim. Pol. 2001, 48, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Pnueli, L.; Hareven, D.; Rounsley, S.D.; Yanofsky, M.F.; Lifschitz, E. Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell 1994, 6, 163–173. [Google Scholar] [PubMed]
- Pan, I.L.; McQuinn, R.; Giovannoni, J.J.; Irish, V.F. Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J. Exp. Bot. 2010, 61, 1795–1806. [Google Scholar] [CrossRef]
- Itkin, M.; Seybold, H.; Breitel, D.; Rogachev, I.; Meir, S.; Aharoni, A. TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 2009, 60, 1081–1095. [Google Scholar] [CrossRef]
- Vrebalov, J.; Pan, I.L.; Arroyo, A.J.M.; McQuinn, R.; Chung, M.; Poole, M.; Rose, J.; Seymour, G.; Grandillo, S.; Giovannoni, J.; et al. Fleshy fruit expansion and ripening are regulated by the Tomato SHATTERPROOF gene TAGL1. Plant Cell 2009, 21, 3041–3062. [Google Scholar] [CrossRef]
- Giménez, E.; Pineda, B.; Capel, J.; Antón, M.T.; Atarés, A.; Pérez-Martín, F.; García-Sogo, B.; Angosto, T.; Moreno, V.; Lozano, R. Functional analysis of the Arlequin mutant corroborates the essential role of the Arlequin/TAGL1 gene during reproductive development of tomato. PLoS ONE 2010, 5, e14427. [Google Scholar] [CrossRef] [PubMed]
- Busi, M.V.; Bustamante, C.; D’angelo, C.; Hidalgo-Cuevas, M.; Boggio, S.B.; Valle, E.M.; Zabaleta, E. MADS-box genes expressed during tomato seed and fruit development. Plant Mol. Biol. 2003, 52, 801–815. [Google Scholar]
- Seymour, G.B.; Manning, K.; Eriksson, E.M.; Popovich, A.H.; King, G.J. Genetic identification and genomic organization of factors affecting fruit texture. J. Exp. Bot. 2002, 53, 2065–2071. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, M.; Shima, Y.; Higuchi, N.; Nakano, T.; Koyama, Y.; Kasumi, T.; Ito, Y. Direct targets of the tomato-ripening regulator RIN identified by transcriptome and chromatin immunoprecipitation analyses. Planta 2012, 235, 1107–1122. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Schilling, S.; Kennedy, A.; Pan, S.; Jermiin, L.S.; Melzer, R. Genome-wide analysis of MIKC-type MADS-box genes in wheat: Pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 2020, 225, 511–529. [Google Scholar] [CrossRef]
- Kaufmann, K.; Melzer, R.; Theissen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Wang, M.; Meyerowitz, E.M. DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 1996, 24, 3134–3141. [Google Scholar] [CrossRef]
- Immink, R.G.; Gadella, T.W., Jr.; Ferrario, S.; Busscher, M.; Angenent, G.C. Analysis of MADS box protein-protein interactions in living plant cells. Proc. Natl. Acad. Sci. USA 2002, 99, 2416–2421. [Google Scholar] [CrossRef]
- Yang, Y.; Jack, T. Defining subdomains of the K domain important for protein-protein interactions of plant MADS proteins. Plant Mol. Biol. 2004, 55, 45–59. [Google Scholar] [CrossRef]
- Melzer, R.; Verelst, W.; Theissen, G. The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in ‘floral quartet’-like complexes in vitro. Nucleic Acids Res. 2009, 37, 144–157. [Google Scholar] [CrossRef] [PubMed]
- Theißen, G.; Melzer, R.; Rümpler, F. MADS-domain transcription factors and the floral quartet model of flower development: Linking plant development and evolution. Development 2016, 143, 3259–3271. [Google Scholar] [CrossRef]
- Honma, T.; Goto, K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 2001, 409, 525–529. [Google Scholar] [CrossRef]
- Gramzow, L.; Theissen, G. A hitchhiker’s guide to the MADS world of plants. Genome Biol. 2010, 11, 214. [Google Scholar] [CrossRef]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, 242. [Google Scholar] [CrossRef]
- Jia, J.; Zhao, P.; Cheng, L.; Yuan, G.; Yang, W.; Liu, S.; Chen, S.; Qi, D.; Liu, G.; Li, X. MADS-box family genes in sheepgrass and their involvement in abiotic stress responses. BMC Plant Biol. 2018, 18, 42. [Google Scholar] [CrossRef]
- Wei, M.; Wang, Y.; Pan, R.; Li, W. Genome-wide identification and characterization of MADS-box family genes related to floral organ development and stress resistance in Hevea brasiliensis Müll. Arg. Forests 2018, 9, 304. [Google Scholar] [CrossRef]
- Dong, T.; Hu, Z.; Deng, L.; Wang, Y.; Zhu, M.; Zhang, J.; Chen, G. A tomato MADS-box transcription factor, SlMADS1, acts as a negative regulator of fruit ripening. Plant Physiol. 2013, 163, 1026–1036. [Google Scholar] [CrossRef]
- Lu, W.; Chen, J.; Ren, X.; Yuan, J.; Han, X.; Mao, L.; Ying, T.; Luo, Z. One novel strawberry MADS-box transcription factor FaMADS1a acts as a negative regulator in fruit ripening. Sci. Hortic. 2018, 227, 124–131. [Google Scholar] [CrossRef]
- Elitzur, T.; Yakir, E.; Quansah, L.; Zhangjun, F.; Vrebalov, J.; Khayat, E.; Giovannoni, J.J.; Friedman, H. Banana MaMADS transcription factors are necessary for fruit ripening and molecular tools to promote shelf-life and food security. Plant Physiol. 2016, 171, 380–391. [Google Scholar] [PubMed]
- Qi, X.L.; Liu, C.L.; Song, L.L.; Li, M. PaMADS7, a MADS-box transcription factor, regulates sweet cherry fruit ripening and softening. Plant Sci. 2020, 301, 110634. [Google Scholar] [CrossRef]
- Yin, W.; Yu, X.; Chen, G.; Tang, B.; Wang, Y.; Liao, C.; Zhang, Y.; Hu, Z. Suppression of SlMBP15 inhibits plant vegetative growth and delays fruit ripening in tomato. Front. Plant Sci. 2018, 9, 938. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.R.; Cui, J.; He, Q.; Liu, Y.Z.; Lu, X.F.; Qi, J.; Xiong, J.; Yu, W.J.; Li, C.X. Genome-wide identification of HIPP and mechanism of SlHIPP4/7/9/21/26/32 mediated phytohormones response to Cd, osmotic, and salt stresses in tomato. Plant Physiol. 2024, 217, 109220. [Google Scholar] [CrossRef]
- Bi, Y.; Fu, H.; Jiang, Z.; Jiang, Y.; You, L.; Li, C.; Tu, X.; Ahmad, S.; Liu, Z.; Chen, S.; et al. Integrating Genome and Transcriptome-Wide Data to Explore the Expression Dynamics of ABCDE-like MADS-Box Genes in Phoebe bournei Floral Organs. Forests 2025, 16, 313. [Google Scholar] [CrossRef]
- Cui, J.; Xu, J.R.; Qi, J.; Lu, X.F.; Liu, Y.Z.; Xiong, J.; Yu, W.J.; Li, C.X. Genome-wide identification of SlIQMs and the regulatory effect of calcium on tomato seedlings under drought stress and phytohormone treatment. Plant Cell 2025, 44, 70. [Google Scholar]
- Zhou, E.; Zhang, Y.; Wang, H.; Jia, Z.; Wang, X.; Wen, J.; Shen, J.; Fu, T.; Yi, B. Identification and characterization of the MIKC-Type MADS-Box gene family in Brassica napus and its role in floral transition. Int. J. Mol. Sci. 2022, 23, 4289. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, J.; Hu, Z.; Guo, X.; Tian, S.; Chen, G. Genome-Wide analysis of the MADS-Box transcription factor family in Solanum lycopersicum. Int. J. Mol. Sci. 2019, 20, 2961. [Google Scholar] [CrossRef] [PubMed]
- Parenicová, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef]
- Ray, S.; Agarwal, P.; Arora, R.; Kapoor, S.; Tyagi, A.K. Expression analysis of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. Ssp. Indica). Mol. Genet. Genom. 2007, 278, 493–505. [Google Scholar] [CrossRef]
- Qu, Y.; Kong, W.; Wang, Q.; Fu, X. Genome-wide identification MIKC-Type MADS-Box gene family and their roles during development of floral buds in wheel wingnut (Cyclocarya paliurus). Int. J. Mol. Sci. 2021, 22, 10128. [Google Scholar] [CrossRef]
- Chen, S.P.; Sun, W.H.; Xiong, Y.F.; Jiang, Y.T.; Liu, X.D.; Liao, X.Y.; Zhang, D.Y.; Jiang, S.Z.; Li, Y.; Liu, B.; et al. The Phoebe genome sheds light on the evolution of magnoliids. Hortic. Res. 2020, 7, 146. [Google Scholar] [CrossRef]
- Chen, T.T.; Liu, H.; Li, Y.P.; Yao, X.H.; Qin, W.; Yan, X.; Wang, X.Y.; Peng, B.W.; Zhang, Y.J.; Shao, J.; et al. AaSEPALLATA1 integrates jasmonate and light-regulated glandular secretory trichome initiation in Artemisia annua. Plant Physiol. 2023, 192, 1483–1497. [Google Scholar] [CrossRef] [PubMed]
- Mou, Y.; Yuan, C.; Sun, Q.; Yan, C.; Zhao, X.; Wang, J.; Wang, Q.; Shan, S.; Li, C. MIKC-type MADS-box transcription factor gene family in peanut: Genome-wide characterization and expression analysis under abiotic stress. Front. Plant Sci. 2022, 13, 980933. [Google Scholar] [CrossRef] [PubMed]
- Martel, C.; Vrebalov, J.; Tafelmeyer, P.; Giovannoni, J.J. The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol. 2011, 157, 1568–1579. [Google Scholar] [CrossRef]
- Bemer, M.; Karlova, R.; Ballester, A.R.; Tikunov, Y.M.; Bovy, A.G.; Wolters-Arts, M.; de Barros Rossetto, P.; Angenent, G.C.; de Maagd, R.A. The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. The Plant Cell 2012, 24, 4437–4451. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.D.; Yuan, X.; Chen, S.; Fu, D.Q.; Jiang, C.Z. Metabolomic and transcriptomic analyses reveal that a MADS-Box transcription factor TDR4 regulates tomato fruit quality. Front. Plant Sci. 2019, 10, 792. [Google Scholar] [CrossRef]
- Gimenez, E.; Castañeda, L.; Pineda, B.; Pan, I.L.; Moreno, V.; Angosto, T.; Lozano, R. TOMATO AGAMOUS1 and ARLEQUIN/TOMATO AGAMOUS-LIKE1 MADS-box genes have redundant and divergent functions required for tomato reproductive development. Plant Mol. Biol. 2016, 91, 513–531. [Google Scholar] [CrossRef]
- Nakano, T.; Kimbara, J.; Fujisawa, M.; Kitagawa, M.; Ihashi, N.; Maeda, H.; Kasumi, T.; Ito, Y. MACROCALYX and JOINTLESS interact in the transcriptional regulation of tomato fruit abscission zone development. Plant Physiol. 2012, 58, 439–450. [Google Scholar] [CrossRef]
- Zheng, S.; He, J.J.; Lin, Z.S.; Zhu, Y.Y.; Sun, J.Y.; Li, L.G. Two MADS-box genes regulate vascular cambium activity and secondary growth by modulating auxin homeostasis in Populus. Plant Commun. 2020, 2, 100134. [Google Scholar] [CrossRef]
- Li, C.H.; Lei, C.Y.; Wang, K.T.; Tan, M.L.; Xu, F.; Wang, J.S.; Zheng, Y.H. MADS2 regulates priming defence in postharvest peach through combined salicylic acid and abscisic acid signaling. J. Exp. Bot. 2022, 73, 3787–3806. [Google Scholar] [CrossRef]
- Kou, X.; Zhao, Y.; Wu, C.; Jiang, B.; Zhang, Z.; Rathbun, J.R.; He, Y.; Xue, Z. SNAC4 and SNAC9 transcription factors show contrasting effects on tomato carotenoids biosynthesis and softening. Postharvest Biol. Technol. 2018, 144, 9–19. [Google Scholar] [CrossRef]
- Negi, S.; Bhakta, S.; Ganapathi, T.R.; Tak, H. MpSNAC67 transcription factor of banana regulates stress induced senescence through salicylic acid dependent pathway. Environ. Exp. Bot. 2023, 205, 105104. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
