Abstract
Dehydrins (DHNs; late embryogenesis-abundant D11 family) are a class of hydrophilic proteins involved in plant abiotic stress response. However, there is less information regarding DHN gene function in cucurbit crops. Herein, 34 DHN gene family members were identified and characterized in Cucumis sativus, Cucumis melo, Citrullus lanatus, Benincasa hispida, Lagenaria siceraria, and Cucurbita maxima. The DHN genes in the six cucurbit crops exhibited greater collinearity within subfamilies than between different subfamilies. Responses to stress (including low-temperature, salt, cadmium, and aluminum stress) varied among the DHN members, with a significant alteration in the expression of the acidic SnKn-type DHN gene CmDHN3 in response to aluminum stress. Subcellular localization analysis confirmed that CmDHN3 is expressed in the nucleus and cytoplasm. Virus-induced gene silencing (VIGS) revealed a remarkable decrease in CmDHN3 expression, which markedly increased malondialdehyde content, relative conductivity, and proline content in the roots and leaves of plants under aluminum stress. Transcriptome analysis showed that the decreased CmDHN3 expression reduced the expression of water channel protein-encoding genes. Interactions between CmDHN3 and CmAQP1 (MELO3C007188) and between CmDHN3 and CmAQP2 (MELO3C020774) were confirmed using yeast two-hybrid assays. These results clarify the pathway by which dehydrin genes are involved in the transcriptional-level response of melon to aluminum stress and provide a theoretical basis to comprehensively analyze the functions of this gene family in cucurbit crops.
1. Introduction
Dehydrins (DHNs) are a class of highly hydrophilic and intrinsically disordered proteins without distinct secondary or tertiary structures, which results in their functional diversity [1]. DHNs have crucial functions, particularly in defense against abiotic stress in higher plants. Previous reports have suggested that DHNs contribute to abiotic stress response by preventing protein aggregation or inactivation during dehydration by acting as protein-binding chaperones for cryoprotection and scavenging of free radicals [2,3,4]. However, along with their roles as molecular chaperones for protecting organellar structures from abiotic stress by stabilizing biological membranes and binding to reactive oxygen species (ROS) [5,6], DHNs participate in transcriptional regulation during stress response. They also indirectly participate in the modification of histones. During this epigenetic process, H3K4me3 modification shows a positive correlation with the expression of DHN and other drought response-related genes [7].
DHN proteins have evolutionarily conserved sequence motifs, including a lysine-rich (EKKGIMDKIKEKLPG) K segment located at the C-terminus, which forms an amphipathic α-helix structure that serves as the structural basis for the hydrophilicity of DHN [8,9]. Additionally, an S segment containing serine residues 3–13 can be phosphorylated by casein kinase 2 (CK2) and can enter the nucleus under the guidance of a signal peptide [10]. S segment deletion affects the nuclear localization of DHNs [11]. The Y segment (T/VDEYGNP) in the N-terminus is rich in tyrosine and typically contains 1–3 closely spaced copies. According to the number of motifs, DHN proteins are categorized as follows: YnSKn, YnKn, SKn, KnS, and Kn. YnSKn is the most common category; proteins in this category usually contain three or fewer conserved Y segments and several K segments, and they are neutral or alkaline. YnKn is a class of acidic DHNs containing Y and K segments, typically with no more than two segments each. Both SKn and KnS contain one S segment and two to three or one to two K segments and are mostly acidic. Most Kn proteins contain two to nine K segments and are acidic or neutral [11,12].
Different DHN proteins play varying roles in abiotic stress response by stabilizing cell membranes, scavenging ROS, binding metal ions, or acting as molecular chaperones. Silencing of the SKn-type DHN CaDHN1 in Capsicum annuum L. exacerbated wilting under low-temperature stress and remarkably elevated the content of malondialdehyde (MDA) and conductivity [13]. The acidic SKn-type MsDHN1 in Medicago sativa L. enhances tolerance to aluminum (Al) stress by increasing the exudation of oxalate from root tips through interactions with two aquaporin (AQP) water channel proteins. Moreover, an abscisic acid (ABA)-responsive element-binding transcription factor (MsABF2) may regulate the transcription of MsDHN1 [14]. YnSKn-type DHNs have a role in the response of plants to stress due to low temperature and salinity. Silencing of the CaDHN3 gene in Capsicum annuum L. reduces plant resistance to low-temperatures and saline stress [15]. CaDHN4 can slow down the damage caused by low-temperature and saline stress by reducing sensitivity to ABA [16]. CaDHN5 exerts a positive regulatory effect on the saline stress signaling pathway by affecting ROS accumulation [17]. Drought stress induces the expression of the Oryza sativa L. YnSKn-type DHN OsDhn-Rab16D, and overexpression of OsDhn-Rab16D maintains water balance and cellular homeostasis and improves the tolerance of plants to drought stress [18].
Under adverse conditions, some DHNs form complexes with regulatory factors and are involved in plant adaptation. OsDhnRab16D can interact with OsFKBP in the nucleus, and a novel FKBP- and YSK2-type DHN protein complex, OsFKBP-OsDhn-Rab16D, may be related to ABA signaling, thus enhancing the tolerance of Oryza sativa L. to drought [19]. Screening for freezing resistance genes in wild-type woodland strawberries revealed that the DHN protein Xero2 functions as a key marker of freezing tolerance and binds to CBF2 and CBF4 to influence freezing tolerance in plants [20].
Acidic soils are widely distributed globally, and Al stress is considered an important factor that restricts crop growth in acidic soils [21,22]. Al stress hinders plant growth mainly by inhibiting root elongation [23,24]. Skn-type DHNs may affect oxalic acid secretion from plant roots by controlling water channel protein expression, which enhances Al tolerance [14]. However, it remains unclear whether Skn-type DHNs in plants of the Cucurbitaceae family have similar functions. In the present study, DHN proteins in cucurbit crops were identified and compared, and the functions of melon Skn-type CmDHN3 under Al stress were explored. This study elucidates the regulatory mechanism of cucurbit DHN proteins in Al stress response and offers a theoretical basis for comprehensively analyzing the biological functions of DHNs in Cucurbitaceae.
2. Materials and Methods
2.1. Genome-Wide Identification of the Dehydrin Gene Family in Six Cucurbit Crops
Genome sequence data for six cucurbit crops were acquired from http://cucurbitgenomics.org. Furthermore, the DHN proteins of six Cucurbit crops (CsDHNs, CmDHNs, ClaDHNs, BhiDHNs, LsiDHNs, and CmaDHNs) were predicted with the HLH hidden Markov model (HMM) profile available in the Pfam database (http://pfam.xfam.org, PF00257).
Multiple sequence alignments of the full-length DHN domains of Arabidopsis (AtDHN), Solanum lycopersicum (SlDHN), Cucumis sativus (CsDHN), Cucumis melo (CmDHN), Citrullus lanatus (ClaDHN), Benincasa hispida (BhiDHN), Lagenaria siceraria (LsiDHN), and Cucurbita maxima (CmaDHN) were generated with ClustalW by utilizing default parameters [25]. By using multiple sequence alignments for all six cucurbit crops, Arabidopsis thaliana, and Solanum lycopersicum, we constructed a Neighbor-Joining (NJ) tree with MEGAv 7.0 and evaluated it through bootstrapping [26].
The conserved motifs in the protein sequences of candidate DHNs were analyzed using MEME 5.1.1 online software (Multiple EM for Motif Elicitation—http://memesuite.org/tools/meme) [27]. The GFF/GTF annotation files of cucurbit crop genomes (http://cucurbitgenomics.org) were utilized for examining the exon–intron organization and splicing phase of the predicted DHNs; the Gene Structure Display Server (GSDS—http://gsds.cbi.pku.edu.cn) was employed to visualize the results [28].
The Cucurbit crop genome sequences and GFF annotation files (FASTA suffix files) were retrieved from http://cucurbitgenomics.org. To perform synteny analysis, we obtained the gene duplication landscape by using MCScanX [29]. Next, we utilized TBtools(11 January 2024) to generate and visualize a syntenic map. Connector lines were used to highlight the putative duplicated genes.
2.2. Plant Materials and Stress Treatments
Cucumber (Cucumis sativus L. cv. Chinese long No. 9930), melon (Cucumis melo L. cv. balixiang), watermelon (Citrullus lanatus Thunb. cv. Qilin), wax gourd (Benincase hispida L. cv. dafenpi), pumpkin (Cucurbita maxima L. cv. baby), and bottle gourd (Lagenaria siceraria) cultivars were obtained from the Liaoning Provincial Key Laboratory of Biological System Evolution and Agricultural Ecology, College of Life Sciences, Shenyang Normal University, Shenyang, China. Plant tissues, including roots, stems, leaves, flowers, and fruits, were rapidly frozen in liquid nitrogen and stored at −80 °C until RNA extraction.
The Cucumis melo seedlings were cultured in Hoagland solution, and almost two-week-old seedlings (two true leaves fully unfolded) were used for abiotic stress treatment. The effects of cold stress (4 °C), salt stress (100 mmol/L NaCl), cadmium stress (500 μmol/L CdCl2), and Al stress (500 μmol/L AlCl3) were evaluated. Root and leaf collection was performed at 0, 12, 24, and 48 h for cold-, NaCl-, cadmium-, and Al-treated seedlings. All samples were stored at −80 °C by freezing them in liquid nitrogen before total RNA extraction.
2.3. Silencing of the CmDHN3 Gene in Melon
Cucumis melo L. cultivar “M1-23” was used for gene silencing. pTRV2:CmDHN3 and pTRV2:CmPDS were generated using two virus-induced gene silencing (VIGS) techniques, in accordance with a previously documented procedure [30]. Briefly, two 300 bp segments from CmDHN3 and CmPDS were inserted into a pTRV2 vector to develop the recombinant plasmids pTRV2:CmDHN3 and pTRV2:CmPDS, respectively. Subsequently, the freeze–thaw method was utilized for transforming the plasmids pTRV2:00 (negative control), pTRV2:CmDHN3, and pTRV2:CmPDS (positive control) in the Agrobacterium tumefaciens GV3101 strain. All vectors were injected into melon cotyledons after GV3101 was combined with pTRV1, and the plants were cultured as reported earlier [30].
2.4. Gene Expression Analysis
Total RNA was isolated from tissues by using TRIzol reagent (Takara, Kusatsu, Japan). Real-time quantitative PCR (qRT-PCR) and data analyses were conducted by following previously described methods [31]. We employed the comparative ΔΔCt method to calculate relative gene expression levels, using untreated samples or wild-type samples as the control group and 18SrRNA as the reference gene for normalization. Primer3 (http://frodo.wi.mit.edu/) was utilized for generating primers. All primers are shown in Table S1. A heatmap of expression profiles was created using the gene expression heatmap package in R.
After 2 weeks of Al stress, the roots of transgenic plants and control plants were subjected to total RNA extraction by using an Ultrapure RNA Kit (Kangwei, Beijing, China). RNA from each of the three biological replicates was reverse transcribed into cDNA by utilizing random primers. The obtained cDNA was sequenced at the Beijing Genomics Institute (BGI) (Guangzhou, China) with the Illumina HiSeq 2000 instrument. The raw reads were filtered using TopHat2 (version 2.0.3.12) to exclude adapters and low-quality reads, followed by alignment to the genome sequences (http://melon-genome-hub.southgreen.fr/) [32].
2.5. Malondialdehyde (MDA) Content, Proline Content, and Relative Electrolyte Leakage
Approximately 0.5 g of melon roots and leaves were used for determining MDA and proline contents and relative electrolyte leakage. MDA content was estimated by the thiobarbituric acid method as reported earlier [33]. The proline content was measured to assess membrane permeability based on a previously described method [34]. Relative electrolyte leakage was estimated using a previously described method [35]. All measurements were performed in triplicate (biological replicates).
2.6. Subcellular Localization
The positive control plasmids pBI221-GFP and pBI221-CmDHN3-GFP were mixed with gold powder after cleaning and disinfection. Gene gun technology was utilized to inject this gold powder into Arabidopsis protoplasts. The protoplasts were then cultured at 28 °C for 8–12 h in the dark. Confocal laser microscopy was used to observe green fluorescence [36].
2.7. Yeast Two-Hybrid Validation Assay
By using the yeast two-hybrid (Y2H) antibody optimization system (Y2H-AOS) constructed by ProNet Biotech (Nanjing, China), we simulated and analyzed the binding mode between antibodies and antigens and constructed a three-dimensional structural interaction model for CmDHN3-CmAQP1/CmAQP2 and CmDHN3-CmZIP proteins. To conduct the Y2H assay, CmDHN3, CmAQP1, or CmZIP coding sequences were amplified and cloned into the pGADT7 or pGBKT7 vector (Clontech, San Francisco, CA, USA). Y2H assays were performed using the Yeastmaker Yeast Transformation System 2 (Clontech).
2.8. Statistical Analysis
The experimental data were organized using Microsoft Excel 2010, and the physiological measurements were subjected to one-way ANOVA followed by Duncan’s multiple range test (p < 0.05) using SPSS 22.0 software. Data visualization was performed using Origin 8.5 (Microcal Software Inc., Northampton, MA, USA).
3. Results
3.1. DHN Gene Family Member Identification in Cucurbit Genomes
Thirty-four candidate DHN genes were identified by searching the whole genomes of six Cucurbit crops using the hmmsearch command, based on the HMM of the DHN (PF00257). The protein sequences of all candidate genes contained the conserved DHN structural domain (Figure S1). Therefore, 34 DHN members were identified in six Cucurbit crops, including four members in Cucumis sativus, Citrullus lanatus, Cucumis melo, and Cucurbita maxima; five members in Lagenaria siceraria; and thirteen members in Benincasa hispida.
3.2. Analyses of Phylogenetic Relationships, Conserved Structural Domains, and Physicochemical Properties
A phylogenetic tree was constructed with the protein sequences of thirty-four DHN members from Cucurbit crops, seven DHN members from A. thaliana, and four DHN members from Solanum lycopersicum. The DHN gene family of Cucurbit crops is classified into three DHN subfamilies: class I, class II, and class III; this is based on the number of K segments. Each subfamily contains varying numbers of DHN members, including YnSKn, YnKn, SnKn, YnKn, and Kn (Figure 1). The A. thaliana genome contains only one member of the subfamily YnSKn. Two members were detected in watermelons, cucumbers, and melons, and three members were detected in Cucurbita maxima, Lagenaria siceraria, and Benincasa hispida. In the subfamily YnKn, we detected one member in Lagenaria siceraria, Cucumis sativus, and Benincasa hispida and three members in Solanum lycopersicum. Two and eight members of the subfamily Kn were detected in A. thaliana and Benincasa hispida, respectively. In the SnKn subfamily, we detected one member in each of the six cucurbit crops and Solanum lycopersicum and four members in A. thaliana. Thus, in comparison with A. thaliana, the YnSKn-type subfamily in cucurbit crops expanded to a certain extent during evolution, whereas the Kn-type DHNs in Benincasa hispida expanded to a certain extent. Physicochemical analysis (Table 1) showed that the number of amino acids in the DHN family members of Cucurbit crops was 49–714, and the isoelectric points (pIs) ranged from 4.95 to 10.27. The number of amino acid residues varied substantially among the DHN members of different subfamilies.
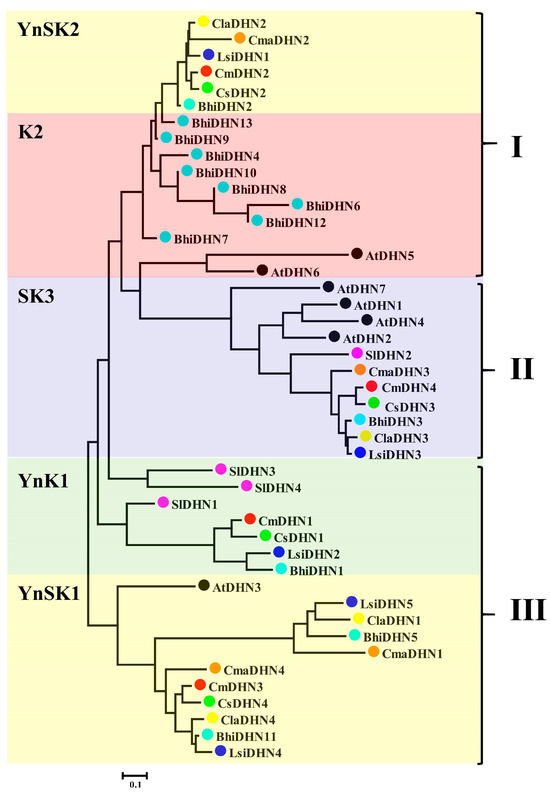
Figure 1.
The phylogenetic tree of Cucumis sativus, watermelon, melon, wax gourd, bottle gourd, pumpkin, and Arabidopsis and Solanum lycopersicum DHN proteins was constructed from a complete alignment of DHN proteins with MEGAv 7.0 using the Neighbor-Joining (NJ) method with 1000 bootstrap replicates. The groups are marked by colorful backgrounds. The protein names and corresponding accession numbers used to build the phylogenetic tree are listed in Table 1.

Table 1.
Physicochemical properties of DHN gene family members in Cucurbitaceae crops.
3.3. Analyses of Gene Structure and Conserved Motifs
To determine the evolutionary relationships among the DHN gene family members in Cucurbit crops, the number and arrangement of exons and introns in 34 DHN members of the Cucurbitaceae family were analyzed. The number of exons and introns, as well as their arrangement, differed significantly among the subfamilies, whereas the intron–exon arrangement of DHN members within each subfamily was generally conserved (Figure 2).
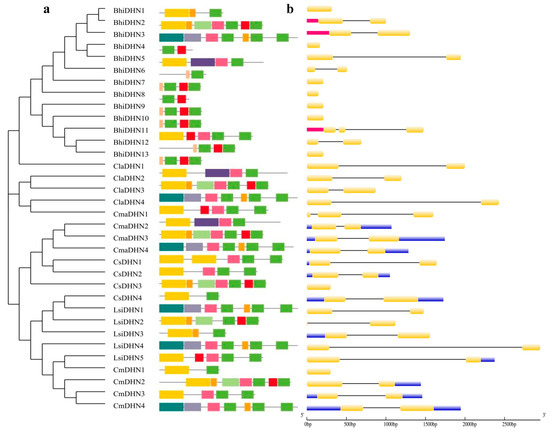
Figure 2.
(a) Conserved motifs of DHN proteins in Cucurbitaceae crops; (b) exon–intron structures. Light blue boxes, yellow boxes, and black horizontal lines indicate exons, UTRs, and introns, respectively.
By using the MEME tool, 15 conserved motifs were identified in the DHN members of Cucurbit crops. Each DHN member contains approximately five to nine conserved motifs. The number and distribution of the conserved motifs were generally conserved within the same subfamily (Figure 2), suggesting that DHNs of the same subfamily perform identical functions. Conserved motif distribution varied among DHN members in different subfamilies, which may contribute to functional differences.
3.4. Evolutionary Relationships of DHN Gene Family Members in Cucurbitaceae
To determine the evolutionary history of the DHN gene family within the Cucurbitaceae family, we conducted a pairwise collinearity analysis based on the phylogenetic tree. The closer the relationship, the higher the number of collinear pairs of DHN. Three collinear pairs were observed between Cucumis sativus and Cucumis melo, Cucumis melo and Cucurbita maxima, and Citrullus lanatus and Lagenaria siceraria, while two collinear pairs were observed between Cucurbita maxima and Cucumis melo. A certain degree of collinearity was observed among members of the same DHN subfamily in different Cucurbit crops. The absence of collinearity between different DHN subfamilies suggests that an ancient duplication event occurred in the ancestral species of Cucurbitaceae, resulting in different types of DHNs (Figure 3).
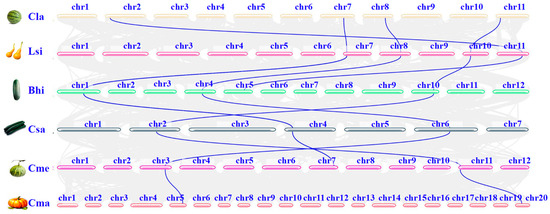
Figure 3.
Synteny analysis of DHN genes among the six cucurbit crops. Light blue lines display the collinear DHN genes among the six Cucurbit crop genomes. The light blue lines denote collinear blocks.
3.5. Expression Patterns of DHN Gene Family Members in Different Plant Tissues
To determine the potential functions of DHN family members in Cucurbitaceae, DHNs of six common Cucurbit species were analyzed for their expression levels in the roots, stems, leaves, flowers, and fruits under normal physiological conditions. The different DHN genes showed remarkable variations in their expression patterns. Overall, the DHN genes were constitutively expressed in all tissues. High expression of most genes was observed in fruits. However, CmDHN3 and CmDHN4 expression levels in the vegetative organs (roots, stems, and leaves) were markedly higher than those in the reproductive organs (fruits and seeds). CmDHN2 showed high expression in all tissues, except the roots (Figure 4).
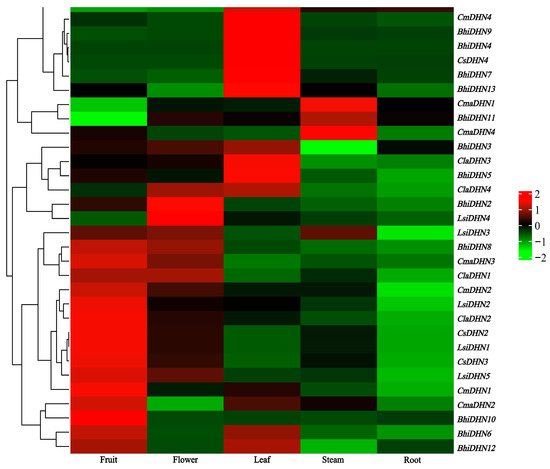
Figure 4.
The DHN gene expression heatmap of the six cucurbit crops in different tissues.
3.6. Expression Patterns of CmDHN Gene Family Members in Abiotic Stress Response
To determine whether Cucumis melo DHNs are involved in stress response, the expression levels of all DHN genes in Cucumis melo roots and leaves under various stresses were analyzed (Figure 5). The mRNA levels also exhibited variations under Cd stress. CmDHN1 was the most affected gene, with a 16-fold increase in the number of roots, whereas CmDHN3 expression was altered to a lesser extent. Under salt stress, the CmDHN1 and CmDHN2 expression levels in the roots increased 2000-fold and 2100-fold, respectively, whereas CmDHN1 and CmDHN2 expression levels in the leaves increased by more than 38-fold and 16-fold, respectively. Under Al stress, CmDHN3 was upregulated 22-fold in the roots and 4.8-fold in the leaves, respectively, after 48 h. The mRNA levels increased following low-temperature treatment (4 °C) and increased with prolonged low-temperature stress. At 48 h, CmDHN1 and CmDHN2 levels were 1300-fold and 2100-fold higher than those in the control roots, respectively, and 1800-fold and 2600-fold higher in the leaves, respectively.
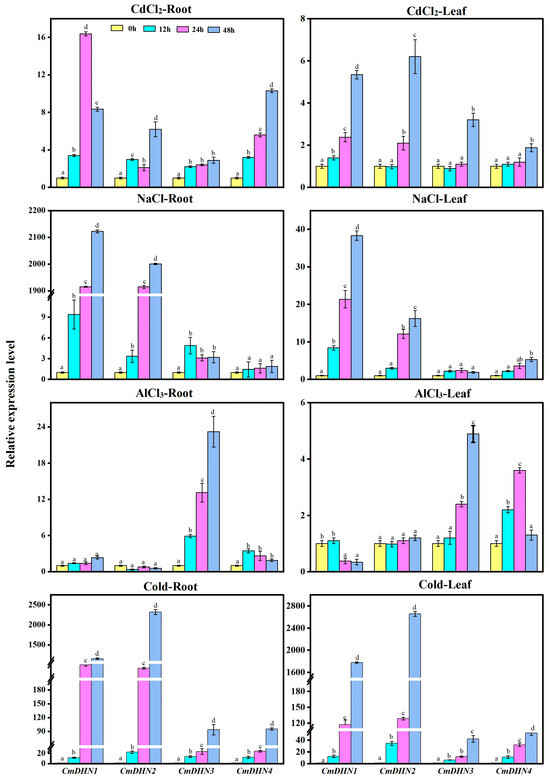
Figure 5.
qRT-PCR analysis of CmDHN expression in the roots and leaves of oriental melon plants following abiotic stresses. The value for each sample is the mean ± standard error (SE), replicated thrice. The expression levels are relative to 0 h. Different lowercase letter (a–d) indicate significant differences (Student’s test, p < 0.05).
3.7. Subcellular Localization of CmDHN3 in Cucumis melo
Gene function is often closely associated with the location of protein expression in cells. Therefore, these functions can be evaluated through subcellular localization analysis. To preliminarily investigate the location of CmDHN3 expression in Cucumis melo cells, a subcellular localization vector, pBI221-CmDHN3-GFP (Figure 6), was constructed using pBI221-GFP as a control. The localization and control vectors were jointly transformed into protoplasts of A. thaliana by using the gene gun technique. After gene expression, the CmDHN3 expression site was identified based on the green fluorescent protein signal (Figure 6). CmDHN3 is expressed in both the nucleus and the cytoplasm.
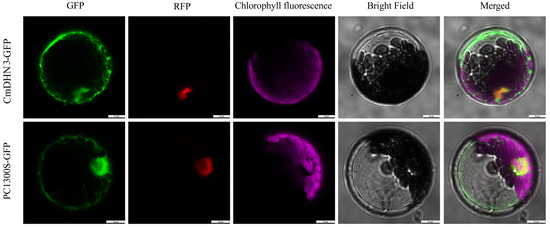
Figure 6.
The subcellular localization of Cucumis melo CmDHN3 in Arabidopsis leaf protoplasts.
3.8. Effect of Cucumis melo CmDHN3 on Tolerance to Al Stress
VIGS is widely used to determine gene function. In a previous study, the authors utilized VIGS to determine the function of CmDHN3 in Al stress response in Cucumis melo [28]. An approximately 300-base-pair (bp) sequence with high specificity for the CmDHN3 gene was selected and inserted into the pTRV2 vector to prepare the recombinant plasmid pTRV2-CmDHN3 (using pTRV2 as a negative control). pTRV2-CmPDS silences the CmPDS gene and induces leaf albinism (a phenotype observed in approximately 70% of plants after gene silencing). This gene is frequently used as a positive control for gene silencing. Two true leaves after induction by Nicotiana tabacum L. rattle virus (TRV)-mediated gene silencing, Cucumis melo plants injected with pTRV2-CmDHN3, and the control plants displayed no visible phenotypic differences. However, the CmPDS-silenced plants showed significant leaf albinism (Figure 7a). After two weeks of Al stress treatment, pTRV2-CmDHN3 plants and negative control plants exhibited remarkable differences in their phenotypes. In particular, CmDHN3-silenced plants showed more prominent growth inhibition and significant suppression of root system growth (Figure 7b). Additionally, under normal growth conditions, CmDHN3 expression in the roots and new leaves of gene-silenced melon plants was markedly lower than that in control plants, with no remarkable alterations in the expression of the remaining DHN genes; this finding suggests that CmDHN3 silencing using VIGS was effective (Figure 7c).
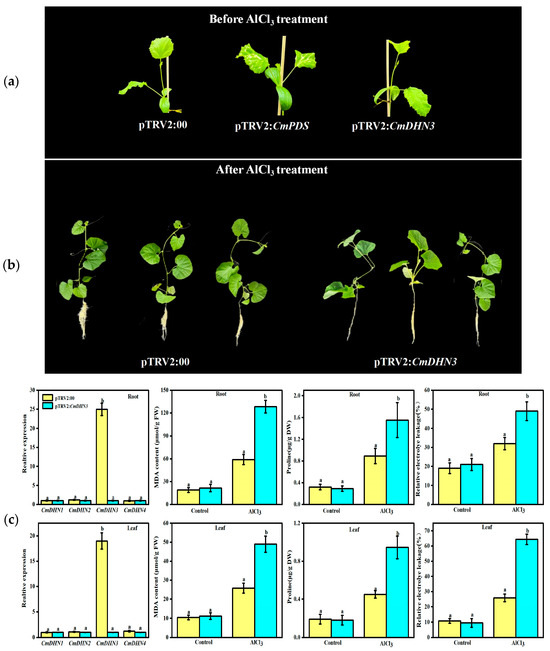
Figure 7.
Effects of Al stress on plant phenotypes. (a) The phenotype empty vector (negative control), CmPDS-silenced (albino gene–positive control), and CmDHN3-silenced Cucumis melo plants when growing to two true leaves; (b) the phenotypes of the negative control and CmDHN3-silenced Cucumis melo plants under Al stress on day 14; (c) expression levels of the DHN gene, MDA content, proline levels, and relative electrolytic leakage in the roots and leaves of CmDHN3-silenced melon plants. Data that are significantly different are indicated with letters above the error bars (±S.D.).
Physiological parameters were measured under Al stress. In normal physiological conditions, the control and silenced plants did not differ significantly in their MDA content. However, after AlCl3 treatment, both plants showed a remarkable increase in MDA levels. This increase in MDA content was 2.3-fold and 1.9-fold greater in the roots and leaves of silenced plants than in the roots and leaves of control plants. Following AlCl3 treatment, the proline content increased by 1.4-fold and 2.1-fold in the roots and leaves of the silenced Cucumis melo plants, respectively, in comparison with that in the control plants. The relative conductivity values were 1.3-fold and 2.5-fold greater in the roots and leaves of the silenced plants, respectively, than those in the control plants (Figure 7c).
Following Al stress for two weeks, the root tips of the silenced and control plants were subjected to transcriptomic analyses. A total of 1284 differentially expressed genes (DEGs) were identified; of these genes, 575 were downregulated and 708 were upregulated under Al stress (Figure 8a). According to the KEGG enrichment analysis, the DEGs were enriched in various metabolic pathways, including mitogen-activated protein kinase (MAPK) signaling, hormone signaling, and glutathione metabolism (Figure 8b). Moreover, various genes, such as CmAQP1, CmAQP2, and CmZIP, were significantly downregulated in plants with silenced CmDHN3 (Figure 8c,d).
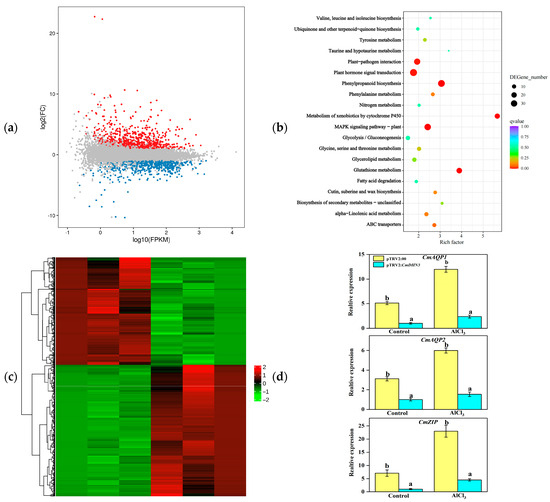
Figure 8.
Overview of CmDHN3-silenced Cucumis melo plants transcriptomes of roots under Al stress on day 14. (a) Volcano diagram of gene expression. Red represents up-regulated genes, and blue represents down-regulated genes.(b) KEGG enrichment of genes. (c) Differential gene expression heatmap. (d) Expression levels of CmAQP1, CmAQP2, and CmZIP genes in the roots of CmDHN3-silenced Cucumis melo plants. The lowercase letters denote statistically significant differences (p < 0.05).
Three-dimensional protein structural interaction models of CmDHN3 with CmZIP, CmDHN3 with CmAQP1, and CmDHN3 with CmAQP2 were constructed using the Y2H-AOS three-dimensional protein structural simulation technique developed by Nanjing ProNet Biotech Co., Ltd. (Figure 9a). The confidence levels of these models were as high as 0.91, 0.97, and 0.80, respectively, thus indicating interactions between each group of proteins. Y2H assays further showed that CmDHN3 and CmZIP (MELO3C03794), CmDHN3 and CmAQP1 (MELO3C007188), and CmDHN3 and CmAQP2 (MELO3C020774) exhibited growth on the deficient culture medium, indicating the presence of interactions within all three groups of proteins (Figure 9b).
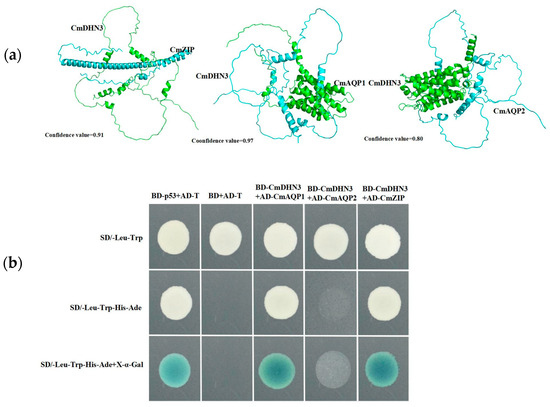
Figure 9.
Validation of the protein that interacts with CmDHN3. (a) A three-dimensional model of the interaction protein between CmDHN3 vs. CmAQP1, CmDHN3 vs. CmAQP2, and CmDHN3 vs. CmZIP; (b) Interaction between CmDHN3 and CmAQP1, CmAQP2, and CmZIP determined in a yeast-two-hybrid (Y2H) assay. CmDHN3 was inserted into pGBKT7 and CmAQP1; CmAQP2 and CmZIP were inserted into pGADT7. Different co-transformed yeast cells were dropped onto synthetic dropout medium (SD/-Trp/-Leu/-Ade/-His) containing 20 mg mL−1 X-a-gal.
4. Discussion
DHNs have pivotal roles in plant responses to different abiotic stressors [2,37,38]. The functions of DHN family members in many plants have been reported, such as A. thaliana [39], Solanum lycopersicum [40], barley [41], Oryza sativa L. [42], and chili pepper [13]. However, few studies have evaluated the functions of DHNs in Cucurbit crops under abiotic stress; to date, only four DHN members are known in Cucurbits [43]. In the study presented herein, we identified 34 DHN family members in cucurbit crops on the basis of the Cucurbit genome [44] (Table 1).
DHN protein sequences in the family Cucurbitaceae are disordered, and DHNs are highly hydrophilic but show significant variations in pI, kinases, and a number of conserved motifs. The pI values of YnSKn- and SKn-type DHNs are higher than those of SKn-type DHNs, suggesting that YnSKn-type DHNs bind more easily to negatively charged membranes [9]. Furthermore, DHN phosphorylation may be involved in the functional regulation of plant cells under stress and in the membrane binding of DHNs43. SKn-type DHNs of the Cucurbitaceae family contain more CK2 phosphorylation sites than YnSKn-type DHNs (Table 1). These findings agree with those of studies on DHN gene families in Capsicum annuum and Vitis vinifera, thus showing that YnSKn-type DHNs and SKn-type DHNs are primarily phosphorylated by PKC and CK2, respectively [45,46,47,48].
We constructed a phylogenetic tree based on the DHN proteins of Cucumis sativus, Cucumis melo, Citrullus lanatus, Benincasa hispida, Lagenaria siceraria, Cucurbita maxima, A. thaliana, and Solanum lycopersicum (Figure 1 and Figure 2). The Cucurbit crops had a similar number of DHN members (4–5), except for the 13 DHN members in the Lagenaria siceraria. However, the DHN families of Cucumis sativus, Cucurbita maxima, Citrullus lanatus, Cucumis melo, and Benincasa hispida included only five YnSKn-, YnKn-, and SKn-type DHNs, whereas Cucurbita maxima contained only Kn-type DHNs and lacked KS-type DHNs, suggesting that these DHN types were lost in Cucurbit crops. Moreover, in the phylogenetic tree, YnSK2-type DHNs constituted the largest fraction of total DHN members, accounting for a proportion of >37.78% (Figure 3). YnSK2-type DHN proteins are among the most abundant neutral and basic proteins found in nature [38]. DHN genes in Cucurbit crops showed largely distinct expression patterns, despite some overlaps (Figure 4). Rorat (2006) [38] showed that DHNs were expressed in almost all tissues under normal physiological conditions. Cucurbit DHN genes are also expressed in all tissues, suggesting that they participate in crop growth and development.
DHN genes are present in various plants of a multigene family and have been extensively investigated. As suggested previously, SKn- and Kn-type proteins are primarily involved in protection against cold stress, whereas YnSKn-type DHNs are primarily involved in plant adaptation to ABA, desiccation, and salt stresses [4]. Different expression patterns have been observed for different types of DHN family members. In Capsicum annuum, most DHN genes (including SKn and YnSKn) are upregulated under low temperature, NaCl, and mannitol stress. In particular, the SKn-type CaDHN7 is highly upregulated, suggesting its critical function in response to abiotic stress [14]. YnSKn-type DHNs in Hordeum vulgare L. were almost entirely induced by ABA and drought stresses but not by low temperatures. In contrast, low temperatures upregulated the expression of DHN5 (K9) and DHN8 (Sk3). Furthermore, low temperatures induce DHN gene expression in A. thaliana and Triticum aestivum L. [49,50]. In Cucumis melo, almost all DHN genes displayed upregulated expression in stress due to low temperature, NaCl, and CdCl2. In particular, CmDHN1 (YnKn-type) and CmDHN2 (YnSKn-type) were highly upregulated, which suggests their involvement in abiotic stress response. Notably, both YnSKn-type CmDHN3 and SKn-type CmDHN4 were upregulated under AlCl3 stress, especially CmDHN3, which was upregulated to a greater extent, suggesting that CmDHN3 has a crucial function in Al stress response (Figure 5).
In this study, CmDHN3 silencing significantly reduced Al tolerance in Cucumis melo and led to substantial MDA and proline accumulation in the roots and leaves, as well as a significant elevation in relative conductivity (Figure 7). Transcriptome analysis showed that CmDHN3 might contribute to Al tolerance by affecting the expression levels of Al stress-related genes, such as aquaporin (AQP) and aluminum-activated malate transporter (ALMT). The promoter of CmDHN3 had an ABA-binding site, and CmDHN3 expression levels showed a significant correlation with CmZIP levels (Figure 8). Therefore, interactions between these genes and CmDHN3 were predicted using the yeast two-hybrid antibody optimization system (Y2H-AOS). Finally, Y2H assays confirmed that CmDHN3, CmZIP, CmDHN3, and CmAQP1, as well as CmDHN3 and CmAQP2, interacted to varying degrees (Figure 9). These findings suggest that CmDHN1 interacts with two AQP water channel proteins and is regulated by the ABA response factor CmZIP at the transcriptional level. The acidic SnKn-type MsDHN1 in Medicago sativa increases the secretion of oxalic acid from the root tips to enhance plant tolerance to Al stress [14]. Whether CmDHN3, which is also an acidic SnKn-type DHN, can increase the oxalic acid content in root tips to improve Al tolerance in plants requires further investigation. These findings suggest that SnKn-type DHNs have similar regulatory pathways in the Al stress response mechanism of plants.
5. Conclusions
We identified 34 DHN genes in the Cucurbitaceae genome database; these genes showed differential expression in various tissues. The DHN gene family members in melons exhibited varying expression patterns in response to different stresses. Al stress induces acidic SnKn-type DHN CmDHN3 gene expression, and silencing of the CmDHN3 gene reduces Al stress tolerance in melon. The knockout of the CmDHN3 gene also suppressed the expression of the water channel proteins CmAQP1 and CmAQP2, as well as the ABA signaling factor CmZIP. The Y2H assay verified the interactions between these groups of proteins. This study offers a theoretical basis for revealing the mechanism through which acidic SnKn-type DHN contributes to the response to Al stress in melons.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae11050480/s1. Figure S1: Protein sequence alignment of DHN protein sequences from six Cucurbitaceae crops. Y, S, and K segments are boxed in pink, yellow, and blue, respectively; Table S1: The primers used in the study.
Author Contributions
C.Z.: Conceptualization, methodology, software, data curation, and writing—original draft; Q.C.: Analysis and interpretation of the data; X.G.: Conceptualization and methodology; H.P.: Funding; Y.Z.: Visualization, funding, investigation, supervision, and writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Natural Science Foundation of Liaoning Province (No. 2024-MS-112), the Liaoning Province Science and Technology Plan Project (No. 2022JH2/101300179), the Liaoning Basic Scientific Research Project (No. LJ232410166068; No. LJ232410166084), and the Special Fund for Innovation Team Support Plan under the Basic Research Business Expenses for Provincial Universities in Liaoning (No. LJ202410166052).
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Materials; further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Smith, M.A.; Graether, S.P. The disordered dehydrin and its role in plant protection: A biochemical perspective. Biomolecules 2022, 12, 294. [Google Scholar] [CrossRef] [PubMed]
- Riyazuddin, R.; Nisha, N.; Singh, K.; Verma, R.; Gupta, R. Involvement of dehydrin proteins in mitigating the negative effects of drought stress in plants. Plant Cell Rep. 2022, 41, 519–533. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Reyes, J.L.; Wei, H.; Yang, Y.; Karlson, D.; Covarrubias, A.A.; Krebs, S.L.; Fessehaie, A.; Arora, R. RcDhn5, a cold acclimation-responsive dehydrin from Rhododendron catawbiense rescues enzyme activity from dehydration effects in vitro and enhances freezing tolerance in RcDhn5-overexpressing Arabidopsis plants. Physiol. Plant. 2008, 134, 583–597. [Google Scholar] [CrossRef]
- Cheng, Y.; He, J.; Feng, Y.; Zhao, J.; Guan, J. Low temperature conditioning reduced the chilling injury by regulating expression of the dehydrin genes in postharvest huangguan pear (pyrus bretschneideri rehd. Cv. Huangguan). Horticulturae 2022, 8, 1022. [Google Scholar] [CrossRef]
- Szlachtowska, Z.; Rurek, M. Plant dehydrins and dehydrin-like proteins: Characterization and participation in abiotic stress responses. Front. Plant Sci. 2023, 14, 1197469. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.P.; Liu, Y.; Kang, Y.; Liu, W.; Li, S.; Wang, Z.H.; Xia, X.Y.; Chen, X.Y.; Qian, L.W.; Xiong, X.H.; et al. Genome-wide characterization of LEA gene family reveals a positive role of BnaA.LEA6.a in freezing tolerance in rapeseed (Brassica napus L.). BMC Plant Biol. 2024, 24, 433. [Google Scholar] [CrossRef]
- Tiwari, P.; Chakrabarty, D. Dehydrin in the past four decades: From chaperones to transcription co-regulators in regulating abiotic stress responses. Curr. Res. Biotechnol. 2021, 3, 249–259. [Google Scholar] [CrossRef]
- Liu, Q.; Zheng, L.; Wang, Y.; Zhou, Y.; Gao, F. AmDHN4, a winter accumulated SKn-type dehydrin from Ammopiptanthus mongolicus, and regulated by AmWRKY45, enhances the tolerance of Arabidopsis to low temperature and osmotic stress. Int. J. Biol. Macromol. 2024, 266, 131020. [Google Scholar] [CrossRef]
- Koag, M.C.; Wilkens, S.; Fenton, R.D.; Resnik, J.; Vo, E.; Close, T.J. The K-segment of maize DHN1 mediates its binding to anionic phospholipid vesicles and the concomitant structural changes. Plant Physiol. 2009, 150, 1503–1514. [Google Scholar] [CrossRef]
- Alsheikh, M.K.; Svensson, J.T.; Randall, S.K. Phosphorylation-regulated ion binding is a property shared by the acidic subclass of dehydrins. Plant Cell Environ. 2005, 28, 1114–1122. [Google Scholar] [CrossRef]
- Hernández-Sánchez, I.E.; Maruri-López, I.; Ferrando, A.; Carbonell, J.; Graether, S.P.; Jiménez-Bremont, J.F. Nuclear localization of the dehydrin opsdhn1 is determined by a histidine-rich motif. Front. Plant Sci. 2015, 6, 702. [Google Scholar] [CrossRef]
- Jia, J.S.; Ge, N.; Wang, Q.Y.; Zhao, L.T.; Chen, C.; Chen, J.W. Genome-wide identification and characterization of LEA gene family members in Panax notoginseng and their transcriptional responses to dehydration in recalcitrant seeds. BMC Genom. 2023, 24, 126. [Google Scholar] [CrossRef]
- Chen, R.G.; Jing, H.; Guo, W.L.; Wang, S.B.; Ma, F.; Pan, B.G.; Gong, Z.H. Silencing of dehydrin CaDHN1 diminishes tolerance to multiple abiotic stresses in Capsicum annuum L. Plant Cell Rep. 2015, 34, 2189–2200. [Google Scholar] [CrossRef] [PubMed]
- Lv, A.M.; Wen, W.W.; Fan, N.N.; Su, L.T.; Peng, Z.; Yuan, A. Dehydrin MsDHN1 improved aluminum tolerance in alfalfa (Medicago sativa L.) by affecting oxalate exudation from root tips. Plant J. 2021, 108, 441–458. [Google Scholar] [CrossRef]
- Jing, H.; Li, C.; Ma, F.; Ma, J.H.; Khan, A.; Wang, X.; Zhao, L.Y.; Gong, Z.H.; Chen, R.G. Genome-wide identification, expression divergence of the dehydrin gene family, and characterization of CaDHN3 in pepper (Capsicum annuum L.). PLoS ONE 2016, 11, e0161073. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.F.; Liu, S.Y.; Ma, J.H.; Wang, X.K.; Haq, S.U.; Meng, Y.C.; Zhang, Y.M.; Chen, R.G. CaDHN4, a salt and cold stress-responsive dehydrin gene from pepper, decreases abscisic acid sensitivity in Arabidopsis. Int. J. Mol. Sci. 2019, 21, 26. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Hou, X.M.; Zhang, Y.M.; Meng, Y.C.; Zhang, H.F.; Liu, S.Y.; Wang, X.K.; Chen, R.G. CaDHN5, a pepper dehydrin gene, plays an important role in salt and osmotic stress responses. Int. J. Mol. Sci. 2019, 20, 1989. [Google Scholar] [CrossRef]
- Verma, G.; Dhar, Y.V.; Srivastava, D.; Kidwai, M.; Chauhan, P.S.; Bag, S.K.; Asif, M.H.; Chakrabarty, D. Genome-wide analysis of the rice dehydrin gene family: Evolutionary conservation and expression patterns in response to PEG-induced dehydration stress. PLoS ONE 2017, 12, e0176399. [Google Scholar] [CrossRef]
- Tiwari, P.; Indoliya, Y.; Singh, P.K.; Chauhan, P.S.; Pande, V.; Chakrabarty, D. Role of the dehydrin-FK506-binding protein complex in enhancing drought tolerance via the ABA-mediated signaling pathway. Environ. Exp. Bot. 2019, 158, 136–149. [Google Scholar] [CrossRef]
- Kanbar, A.; Weinert, C.H.; Kottutz, D.; Thinh, L.; Abuslima, E.; Kabil, F.; Hazman, M.; Egert, B.; Trierweiler, B.; Kulling, S.E.; et al. Cold tolerance of woodland strawberry (Fragaria vesca) is linked to Cold Box Factor 4 and the dehydrin Xero2. J. Exp. Bot. 2024, 75, 5857–5879. [Google Scholar] [CrossRef]
- Chauhan, D.K.; Vaishali, Y.; Marek, V.; Walter, G.; Sharon, P.; Namira, A.; Vijay, P.S.; Rupesh, D.; Shivendra, S.; Durgesh, K.T. Aluminum toxicity and aluminum stress-induced physiological tolerance responses in higher plants. Crit. Rev. Biotechnol. 2021, 41, 715–730. [Google Scholar] [CrossRef] [PubMed]
- Hajiboland, R.; Panda, C.K.; Lastochkina, O.; Gavassi, M.A.; Habermann, G.; Pereira, J.F. Aluminum toxicity in plants: Present and future. J. Plant Growth Regul. 2023, 42, 3967–3999. [Google Scholar] [CrossRef]
- Ofoe, R.; Thomas, R.H.; Asiedu, S.K.; Wang-Pruski, G.; Fofana, B.; Abbey, L. Aluminum in plants: Benefits, toxicity, and tolerance mechanisms. Front. Plant Sci. 2023, 13, 1085998. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Riaz, M.; Liu, J.; Yu, M.; Cuncang, J.; Jiang, C. The Aluminum tolerance and detoxification mechanisms in plants: Recent advances and prospects. Crit. Rev. Environ. Sci. Technol. 2022, 52, 1491–1527. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. ClustalW and ClustalX version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Ma, W.X.; Noble, W.S.; Bailey, T.L. Motif-based analysis of large nucleotide datasets using MEME-ChIP. Nat. Protoc. 2014, 9, 1428–1450. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: Upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for the detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Liao, J.J.; Wang, C.H.; Xing, Q.J.; Li, Y.P.; Liu, X.F.; Qi, H.Y. Overexpression and VIGS for functional gene validation in oriental melon (Cucumis melo var. makuwa Makino). Plant Cell Tissue Organ Cult. 2019, 137, 275–284. [Google Scholar] [CrossRef]
- Zhang, C.; Shao, Q.; Cao, S.X.; Tang, Y.F.; Liu, J.Y.; Jin, Y.Z.; Qi, H.Y. Effects of postharvest treatments on expression of three lipoxygenase genes in oriental melon (Cucumis melo var. makuwa Makino). Postharvest Biol. Technol. 2015, 110, 229–238. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions, and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Dhindsa, R.S.; Plumb-Dhindsa, P.; Thorpe, T.A. Leaf senescence correlated with increased membrane permeability, lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J. Exp. Bot. 1981, 32, 93–101. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline levels in water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Jambunathan, N. Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants. In Plant Stress Tolerance: Methods and Protocols; Sunkar, R., Ed.; Humana Press: Totowa, NJ, USA, 2010; pp. 292–298. [Google Scholar]
- Zhang, C.; Cao, S.X.; Jin, Y.Z.; Ju, L.J.; Chen, Q.; Qi, H.Y. Melon13-lipoxygenase CmLOX18 may be involved in C6 volatile biosynthesis in fruits. Sci. Rep. 2017, 7, 2816. [Google Scholar] [CrossRef]
- Szabała, B.M. The cationic nature of lysine-rich segments modulates the structural and biochemical properties of wild potato FSK3 dehydrin. Plant Physiol. Biochem. 2023, 194, 480–488. [Google Scholar] [CrossRef]
- Mohammadrezakhani, S.; Rezanejad, F. Effect of low-temperature stress on dehydrin protein, GABA, endogenous proline and antioxidant enzymes in Citrus reticulata extract in postharvest conditions. J. Plant Process Funct. 2024, 13, 79–86. [Google Scholar]
- Hundertmark, M.; Hincha, D.K. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef]
- Liu, H.; Yu, C.; Li, H.; Ouyang, B.; Wang, T.; Zhang, J.; Wang, X.; Ye, Z. Overexpression of ShDHN, a dehydrin gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses in tomatoes. Plant Sci. 2015, 231, 198–211. [Google Scholar] [CrossRef]
- Zhang, H.M.; Wu, J.F.; Fu, D.D.; Zhang, M.; Wang, L.J.; Gong, M.G. Prokaryotic expression, purification, and the in vitro and in vivo protection study of dehydrin WDHN2 from Triticum aestivum. Protoplasma 2024, 261, 771–781. [Google Scholar] [CrossRef]
- Wang, X.S.; Zhu, H.B.; Jin, G.L.; Liu, H.L.; Wu, W.R.; Zhu, J. Genome-scale identification and analysis of LEA genes in rice (Oryza sativa L.). Plant Sci. 2007, 172, 414–420. [Google Scholar] [CrossRef]
- Zhou, Y.; Hu, L.F.; Xu, S.Y.; Jiang, L.W.; Liu, S.Q. Identification and transcriptional analysis of dehydrin gene family in cucumber (Cucumis sativus). Acta Physiol. Plant. 2018, 40, 144. [Google Scholar] [CrossRef]
- Zheng, Y.; Wu, S.; Bai, Y.; Sun, H.H.; Jiao, C.; Guo, S.G.; Zhao, K.; Blanca, J.; Zhang, Z.H.; Huang, S.; et al. Cucurbit Genomics Database (CuGenDB): A central portal for the comparative and functional genomics of cucurbit crops. Nucleic Acids Res. 2019, 47, D1128–D1136. [Google Scholar] [CrossRef]
- Eriksson, S.K.; Kutzer, M.; Procek, J.; Gröbner, G.; Harryson, P. The tunable membrane binding of the intrinsically disordered dehydrin Lti30, a cold-induced plant stress protein. Plant Cell 2011, 23, 2391–2404. [Google Scholar] [CrossRef]
- Godoy, J.A.; Lunar, R.; Torres-Schumann, S.; Moreno, J.; Rodrigo, R.M.; Pintor-Toro, J.A. Expression, tissue distribution, and subcellular localization of dehydrin TAS14 in salt-stressed tomato plants. Plant Mol. Biol. 1994, 26, 1921–1934. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; He, M.; Zhu, Z.; Li, S.; Xu, Y.; Zhang, C.; Singer, S.D.; Wang, Y. Identification of the dehydrin gene family from grapevine species and analysis of their responsiveness to various forms of abiotic and biotic stress. BMC Plant Biol. 2012, 12, 140. [Google Scholar] [CrossRef]
- Liang, D.; Xia, H.; Wu, S.; Ma, F.W. Genome-wide identification and expression profiling of the dehydrin gene family in Malus domestica. Mol. Biol. Rep. 2012, 39, 10759–10768. [Google Scholar] [CrossRef] [PubMed]
- Sarhan, F.; Ouellet, F.; Vazquez-Tello, A. The wheat WCS120 gene family. A useful model to understand the molecular genetics of freezing tolerance in cereals. Physiol. Plant. 1997, 101, 439–445. [Google Scholar] [CrossRef]
- Welin, B.V.; Olson, Å.; Nylander, M.; Palva, E.T. Characterization and differential expression of Dhn/lea/rab-like genes during cold acclimation and drought stress in Arabidopsis thaliana. Plant Mol. Biol. 1994, 26, 131–144. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).