The Interaction Between Soil Microorganisms and Understory Ginseng During Its Growth
Abstract
1. Introduction
2. Materials and Methods
2.1. Background of the Study Location
2.2. Quadrat Setup and Sample Collection
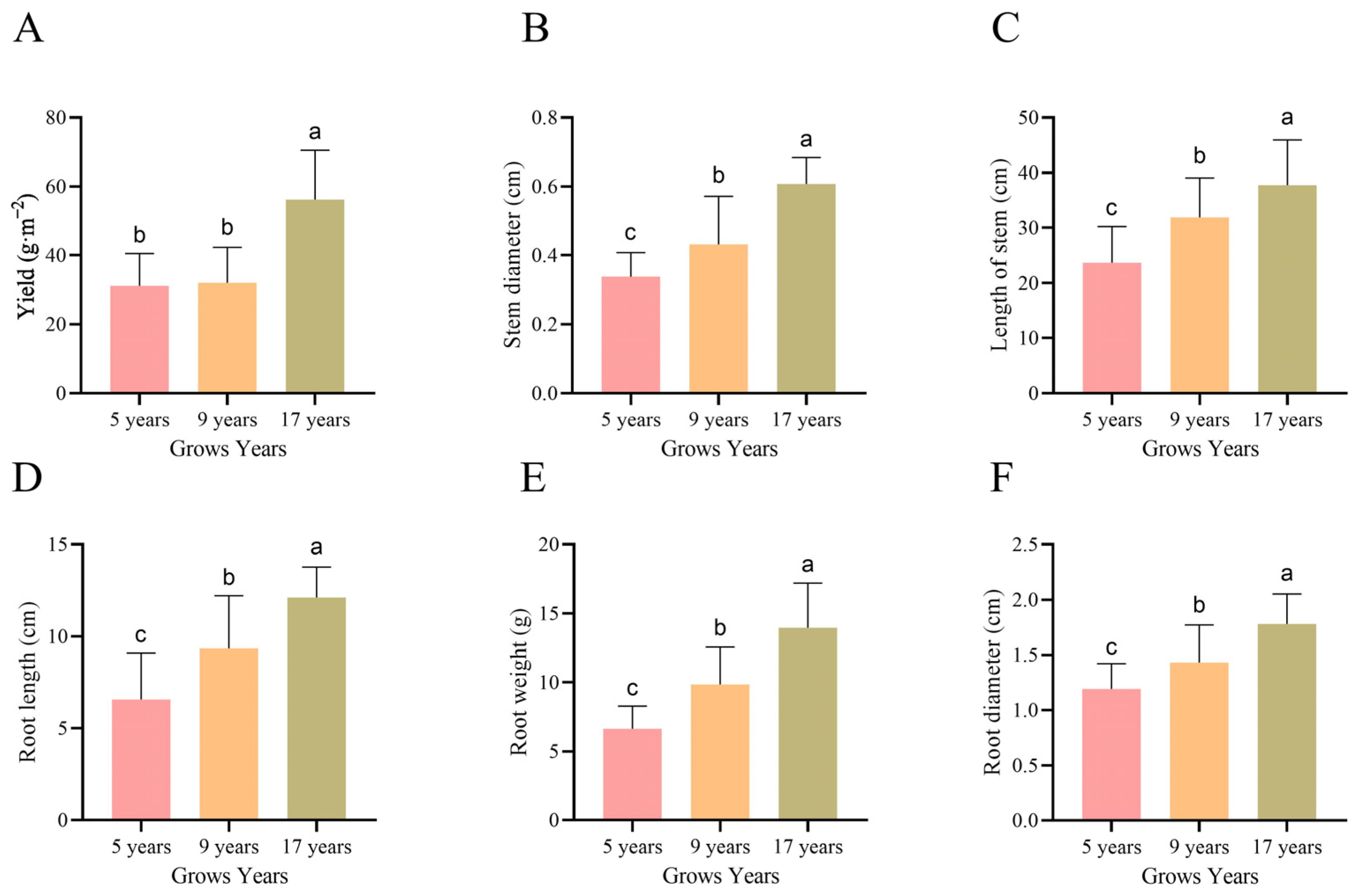
2.3. Determination of Understory Ginseng Yield
2.4. Determination of Ginsenoside Content of Understory Ginseng
2.5. Soil Chemical Analysis
2.6. Biolog EcoPlate Inoculation and Analyses
2.6.1. Extraction of Soil Bacterial Suspension
2.6.2. Determination of Soil Microbial Community Functional Metabolism
2.6.3. Result Calculation and Data Analysis
2.7. Soil DNA Extraction and High-Throughput Sequencing
2.8. Statistical Analysis
3. Results
3.1. Differences in Quality and Growth of Ginseng
3.2. Differences in Soil Chemical Properties and Enzyme Activities
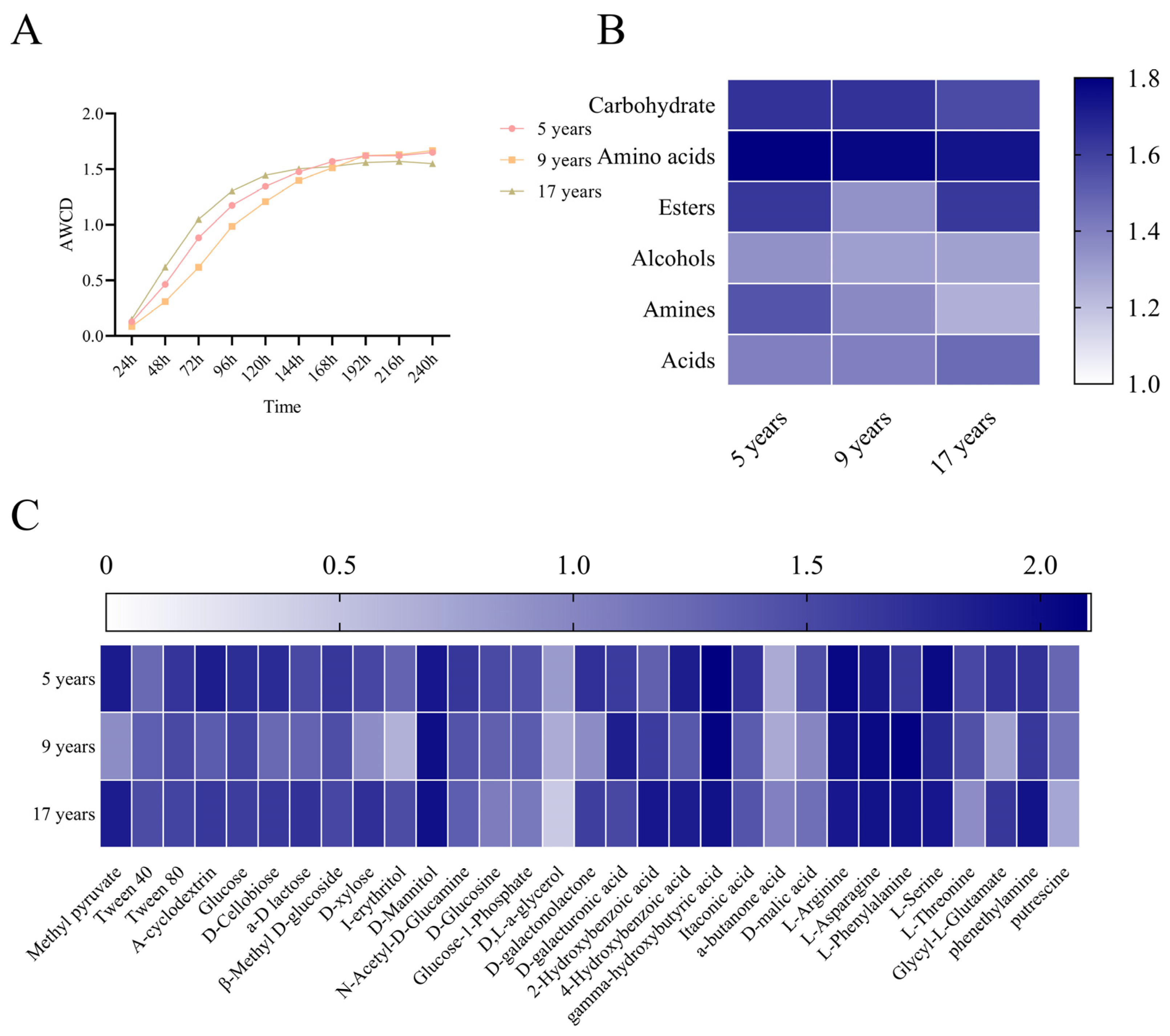
3.3. Analysis of Soil Bacterial Carbon Source Utilization
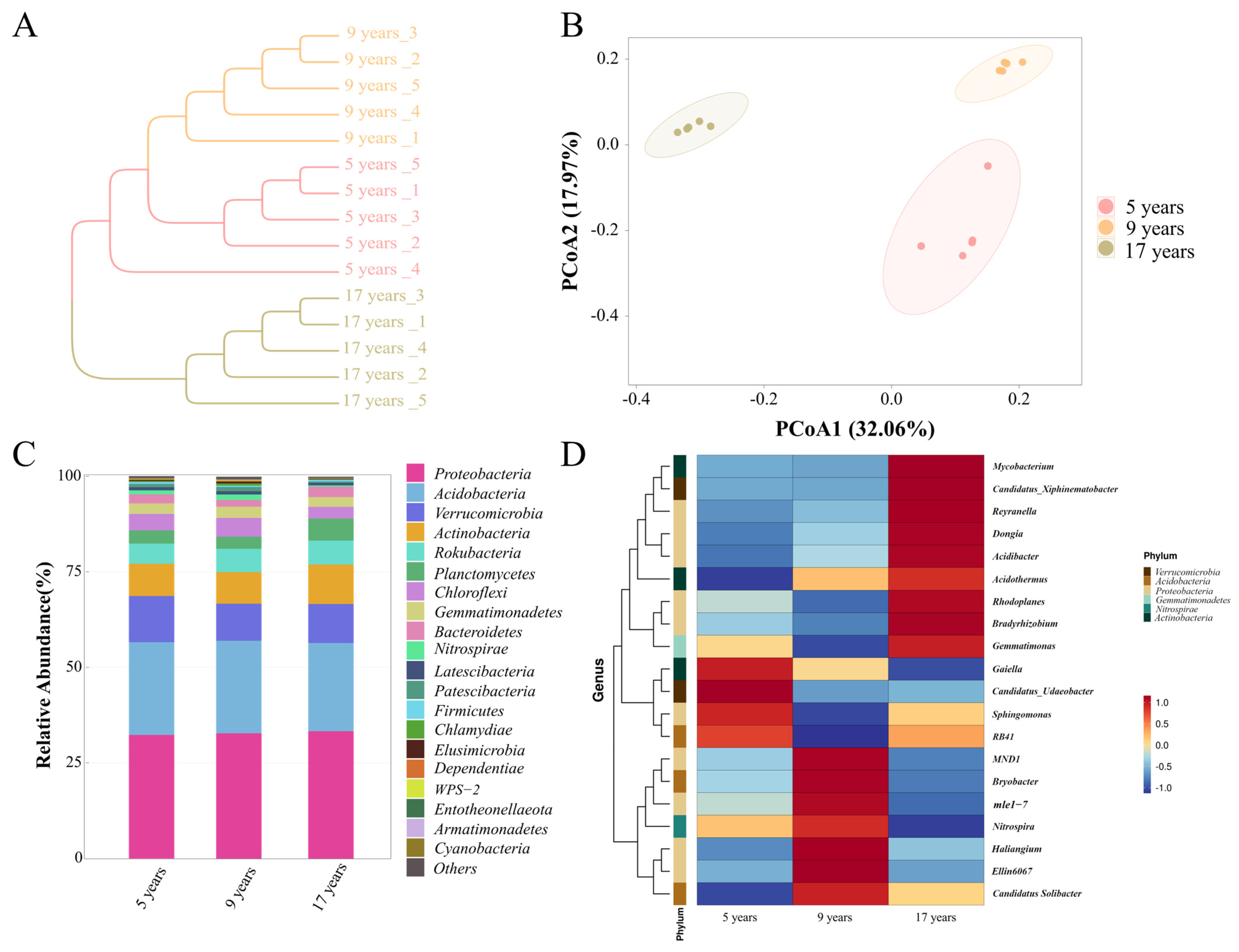
3.4. Analysis of Soil Bacterial Diversity
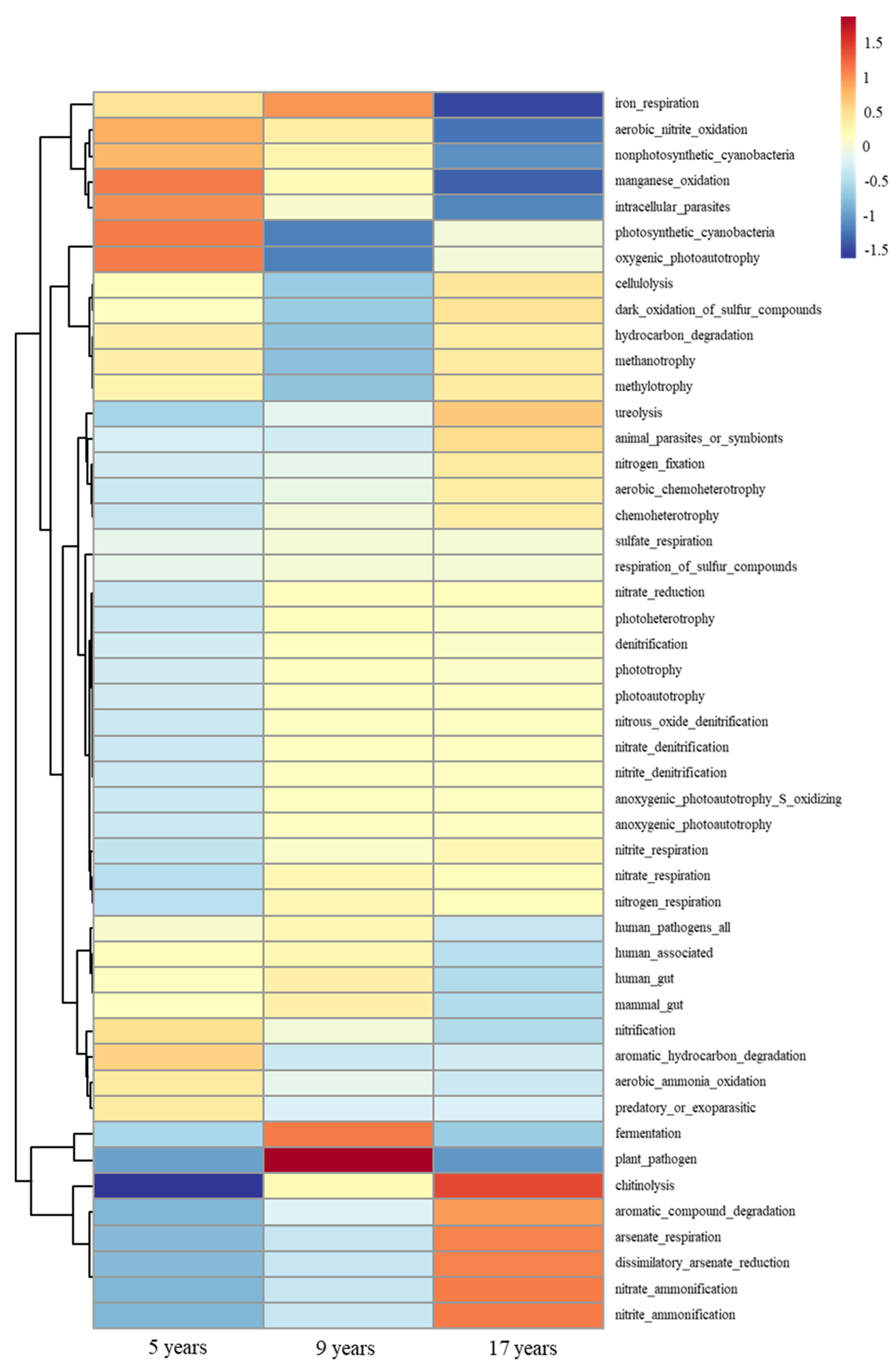
3.5. Soil Bacterial Function Prediction
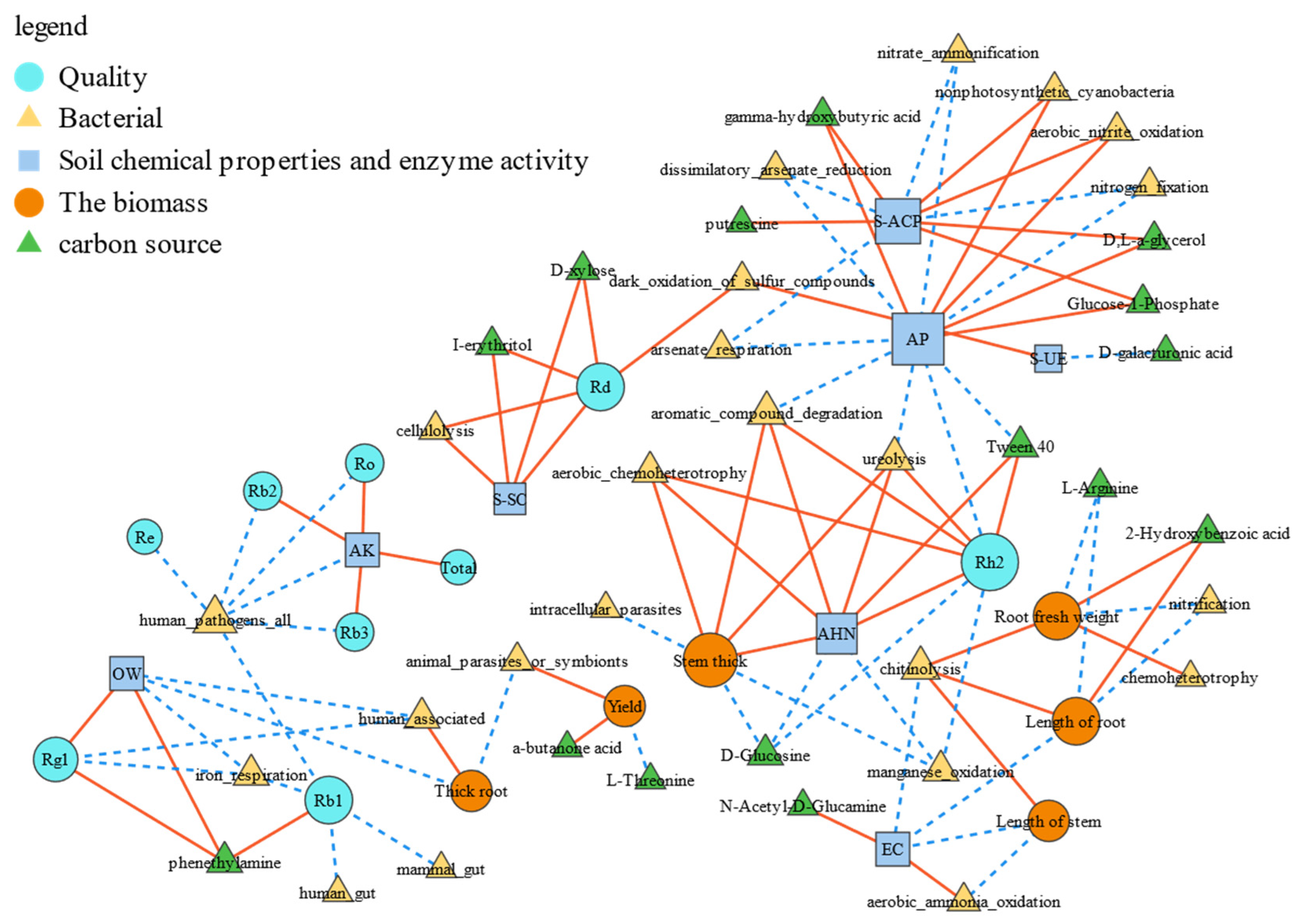
3.6. Analysis of the Correlation Between Ginseng Biomass and Quality and Soil Microecology
4. Discussion
4.1. The Effect of Growth Duration on Ginsenoside Content and Biomass of Understory Ginseng
4.2. Differences in Soil Chemical Properties and Enzyme Activity Among Understory Ginseng Soils of Different Growth Years
4.3. The Differences in Soil Bacteria in Understory Ginseng with Different Growth Years
4.4. The Correlation Between the Quality of Understory Ginseng and Soil Microecology
4.5. Changes in Soil Microecology During the Growth Process of Forest-Grown Ginseng
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen, C.; Lv, Q.; Li, Y.; Jin, Y.H. The Anti-Tumor Effect and Underlying Apoptotic Mechanism of Ginsenoside Rk1 and Rg5 in Human Liver Cancer Cells. Molecules 2021, 26, 3926. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Q.Y.; Shao, M.Y.; Ma, L.; Guo, D.Q.; Wu, Y.; Gao, P.R.; Wang, X.P.; Li, W.L.; Li, C.; et al. Ginsenoside Rb3 regulates energy metabolism and apoptosis in cardiomyocytes via activating PPARα pathway. Biomed. Pharmacother. 2019, 120, 109487. [Google Scholar] [CrossRef] [PubMed]
- Nakhjavani, M.; Smith, E.; Palethorpe, H.M.; Tomita, Y.; Yeo, K.; Price, T.J.; Townsend, A.R.; Hardingham, J.E. Anti-Cancer Effects of an Optimised Combination of Ginsenoside Rg3 Epimers on Triple Negative Breast Cancer Models. Pharmaceuticals 2021, 14, 633. [Google Scholar] [CrossRef]
- Tao, L.; Zhang, J.W.; Lan, W.F.; Liu, H.; Wu, Q.; Yang, S.L.; Song, S.X.; Yu, L.; Bi, Y.F. Neutral oligosaccharides from ginseng residues vs. neutral ginseng polysaccharides: A comparative study of structure elucidation and biological activity. Food Chem. 2025, 464, 141674. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.H.; Hu, Z.Y.; Duan, Y.Q.; Jiang, Y.X.; Xu, W.W.; Yang, P.C.; Zhou, W.; Sun, J.F.; Li, G. Extraction, characterization, and neuroprotective effects of polysaccharides the stems of Panax ginseng C. A. Meyer by three different solvents (water, acid, alkali). Ind. Crops Prod. 2024, 222, 119512. [Google Scholar] [CrossRef]
- Wan, L.; Qian, C.; Yang, C.M.; Peng, S.A.; Dong, G.L.; Cheng, P.; Zong, G.F.; Han, H.K.; Shao, M.Y.; Gong, G.W.; et al. Ginseng polysaccharides ameliorate ulcerative colitis via regulating gut microbiota and tryptophan metabolism. Int. J. Biol. Macromol. 2024, 265, 130822. [Google Scholar] [CrossRef]
- Zhang, H.; Abid, S.; Ahn, J.C.; Mathiyalagan, R.; Kim, Y.J.; Yang, D.C.; Wang, Y.P. Characteristics of Cultivars in Korea and China. Molecules 2020, 25, 2635. [Google Scholar] [CrossRef]
- Zhu, L.L.; Xu, L.; Dou, D.Q.; Huang, L.Q. The distinct of chemical profiles of mountainous forest cultivated ginseng and garden ginseng based on ginsenosides and oligosaccharides. J. Food Compos. Anal. 2021, 104, 104165. [Google Scholar] [CrossRef]
- Shin, S.; Park, M.S.; Lee, H.; Lee, S.; Lee, H.; Kim, T.H.; Kim, H.J. Global Trends in Research on Wild-Simulated Ginseng: Quo Vadis? Forests 2021, 12, 664. [Google Scholar] [CrossRef]
- Liu, D.F.; Sun, H.J.; Ma, H.W. Deciphering Microbiome Related to Rusty Roots of Panax ginseng and Evaluation of Antagonists Against Pathogenic Ilyonectria. Front. Microbiol. 2019, 10, 1350. [Google Scholar] [CrossRef]
- Li, Q.; Zhan, Y.; Xu, Y.H.; Zhang, L.X.; Di, P.; Lu, B.H.; Chen, C.B. Deciphering the transcriptomic response of Ilyonectria robusta in relation to ginsenoside Rg1 treatment and the development of ginseng rusty root rot. FEMS Microbiol. Lett. 2022, 369, fnac075. [Google Scholar] [CrossRef] [PubMed]
- Di, P.; Sun, Z.; Cheng, L.; Han, M.; Yang, L.; Yang, L.M. LED Light Irradiations Differentially Affect the Physiological Characteristics, Ginsenoside Content, and Expressions of Ginsenoside Biosynthetic Pathway Genes in Panax ginseng. Agriculture 2023, 13, 807. [Google Scholar] [CrossRef]
- Chen, T.T.; Wang, L.Q.; Wang, H.T.; Jiang, S.; Zhou, S. Photoperiod and Temperature as Dominant Environmental Drivers Triggering Plant Phenological Development of American ginseng Along With Its Quality Formation. Front. Earth Sci. 2022, 10, 894251. [Google Scholar] [CrossRef]
- Jin, Q.; Zhang, Y.Y.; Ma, Y.Y.; Sun, H.; Guan, Y.M.; Liu, Z.B.; Ye, Q.; Zhang, Y.; Shao, C.; Mu, P.; et al. The composition and function of the soil microbial community and its driving factors before and after cultivation of Panax ginseng in farmland of different ages. Ecol. Indic. 2022, 145, 109748. [Google Scholar] [CrossRef]
- Fang, J.; Xu, Z.F.; Zhang, T.; Chen, C.B.; Liu, C.S.; Liu, R.; Chen, Y.Q. Effects of soil microbial ecology on ginsenoside accumulation in across different cultivation years. Ind. Crops Prod. 2024, 215, 118637. [Google Scholar] [CrossRef]
- Zuppinger-Dingley, D.; Schmid, B.; Petermann, J.S.; Yadav, V.; De Deyn, G.B.; Flynn, D.F.B. Selection for niche differentiation in plant communities increases biodiversity effects. Nature 2014, 515, 108–111. [Google Scholar] [CrossRef]
- Zhao, Y.C.; Wang, Q.Y.; Feng, S.Q.; Zhang, Y.; Dong, W.W.; Ji, W.X. Effects of cultivation duration of the crop and growth stages on rhizosphere soil physicochemical properties, enzyme activities, and microbial communities of ginseng under forest. Plant Soil Environ. 2024, 70, 562–579. [Google Scholar] [CrossRef]
- Piao, X.M.; Huo, Y.; Kang, J.P.; Mathiyalagan, R.; Zhang, H.; Yang, D.U.; Kim, M.; Yang, D.C.; Kang, S.C.; Wang, Y.P. Diversity of Ginsenoside Profiles Produced by Various Processing Technologies. Molecules 2020, 25, 4390. [Google Scholar] [CrossRef]
- Zhang, T.; Xu, Z.F.; Wang, Y.B.; Gao, Q. Effects of soil properties and microbial community composition on ginsenosides accumulation in farmland ginseng. Front. Bioeng. Biotechnol. 2024, 12, 1462342. [Google Scholar] [CrossRef]
- Yeomans, J.C.; Bremner, J.M. A rapid and precise method for routine determination of organic carbon in soil. Commun. Soil Sci. Plant Anal. 1988, 19, 1467–1476. [Google Scholar] [CrossRef]
- Huang, S.H.; Fang, B.; Li, X.; He, S.S. Study on spatial heterogeneity of alkali-hydrolyzable nitrogen in paddy fields at the county scale. J. Ecol. Rural Environ. 2020, 36, 179–185. [Google Scholar] [CrossRef]
- Tang, J.Y.; Zhang, L.H.; Zhang, J.C.; Ren, L.H.; Zhou, Y.Y.; Zheng, Y.Y.; Luo, L.; Yang, Y.; Huang, H.L.; Chen, A.W. Physicochemical features, metal availability and enzyme activity in heavy metal-polluted soil remediated by biochar and compost. Sci. Total Environ. 2020, 701, 134751. [Google Scholar] [CrossRef] [PubMed]
- Papa, S.; Fusco, G.M.; Ciriello, M.; Formisano, L.; Woo, S.L.; De Pascale, S.; Rouphael, Y.; Carillo, P. Microbial and Non-Microbial Biostimulants as Innovative Tools to Increase Macro and Trace Element Mineral Composition of Tomato and Spinach. Horticulturae 2022, 8, 1157. [Google Scholar] [CrossRef]
- van Wyk, D.A.B.; Adeleke, R.; Rhode, O.H.J.; Bezuidenhout, C.C.; Mienie, C. Ecological guild and enzyme activities of rhizosphere soil microbial communities associated with Bt-maize cultivation under field conditions in North West Province of South Africa. J. Basic Microb. 2017, 57, 781–792. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Y.Y.; An, S.S.; Zeng, Q.C. Effects of stem and leaf decomposition in typical herbs on soil enzyme activity and microbial diversity in the south Ningxia loess hilly region of Northwest China. J. Appl. Ecol. 2016, 27, 3182–3188. [Google Scholar]
- Tabatabai, M.A.; Bremner, J.M. Assay of urease activity in soils. Soil Biol. Biochem. 1972, 4, 479–487. [Google Scholar] [CrossRef]
- Sinha, A.K. Colorimetric assay of catalase. Anal. Biochem. 1972, 47, 389–394. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Wang, X.C.C.; Cheng, Z.; Li, Y.Y.; Tang, J.L. Effects of additional fermented food wastes on nitrogen removal enhancement and sludge characteristics in a sequential batch reactor for wastewater treatment. Environ. Sci. Pollut. Res. 2016, 23, 12890–12899. [Google Scholar] [CrossRef]
- Sansupa, C.; Wahdan, S.F.M.; Hossen, S.; Disayathanoowat, T.; Wubet, T.; Purahong, W. Can We Use Functional Annotation of Prokaryotic Taxa (FAPROTAX) to Assign the Ecological Functions of Soil Bacteria? Appl. Sci. 2021, 11, 688. [Google Scholar] [CrossRef]
- Wolinska, A.; Galazka, A.; Kuzniar, A.; Goraj, W.; Jastrzebska, N.; Grzadziel, J.; Stepniewska, Z. Catabolic Fingerprinting and Diversity of Bacteria in Mollic Gleysol Contaminated with Petroleum Substances. Appl. Sci. 2018, 8, 1970. [Google Scholar] [CrossRef]
- Lu, X.H.; Jiao, X.L.; Chen, A.J.; Luo, Y.; Gao, W.W. First Report of Causing Rusty Root of Asian Ginseng in China. Plant Dis. 2015, 99, 156. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.H.; Zhang, X.M.; Jiao, X.L.; Hao, J.J.J.; Zhang, X.S.; Luo, Y.; Gao, W.W. Taxonomy of fungal complex causing red-skin root of Panax ginseng in China. J. Ginseng Res. 2020, 44, 506–518. [Google Scholar] [CrossRef]
- He, J.M.; Zhang, Y.Z.; Luo, J.P.; Zhang, W.J.; Mu, Q. Variation of Ginsenosides in Ginseng of Different Ages. Nat. Prod. Commun. 2016, 11, 739–740. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, S.; Doi, H.; Kasuga, J. Changes over the Years in Soil Chemical Properties Associated with the Cultivation of Ginseng (Panax ginseng Meyer) on Andosol Soil. Agriculture 2022, 12, 1223. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Z.Y.; Niu, J.F.; Dang, K.K.; Zhang, S.K.; Wang, S.Q.; Wang, Z.Z. Changes in physicochemical properties, enzymatic activities, and the microbial community of soil significantly influence the continuous cropping of Panax quinquefolius L. (American ginseng). Plant Soil 2021, 463, 427–446. [Google Scholar] [CrossRef]
- Li, Z.Q.; Mao, C.; Wu, Q.X.; Peng, Y.Y.; Wang, J.; Zhang, B.; Zhang, S.; Liang, X.C.; Yan, W.D.; Chen, X.Y. Temporal Variations in Aboveground Biomass, Nutrient Content, and Ecological Stoichiometry in Young and Middle-Aged Stands of Chinese Fir Forests. Plants 2024, 13, 1877. [Google Scholar] [CrossRef] [PubMed]
- Trasar-Cepeda, C.; Leiros, M.C.; Gil-Sotres, F. Hydrolytic enzyme activities in agricultural and forest soils. Some implications for their use as indicators of soil quality. Soil Biol. Biochem. 2008, 40, 2146–2155. [Google Scholar] [CrossRef]
- Tuo, Y.; Luo, X.Q.; Wang, Z.Y.; Liang, J.P.; Shi, R.; Wang, Z.X.; Wang, S.; Xiang, P.; Yang, Q.L.; He, X.H. Effects of water and fertilizer regulation on soil microbial community, fruit nutrients, and saponin content of Panax notoginseng: A three years field experiment. Ind. Crops Prod. 2024, 220, 119166. [Google Scholar] [CrossRef]
- Guo, C.Q.; Yang, C.; Fu, J.S.; Song, Y.; Chen, S.X.; Li, H.Y.; Ma, C.Q. Effects of crop rotation on sugar beet growth through improving soil physicochemical properties and microbiome. Ind. Crops Prod. 2024, 212, 118331. [Google Scholar] [CrossRef]
- Liu, W.Y.; Wang, F.; Sun, Y.M.; Yang, L.; Chen, H.H.; Liu, W.J.; Zhu, B.; Hui, C.M.; Wang, S.W. Influence of dragon bamboo with different planting patterns on microbial community and physicochemical property of soil on sunny and shady slopes. J. Microbiol. 2020, 58, 906–914. [Google Scholar] [CrossRef]
- Zhang, G.L.; Bai, J.H.; Zhai, Y.J.; Jia, J.; Zhao, Q.Q.; Wang, W.; Hu, X.Y. Microbial diversity and functions in saline soils: A review from a biogeochemical perspective. J. Adv. Res. 2024, 59, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Chenu, C.; Kappler, A.; Rillig, M.C.; Fierer, N. The interplay between microbial communities and soil properties. Nat. Rev. Microbiol. 2024, 22, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.W.; Du, H.J.; Gao, Y.L.; Qiu, W.F. Analysis on Metabolic Functions of Stored Rice Microbial Communities by BIOLOG ECO Microplates. Front. Microbiol. 2018, 9, 1375. [Google Scholar] [CrossRef] [PubMed]
- Koner, S.; Chen, J.S.; Hsu, B.M.; Rathod, J.; Huang, S.W.; Chien, H.Y.; Hussain, B.; Chan, M.W.Y. Depth-resolved microbial diversity and functional profiles of trichloroethylene-contaminated soils for Biolog EcoPlate-based biostimulation strategy. J. Hazard. Mater. 2022, 424, 127266. [Google Scholar] [CrossRef]
- Malik, A.A.; Puissant, J.; Buckeridge, K.M.; Goodall, T.; Jehmlich, N.; Chowdhury, S.; Gweon, H.S.; Peyton, J.M.; Mason, K.E.; van Agtmaal, M.; et al. Land use driven change in soil pH affects microbial carbon cycling processes. Nat. Commun. 2018, 9, 3591. [Google Scholar] [CrossRef]
- Zhou, L.-J.; Li, J.; Zhang, Y.; Kong, L.; Jin, M.; Yang, X.; Wu, Q.L. Trends in the occurrence and risk assessment of antibiotics in shallow lakes in the lower-middle reaches of the Yangtze River basin, China. Ecotoxicol. Environ. Saf. 2019, 183, 109511. [Google Scholar] [CrossRef]
- Stefanowicz, A. The biolog plates technique as a tool in ecological studies of microbial communities. Pol. J. Environ. Stud. 2006, 15, 669–676. [Google Scholar]
- Calbrix, R.; Laval, K.; Barray, S. Analysis of the potential functional diversity of the bacterial community in soil: A reproducible procedure using sole-carbon-source utilization profiles. Eur. J. Soil Biol. 2005, 41, 11–20. [Google Scholar] [CrossRef]
- Moreau, D.; Bardgett, R.D.; Finlay, R.D.; Jones, D.L.; Philippot, L. A plant perspective on nitrogen cycling in the rhizosphere. Funct. Ecol. 2019, 33, 540–552. [Google Scholar] [CrossRef]
- Chen, X.; Wei, H.; Zhang, J.E. Nitrogen and Sulfur Additions Improved the Diversity of nirK- and nirS-Type Denitrifying Bacterial Communities of Farmland Soil. Biology 2021, 10, 1191. [Google Scholar] [CrossRef]
- Strohm, T.O.; Griffin, B.; Zumft, W.G.; Schink, B. Growth yields in bacterial denitrification and nitrate ammonification. Appl. Environ. Microb. 2007, 73, 1420–1424. [Google Scholar] [CrossRef] [PubMed]
- Baggs, E.M. Soil microbial sources of nitrous oxide: Recent advances in knowledge, emerging challenges and future direction. Curr. Opin. Environ. Sustain. 2011, 3, 321–327. [Google Scholar] [CrossRef]
- Sun, J.; Luo, H.; Yu, Q.; Kou, B.; Jiang, Y.; Weng, L.; Xiao, C. Optimal NPK Fertilizer Combination Increases Panax ginseng Yield and Quality and Affects Diversity and Structure of Rhizosphere Fungal Communities. Front. Microbiol. 2022, 13, 919434. [Google Scholar] [CrossRef]
- Wang, M.; Gao, L.M.; Dong, S.Y.; Sun, Y.M.; Shen, Q.R.; Guo, S.W. Role of Silicon on Plant-Pathogen Interactions. Front. Plant Sci. 2017, 8, 701. [Google Scholar] [CrossRef]
- Miao, X.; Wang, E.; Zhou, Y.; Zhan, Y.; Yan, N.; Chen, C.; Li, Q. Effect of ginsenosides on microbial community and enzyme activity in continuous cropping soil of ginseng. Front. Microbiol. 2023, 14, 1060282. [Google Scholar] [CrossRef] [PubMed]
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant Cell Wall–Degrading Enzymes and Their Secretion in Plant-Pathogenic Fungi. Annu. Rev. Phytopathol. 2014, 52, 427–451. [Google Scholar] [CrossRef] [PubMed]
- Tao, F.; Huang, Y.; Hungate, B.A.; Manzoni, S.; Frey, S.D.; Schmidt, M.W.I.; Reichstein, M.; Carvalhais, N.; Ciais, P.; Jiang, L.; et al. Microbial carbon use efficiency promotes global soil carbon storage. Nature 2023, 618, 981–985. [Google Scholar] [CrossRef]
- Schroeter, S.A.; Eveillard, D.; Chaffron, S.; Zoppi, J.; Kampe, B.; Lohmann, P.; Jehmlich, N.; von Bergen, M.; Sanchez-Arcos, C.; Pohnert, G.; et al. Microbial community functioning during plant litter decomposition. Sci. Rep. 2022, 12, 7451. [Google Scholar] [CrossRef]
- Das, P.P.; Singh, K.R.; Nagpure, G.; Mansoori, A.; Singh, R.P.; Ghazi, I.A.; Kumar, A.; Singh, J. Plant-soil-microbes: A tripartite interaction for nutrient acquisition and better plant growth for sustainable agricultural practices. Environ. Res. 2022, 214, 113821. [Google Scholar] [CrossRef]
- Bhat, B.A.; Tariq, L.; Nissar, S.; Islam, S.T.; Ul Islam, S.; Mangral, Z.; Ilyas, N.; Sayyed, R.Z.; Muthusamy, G.; Kim, W.; et al. The role of plant-associated rhizobacteria in plant growth, biocontrol and abiotic stress management. J. Appl. Microbiol. 2022, 133, 2717–2741. [Google Scholar] [CrossRef]
- Zare-Zardini, H.; Alemi, A.; Taheri-Kafrani, A.; Hosseini, S.A.; Soltaninejad, H.; Hamidieh, A.A.; Karamallah, M.H.; Farrokhifar, M.; Farrokhifar, M. Assessment of a New Ginsenoside Rh2 Nanoniosomal Formulation for Enhanced Antitumor Efficacy on Prostate Cancer: An in vitro Study. Drug Des. Dev. Ther. 2020, 14, 3315–3324. [Google Scholar] [CrossRef] [PubMed]
- Hui, C.; Jiang, H.; Liu, B.; Wei, R.; Zhang, Y.P.; Zhang, Q.C.; Liang, Y.C.; Zhao, Y.H. Chitin degradation and the temporary response of bacterial chitinolytic communities to chitin amendment in soil under different fertilization regimes. Sci. Total Environ. 2020, 705, 136003. [Google Scholar] [CrossRef] [PubMed]
- Shahrajabian, M.H.; Chaski, C.; Polyzos, N.; Tzortzakis, N.; Petropoulos, S.A. Sustainable Agriculture Systems in Vegetable Production Using Chitin and Chitosan as Plant Biostimulants. Biomolecules 2021, 11, 819. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Ren, Y.; Bai, X.; Su, Y.; Han, J. Contributions of Beneficial Microorganisms in Soil Remediation and Quality Improvement of Medicinal Plants. Plants 2022, 11, 3200. [Google Scholar] [CrossRef]
- Dong, L.; Xu, J.; Feng, G.; Li, X.; Chen, S. Soil bacterial and fungal community dynamics in relation to Panax notoginseng death rate in a continuous cropping system. Sci. Rep. 2016, 6, 31802. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Zhang, H.; Yang, Y.; Wei, G.; Li, Z. The Composition of Root-Associated Bacteria and Fungi of Astragalus mongholicus and Their Relationship with the Bioactive Ingredients. Front. Microbiol. 2021, 12, 642730. [Google Scholar] [CrossRef]
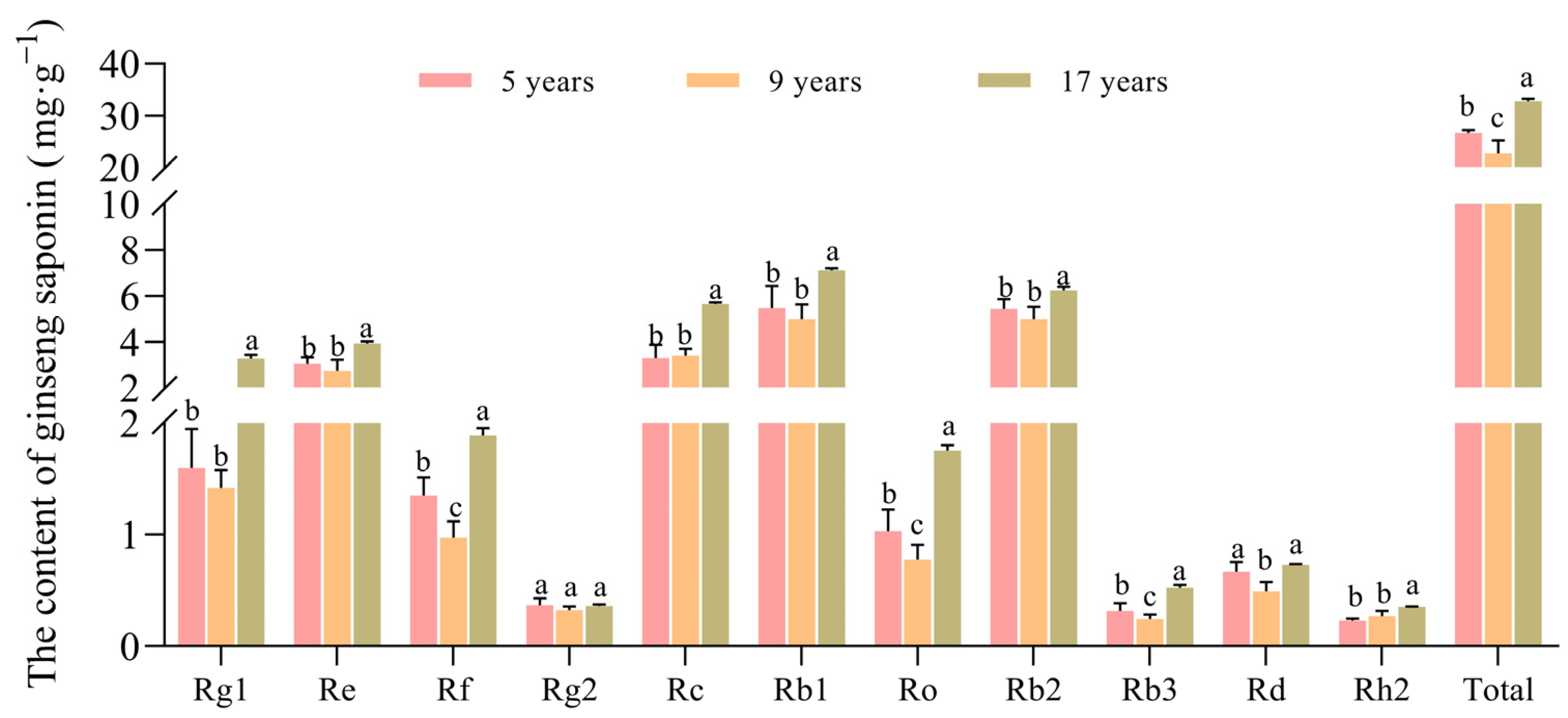

| Time (min) | Elution Gradient |
|---|---|
| 0–40 | 82–79%A |
| 40–42 | 79–75%A |
| 42–48 | 75–68%A |
| 48–72 | 68–63%A |
| 72–79 | 63–51%A |
| 79–82 | 82–82%A |
| Composition | Equation | R2 | Linear Range (μg·mL−1) |
|---|---|---|---|
| Rg1 | Y = 35.572X + 49.594 | 0.9994 | 20~200 |
| Re | Y = 17.156X + 54.901 | 0.9993 | 20~200 |
| Rf | Y = 10.174X + 5.632 | 0.9995 | 10~100 |
| Rg2 | Y = 69.952X − 28.807 | 0.9988 | 40~400 |
| Rc | Y = 48.479X − 10.453 | 0.9994 | 40~400 |
| Rb1 | Y = 39.498X − 27.238 | 0.9994 | 40~400 |
| Rb2 | Y = 16.629X + 5.053 | 0.9983 | 20~200 |
| Rb3 | Y = 11.230X − 7.003 | 0.9993 | 10~100 |
| Rd | Y = 27.398X − 19.852 | 0.9991 | 20~200 |
| Name | 5 Years | 9 Years | 17 Years |
|---|---|---|---|
| pH | 5.46 ± 0.06 a | 4.74 ± 0.13 b | 5.33 ± 0.11 a |
| EC (μS·cm−1) | 40.83 ± 6.47 a | 31.38 ± 2.81 b | 33.82 ± 3.82 b |
| AHN (mg·kg−1) | 269.21 ± 32.49 c | 328.98 ± 6.56 b | 438.32 ± 24.43 a |
| AP (mg·kg−1) | 12.97 ± 1.4 a | 12.83 ± 1.14 a | 10.84 ± 0.51 b |
| AK (mg·kg−1) | 128.08 ± 11.92 b | 101.11 ± 12.3 c | 229.52 ± 12.93 a |
| OM (%) | 10.71 ± 0.65 b | 10.41 ± 0.9 b | 15.28 ± 1.73 a |
| S-SC (mg·g−1·d−1) | 7.89 ± 0.44 b | 3.58 ± 0.3 c | 10.7 ± 1.85 a |
| S-UE (mg·g−1·d−1) | 265.5 ± 41.8 b | 89.65 ± 13.72 c | 594.09 ± 13.15 a |
| S-CAT (μg·g−1·d−1) | 102.02 ± 0.39 a | 86.68 ± 0.49 c | 93.16 ± 1.23 b |
| S-ACP (μmol·g−1·d−1) | 19.78 ± 1.11 a | 17.09 ± 0.23 b | 15.75 ± 0.87 c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lan, Y.; Shen, Y.; Sun, Y.; Han, M.; Wan, M.; Yang, L. The Interaction Between Soil Microorganisms and Understory Ginseng During Its Growth. Horticulturae 2025, 11, 467. https://doi.org/10.3390/horticulturae11050467
Lan Y, Shen Y, Sun Y, Han M, Wan M, Yang L. The Interaction Between Soil Microorganisms and Understory Ginseng During Its Growth. Horticulturae. 2025; 11(5):467. https://doi.org/10.3390/horticulturae11050467
Chicago/Turabian StyleLan, Yiming, Yumu Shen, Yingxin Sun, Mei Han, Mingming Wan, and Limin Yang. 2025. "The Interaction Between Soil Microorganisms and Understory Ginseng During Its Growth" Horticulturae 11, no. 5: 467. https://doi.org/10.3390/horticulturae11050467
APA StyleLan, Y., Shen, Y., Sun, Y., Han, M., Wan, M., & Yang, L. (2025). The Interaction Between Soil Microorganisms and Understory Ginseng During Its Growth. Horticulturae, 11(5), 467. https://doi.org/10.3390/horticulturae11050467






