Identification of SNAT Gene Family and Their Response to Abiotic Stress in Citrus
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of SNAT Genes from Five Citrus Genome
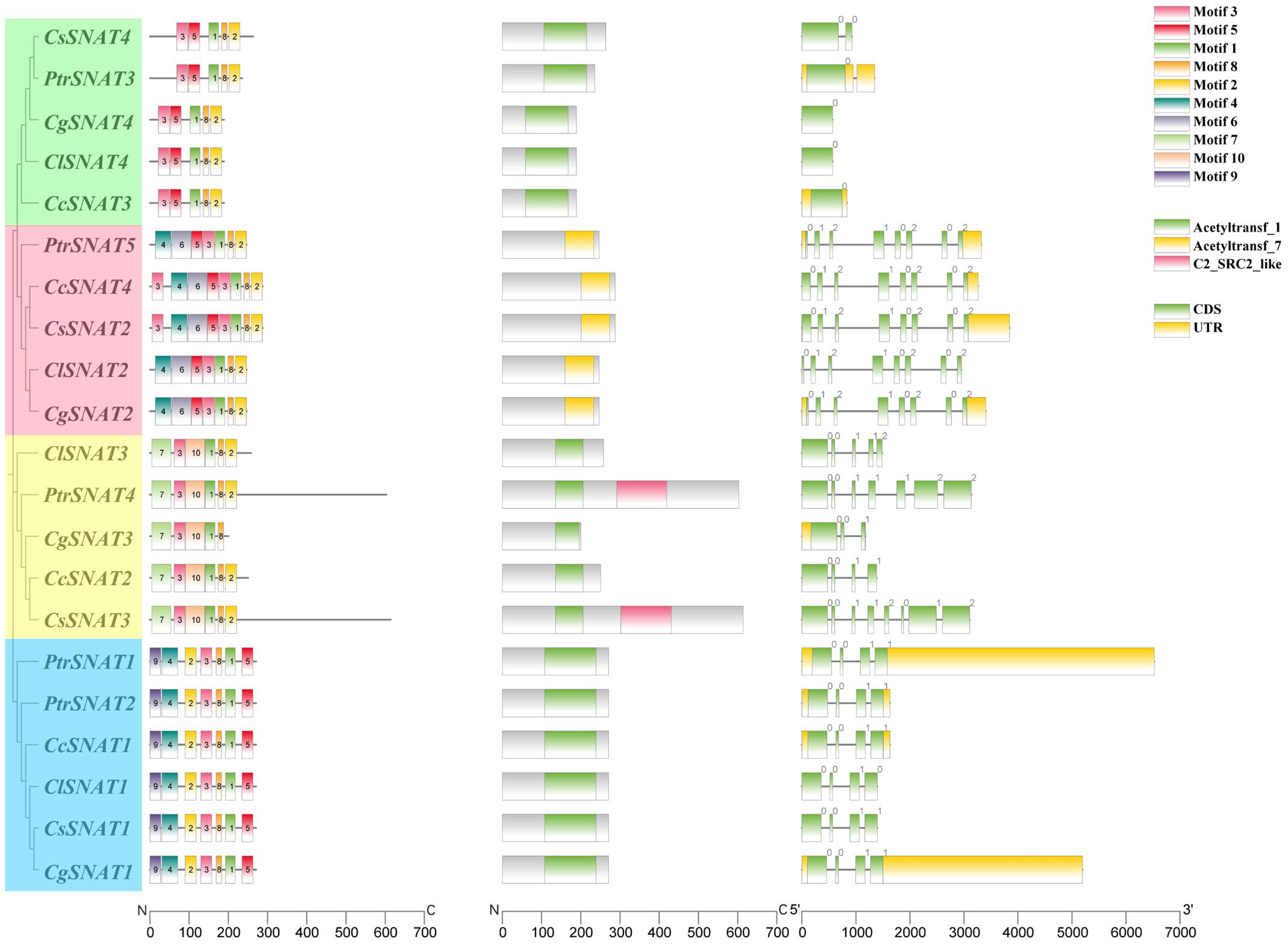
2.2. Gene Structure, Conserved Domains, and Motif Analysis
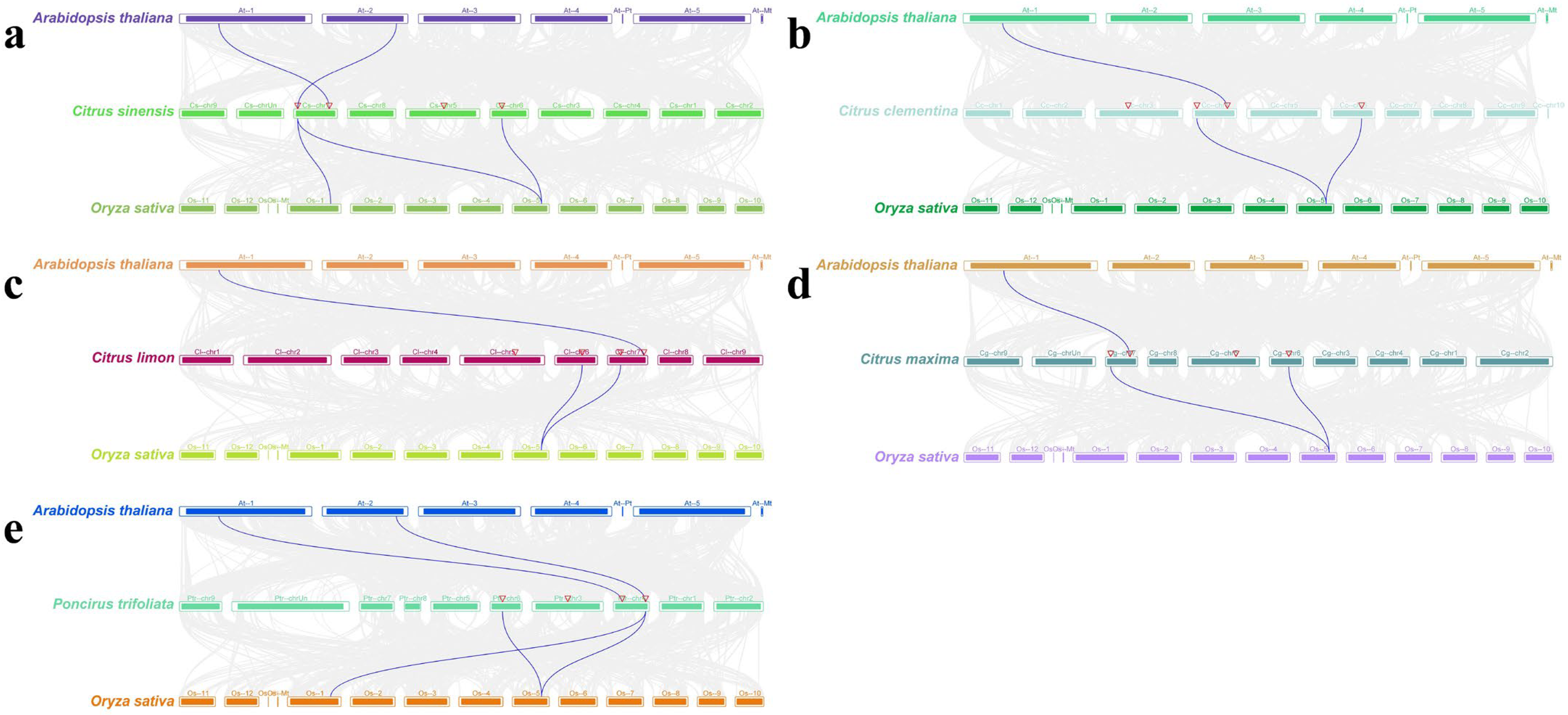
2.3. Phylogenetic Tree Construction and Collinearity Analysis
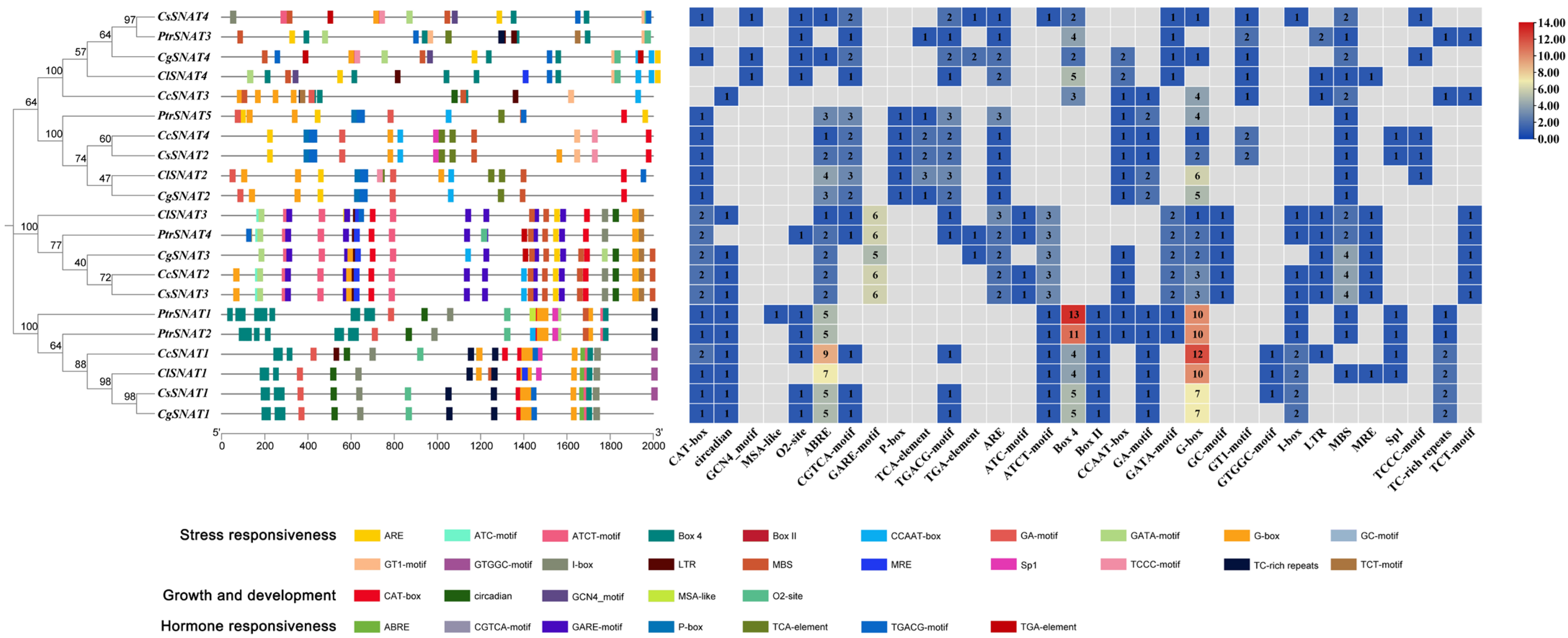
2.4. Identification of Cis-Acting Elements
2.5. Tissue-Specific Expression Analysis
2.6. Plant Material and Adverse Stress Treatments
2.7. RNA Extraction and qRT-PCR Assays
3. Results
3.1. Identification of Citrus SNAT Gene Family Members and Prediction of Physicochemical Properties of Their Corresponding Proteins
3.2. Gene Structure, Conserved Domains, and Motif Analysis of the SNAT Genes in Citrus
3.3. Phylogenetic Tree and Collinearity Analysis
3.4. Analysis of Cis-Acting Elements in the Promoter Regions of Citrus SNAT Genes
3.5. Expression Analysis of PtrSNAT Genes in Different Tissues
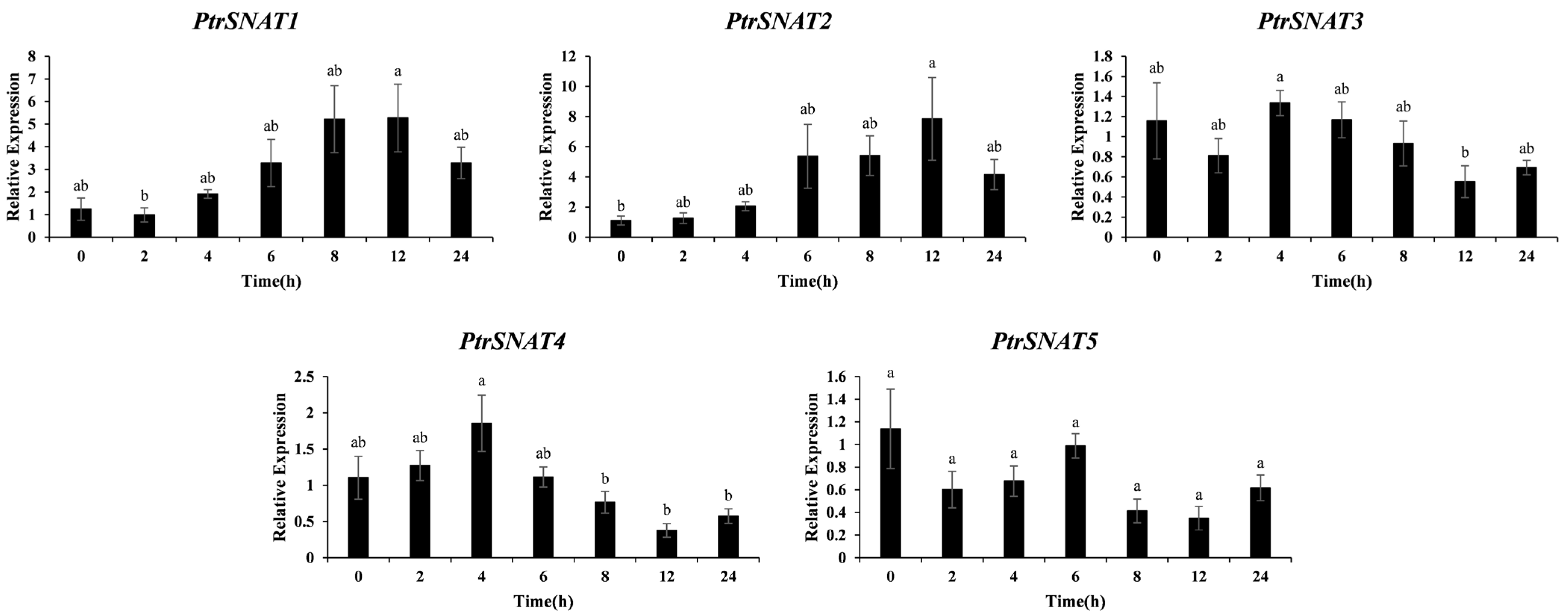
3.6. Expression Profiling of PtrSNAT Genes Under Different Abiotic Stress Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lerner, A.B.; Case, J.D.; Takahashi, Y.; Lee, T.H.; Mori, W. Isolation of melatonin, the pineal gland factor that lightens melanocytes. J. Am. Chem. Soc. 1958, 80, 2587. [Google Scholar] [CrossRef]
- Hattori, A.; Migitaka, H.; Iigo, M.; Itoh, M.; Yamamoto, K.; Ohtani-Kaneko, R.; Hara, M.; Suzuki, T.; Reiter, R.J. Identification of melatonin in plants and its effects on plasma melatonin levels and binding to melatonin receptors in vertebrates. Biochem. Mol. Biol. Int. 1995, 35, 627–634. [Google Scholar] [PubMed]
- Cano, A.; Hernández-Ruiz, J.; Arnao, M.B. Role of exogenous melatonin in plant biotechnology: Physiological and applied aspects. Crit. Rev. Plant Sci. 2024, 43, 395–404. [Google Scholar] [CrossRef]
- Shi, D.W.; Zhao, L.J.; Zhang, R.J.; Song, Q.F. Regulation of plant growth and development by melatonin. Life 2024, 14, 1606. [Google Scholar] [CrossRef]
- Ikram, M.; Mehran, M.; Rehman, H.U.; Ullah, S.; Bakhsh, M.Z.M.; Tahira, M.; Maqsood, M.F.K.; Rauf, A.; Ghafar, S.; Haider, K.; et al. Mechanistic review of melatonin metabolism and signaling pathways in plants: Biosynthesis, regulation, and roles under abiotic stress. Plant Stress 2024, 14, 100685. [Google Scholar] [CrossRef]
- Huang, S.X.; Jin, S.H. Enhancing drought tolerance in horticultural plants through plant hormones: A strategic coping mechanism. Front. Plant Sci. 2024, 15, 1502438. [Google Scholar] [CrossRef]
- Ameen, M.; Zafar, A.; Mahmood, A.; Zia, M.A.; Kamran, K.; Javaid, M.M.; Yasin, M.; Khan, B.A. Melatonin as a master regulatory hormone for genetic responses to biotic and abiotic stresses in model plant Arabidopsis thaliana: A comprehensive review. Funct. Plant Biol. 2024, 51, FP23248. [Google Scholar] [CrossRef]
- Kang, K.; Lee, K.; Park, S.; Byeon, Y.; Back, K. Molecular cloning of rice serotonin N-acetyltransferase, the penultimate gene in plant melatonin biosynthesis. J. Pineal Res. 2013, 55, 7–13. [Google Scholar] [CrossRef]
- Byeon, Y.; Lee, H.J.; Lee, H.Y.; Back, K. Cloning and functional characterization of the Arabidopsis N-acetylserotonin O-methyltransferase responsible for melatonin synthesis. J. Pineal Res. 2016, 60, 65–73. [Google Scholar] [CrossRef]
- Bhowal, B.; Bhattacharjee, A.; Goswami, K.; Sanan-Mishra, N.; Singla-Pareek, S.L.; Kaur, C.; Sopory, S. Serotonin and melatonin biosynthesis in plants: Genome-wide identification of the genes and their expression reveal a conserved role in stress and development. Int. J. Mol. Sci. 2021, 22, 11034. [Google Scholar] [CrossRef]
- Zhang, J.M.; Yao, Z.P.; Zhang, R.J.; Mou, Z.M.; Yin, H.H.; Xu, T.Y.; Zhao, D.K.; Chen, S.Y. Genome-wide identification and expression profile of the SNAT gene family in tobacco (Nicotiana tabacum). Front. Genet. 2020, 11, 591984. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.; Zhang, S.W.; Wang, Y.D.; Feng, Y.Q.; Chen, R.Y.; Su, W.; Song, S.W. Genome-wide identification and expression profiling of the BcSNAT family genes and their response to hormones and abiotic stresses in flowering Chinese cabbage. Sci. Hortic. 2023, 322, 112445. [Google Scholar] [CrossRef]
- Zhou, W.; Yang, S.; Zhang, Q.; Xiao, R.Y.; Li, B.; Wang, D.H.; Niu, J.F.; Wang, S.Q.; Wang, Z.Z. Functional characterization of serotonin N-acetyltransferase genes (SNAT1/2) in melatonin biosynthesis of Hypericum perforatum. Front. Plant Sci. 2021, 12, 781717. [Google Scholar] [CrossRef]
- Guo, X.H.; Ran, L.; Huang, X.Y.; Wang, X.C.; Zhu, J.T.; Tan, Y.Y.; Shu, Q.Y. Identification and functional analysis of two serotonin N-acetyltransferase genes in maize and their transcriptional response to abiotic stresses. Front. Plant Sci. 2024, 15, 1478200. [Google Scholar] [CrossRef]
- Kumar, G.; Arya, M.; Radhika, P.; Giridhar, P. Genome-wide identification, characterization of serotonin N-acetyltransferase and deciphering its importance under development, biotic and abiotic stress in soybean. Int. J. Biol. Macromol. 2022, 220, 942–953. [Google Scholar] [CrossRef]
- Wu, Y.D.; Fan, X.C.; Zhang, Y.; Jiang, J.F.; Sun, L.; Rahman, F.U.; Liu, C.H. VvSNAT1 overexpression enhances melatonin production and salt tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2021, 166, 485–494. [Google Scholar] [CrossRef]
- Wang, L.; Feng, C.; Zheng, X.D.; Guo, Y.; Zhou, F.F.; Shan, D.Q.; Liu, X.; Kong, J. Plant mitochondria synthesize melatonin and enhance the tolerance of plants to drought stress. J. Pineal Res. 2017, 63, e12429. [Google Scholar] [CrossRef]
- Byeon, Y.; Back, K. Low melatonin production by suppression of either serotonin N-acetyltransferase or N-acetylserotonin methyltransferase in rice causes seedling growth retardation with yield penalty, abiotic stress susceptibility, and enhanced coleoptile growth under anoxic conditions. J. Pineal Res. 2016, 60, 348–359. [Google Scholar] [CrossRef]
- Xu, X.Y.; Miao, X.C.; Deng, N.Y.; Liang, M.G.; Wang, L.; Jiang, L.J.; Zeng, S.H. Identification of ascorbate oxidase genes and their response to cold stress in Citrus sinensis. Agriculture 2024, 14, 1643. [Google Scholar] [CrossRef]
- Song, X.; Zhang, M.; Wang, T.T.; Duan, Y.Y.; Ren, J.; Gao, H.; Fan, Y.J.; Xia, Q.M.; Cao, H.X.; Xie, K.D.; et al. Polyploidization leads to salt stress resilience via ethylene signaling in citrus plants. New. Phytol. 2025, 246, 176–191. [Google Scholar] [CrossRef]
- Chen, C.J.; Wu, Y.; Li, J.W.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.H.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Tibor, K.; Zoltán, K.; Adrienn, G.T.; Lilla, S.K.; Zsolt, S.; Júlia, H.K.; Thomas, C.; Margot, O.; Richárd, G.; István, H.Á.; et al. A modified CTAB method for the extraction of high-quality RNA from mono-and dicotyledonous plants rich in secondary metabolites. Plant Methods 2024, 20, 62. [Google Scholar]
- Zhou, W.; Xiao, R.-Y.; Yang, Y.-X.; Wang, X.; Wang, D.-H.; Wang, Z.-Z. Clock protein LHY targets SNAT1 and negatively regulates the biosynthesis of melatonin in Hypericum perforatum. Sci. Adv. 2024, 10, eadq6505. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernandez-Ruiz, J. The physiological function of melatonin in plants. Plant Signal. Behav. 2006, 1, 89–95. [Google Scholar] [CrossRef]
- Posmyk, M.M.; Janas, K.M. Melatonin in plants. Acta Physiol. Plant. 2009, 31, 1–11. [Google Scholar] [CrossRef]
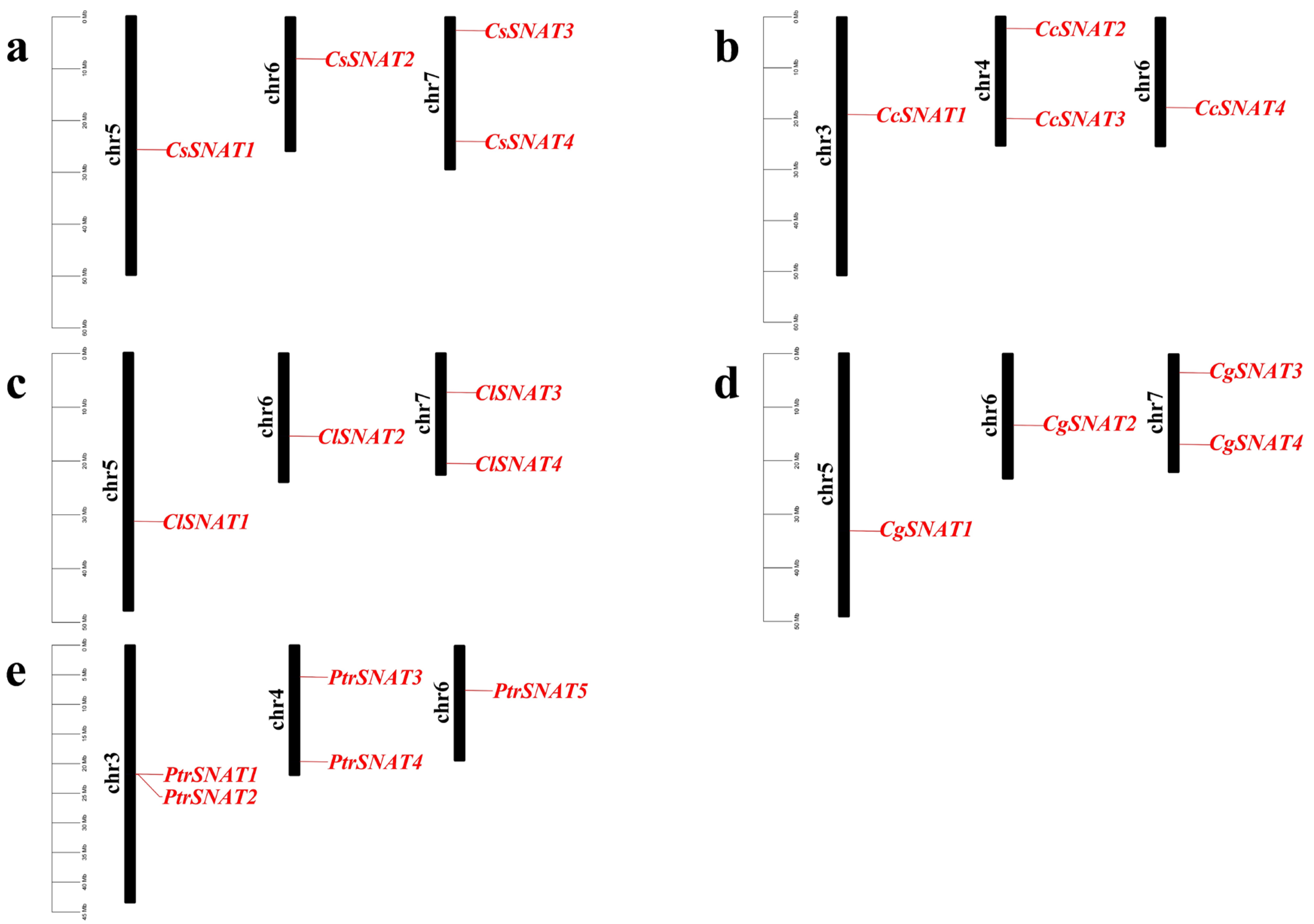




| Gene Name | CPBD Registration Number | Chromosomes | Amino Acid Length | Molecular Weight (kDa) | Isoelectric Point (pI) | Instability Index (II) | Chloroplast Transit Peptide | Subcellular Localization Predicted |
|---|---|---|---|---|---|---|---|---|
| CsSNAT1 | Cs_ont_5g027410.1 | Chr5 | 271 | 31.15 | 9.25 | 44.99 | absence | chloroplast |
| CsSNAT2 | Cs_ont_6g012680.2 | Chr6 | 288 | 32.67 | 8.70 | 45.44 | absence | chloroplast |
| CsSNAT3 | Cs_ont_7g003510.1 | Chr7 | 614 | 67.53 | 8.72 | 50.06 | absence | cytoplasm |
| CsSNAT4 | Cs_ont_7g020400.1 | Chr7 | 264 | 29.19 | 9.35 | 44.25 | presence | cell membrane |
| CcSNAT1 | Ciclev10021647m.1 | Chr3 | 271 | 31.07 | 9.26 | 44.68 | absence | chloroplast |
| CcSNAT2 | Ciclev10033598m.1 | Chr4 | 251 | 27.50 | 8.97 | 44.77 | absence | chloroplast |
| CcSNAT3 | Ciclev10032841m.1 | Chr4 | 180 | 20.11 | 9.13 | 37.93 | absence | chloroplast |
| CcSNAT4 | Ciclev10012371m.1 | Chr6 | 288 | 32.67 | 8.70 | 45.44 | absence | chloroplast |
| ClSNAT1 | CL5G054858012.t1_alt | Chr5 | 271 | 31.11 | 9.32 | 44.68 | presence | chloroplast |
| ClSNAT2 | CL6G059010012.t1_alt | Chr6 | 240 | 26.65 | 5.38 | 43.98 | presence | chloroplast |
| ClSNAT3 | CL7G061361012.t1_alt | Chr7 | 258 | 28.58 | 6.33 | 40.87 | absence | chloroplast |
| ClSNAT4 | CL7G063246012.t1_alt | Chr7 | 189 | 21.23 | 9.78 | 39.34 | absence | chloroplast |
| CgSNAT1 | Cg5g025450.1 | Chr5 | 271 | 31.11 | 9.32 | 44.68 | presence | chloroplast |
| CgSNAT2 | Cg6g010930.1 | Chr6 | 247 | 27.57 | 5.73 | 45.14 | presence | chloroplast |
| CgSNAT3 | Cg7g004160.1 | Chr7 | 200 | 22.14 | 7.69 | 43.33 | absence | nucleus |
| CgSNAT4 | Cg7g015440.1 | Chr7 | 189 | 21.22 | 9.78 | 39.89 | absence | mitochondria |
| PtrSNAT1 | Pt3g018760.1 | Chr3 | 271 | 30.98 | 9.20 | 48.23 | presence | chloroplast |
| PtrSNAT2 | Pt3g018810.1 | Chr3 | 271 | 30.96 | 9.13 | 47.35 | absence | chloroplast |
| PtrSNAT3 | Pt4g009400.1 | Chr4 | 236 | 26.27 | 10.12 | 40.40 | presence | chloroplast |
| PtrSNAT4 | Pt4g020200.1 | Chr4 | 603 | 65.92 | 7.96 | 47.99 | absence | chloroplast |
| PtrSNAT5 | Pt6g011220.2 | Chr6 | 247 | 27.57 | 5.73 | 48.72 | presence | chloroplast |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, Q.; Gu, M.; Song, C.; Jiang, L.; Wang, L.; Xu, X. Identification of SNAT Gene Family and Their Response to Abiotic Stress in Citrus. Horticulturae 2025, 11, 399. https://doi.org/10.3390/horticulturae11040399
Yao Q, Gu M, Song C, Jiang L, Wang L, Xu X. Identification of SNAT Gene Family and Their Response to Abiotic Stress in Citrus. Horticulturae. 2025; 11(4):399. https://doi.org/10.3390/horticulturae11040399
Chicago/Turabian StyleYao, Qian, Mingzhou Gu, Chengyang Song, Lijuan Jiang, Lun Wang, and Xiaoyong Xu. 2025. "Identification of SNAT Gene Family and Their Response to Abiotic Stress in Citrus" Horticulturae 11, no. 4: 399. https://doi.org/10.3390/horticulturae11040399
APA StyleYao, Q., Gu, M., Song, C., Jiang, L., Wang, L., & Xu, X. (2025). Identification of SNAT Gene Family and Their Response to Abiotic Stress in Citrus. Horticulturae, 11(4), 399. https://doi.org/10.3390/horticulturae11040399







