Genome-Wide Identification of Watermelon Trihelix Genes and Their Expression Patterns Under Biotic and Abiotic Stresses
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Physicochemical Property Analysis of Watermelon Trihelix Genes
2.2. Structural Analysis and Phylogenetic Tree Construction of Watermelon Trihelix Genes
2.3. Synteny Analyses of Trihelix Genes
2.4. RNA-Seq Re-Analysis of Transcriptome Sequencing Data
2.5. Tissue-Specific Expression Analysis of Watermelon Trihelix Genes
2.6. Analysis of Expression Patterns Under Abiotic and Biotic Stresses
2.7. RNA Extraction and Gene Expression Analysis
3. Results
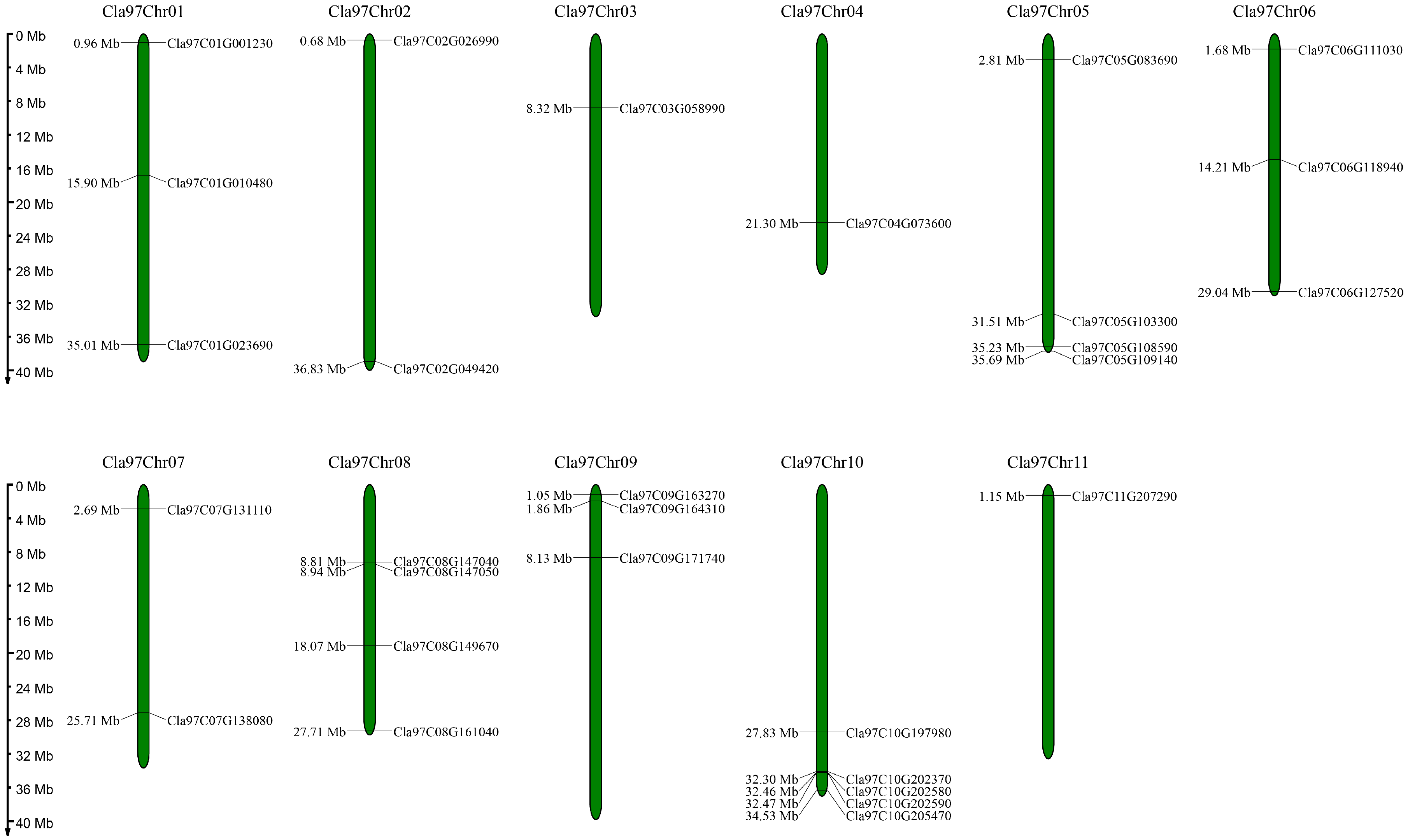
3.1. Identification and Physicochemical Characteristics of Watermelon Trihelix Genes
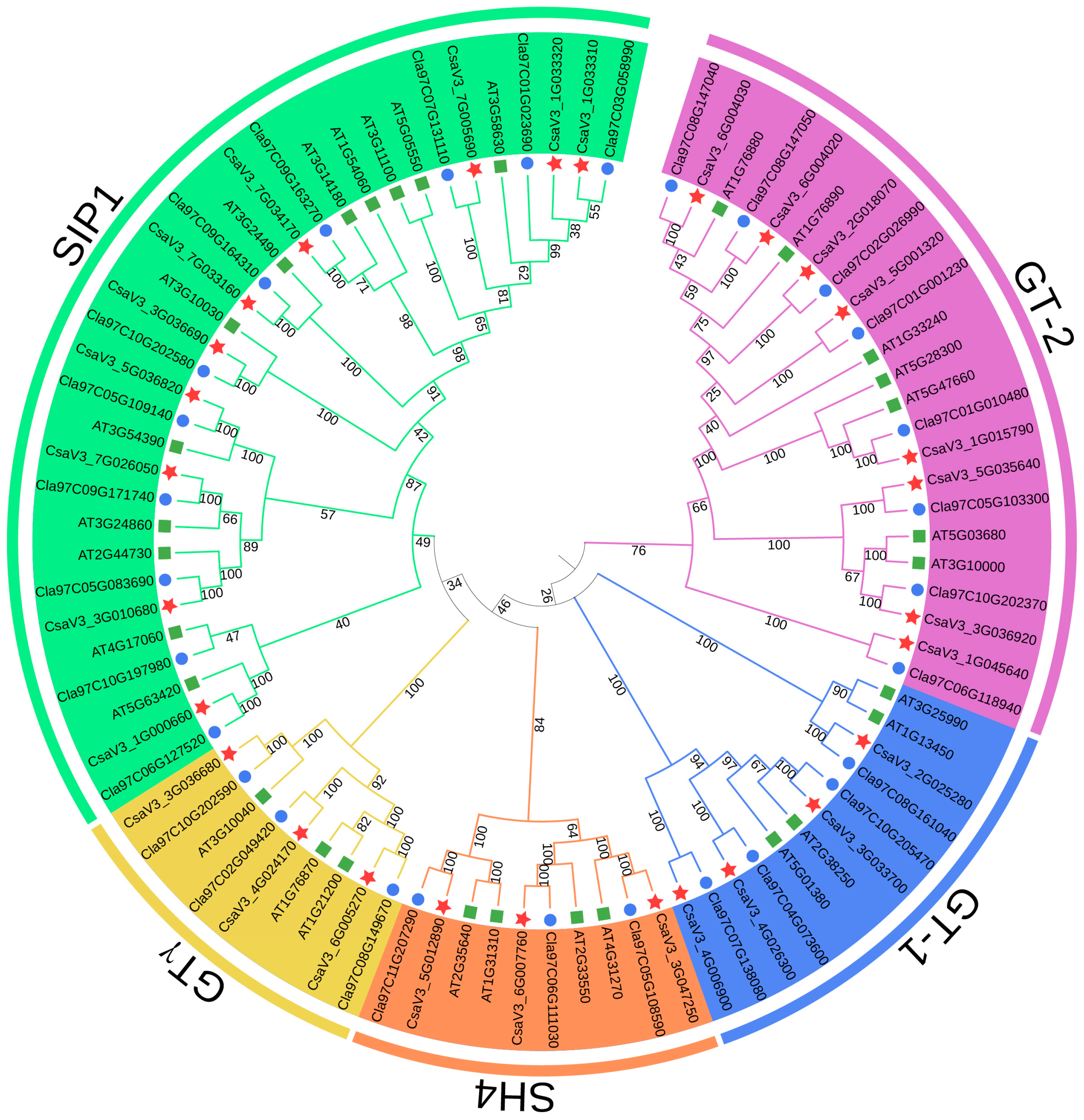
3.2. Evolutionary Analysis of Trihelix Genes Among Different Species
3.3. Structure and Conserved Sequence Analysis of Trihelix Genes
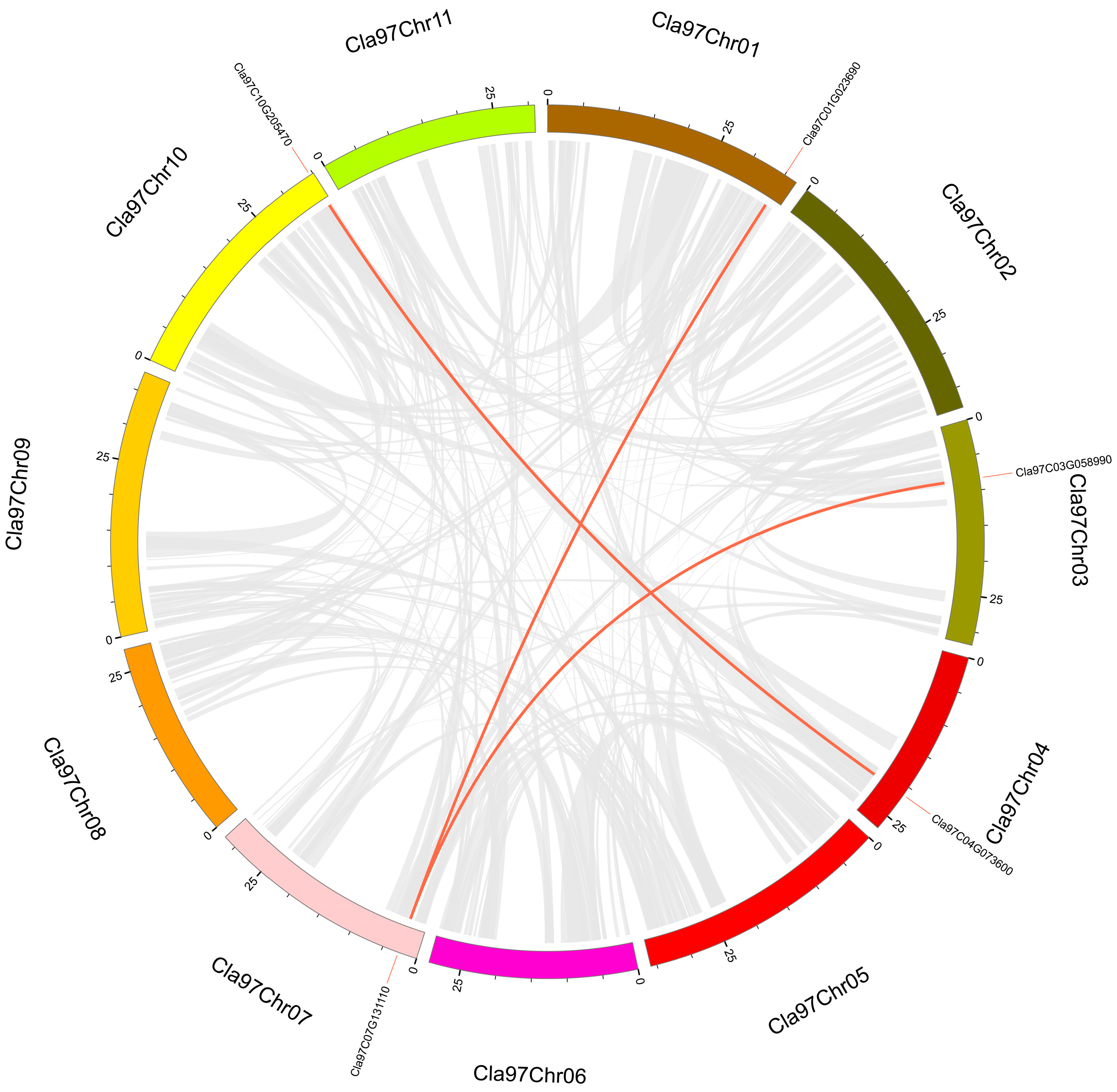
3.4. Intraspecies and Interspecies Synteny Analysis of Trihelix Genes
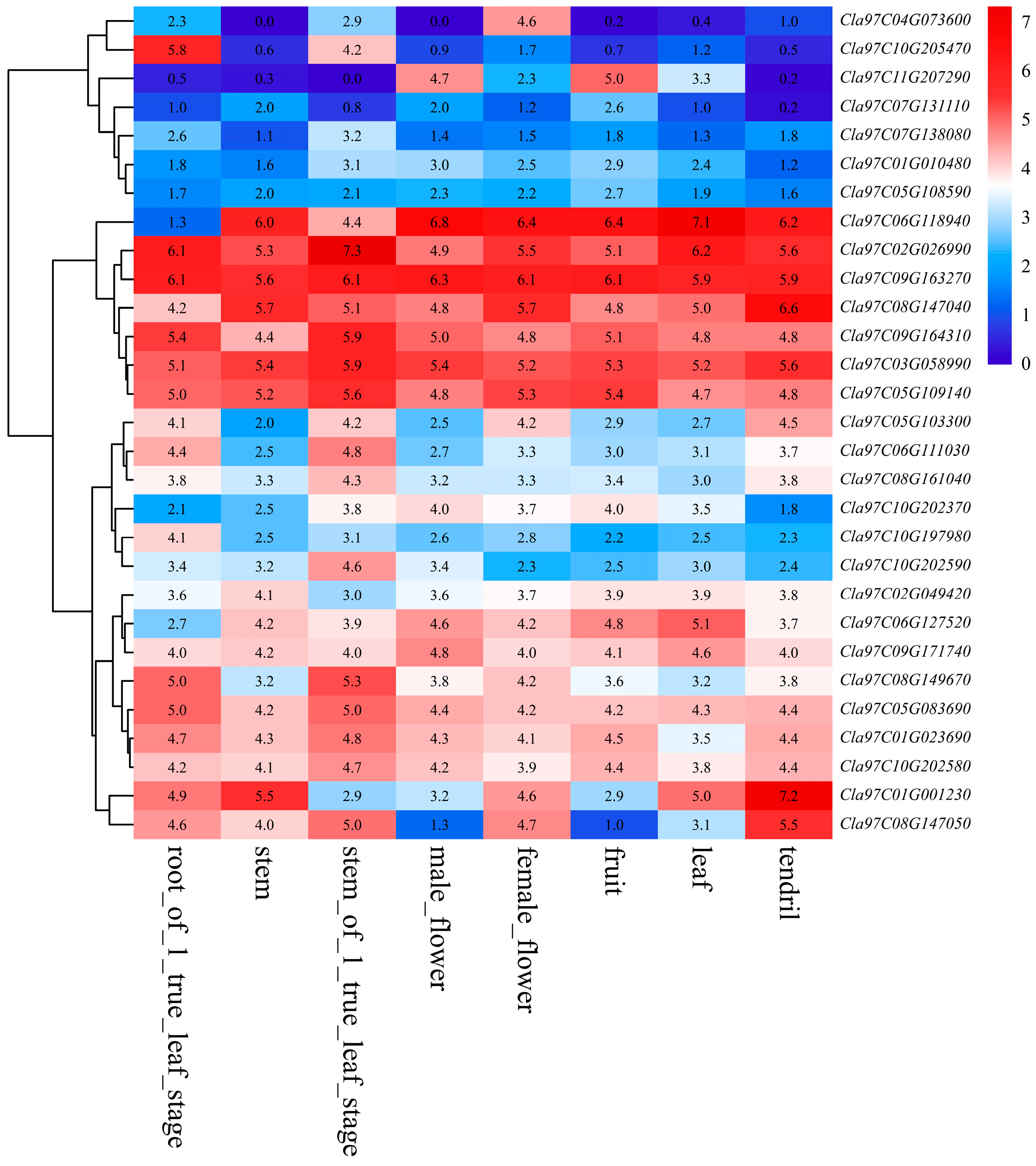
3.5. Tissue-Specific Expression Patterns of Watermelon Trihelix Genes
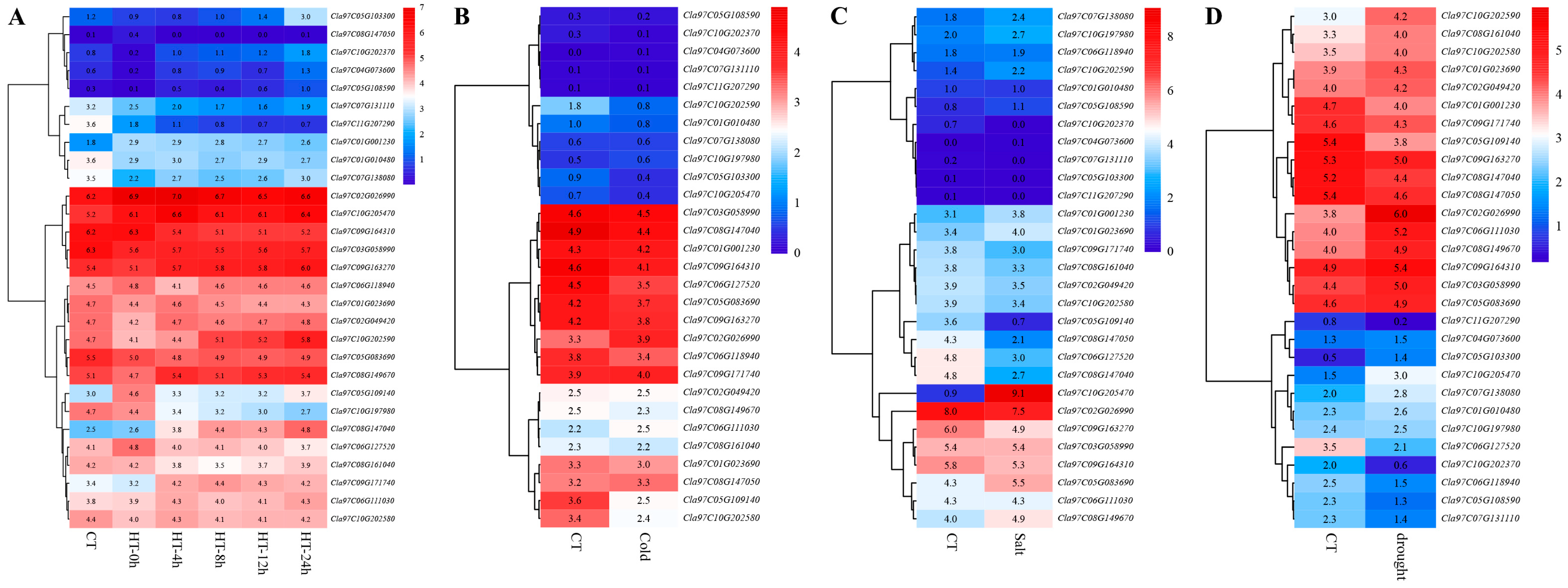
3.6. Expression Patterns of Watermelon Trihelix Genes Under Different Abiotic Stresses
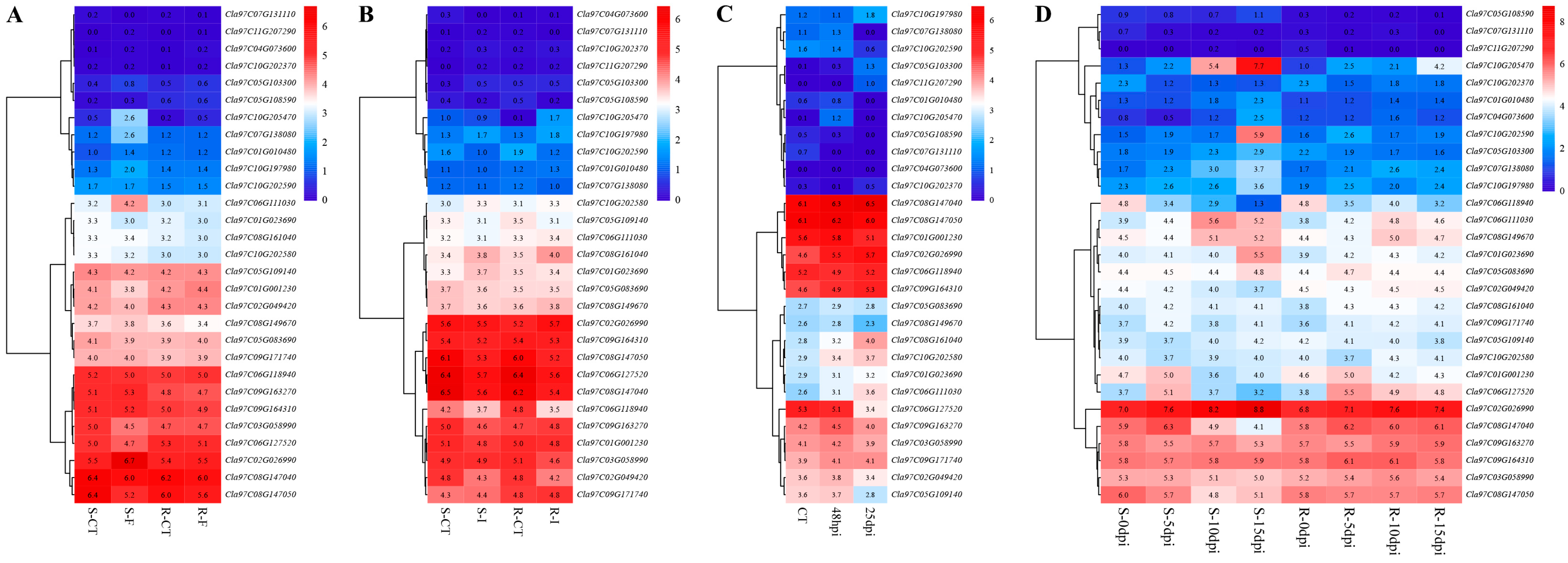
3.7. Expression Patterns of Watermelon Trihelix Genes Under Different Biotic Stresses
3.8. Validation of Gene Transcription Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.X. Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci. 1999, 4, 210–214. [Google Scholar] [CrossRef] [PubMed]
- Ayadi, M.; Delaporte, V.; Li, Y.F.; Zhou, D.X. Analysis of GT-3a identifies a distinct subgroup of trihelix DNA-binding transcription factors in Arabidopsis. FEBS Lett. 2004, 562, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Green, P.J.; Kay, S.A.; Chua, N.H. Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J. 1987, 6, 2543–2549. [Google Scholar] [CrossRef]
- Hiratsuka, K.; Wu, X.; Fukuzawa, H.; Chua, N.H. Molecular dissection of GT-1 from Arabidopsis. Plant Cell 1994, 6, 1805–1813. [Google Scholar]
- Perisic, O.; Lam, E. A tobacco DNA binding protein that interacts with a light-responsive box II element. Plant Cell 1992, 4, 831–838. [Google Scholar]
- Gao, M.J.; Lydiate, D.J.; Li, X.; Lui, H.; Gjetvaj, B.; Hegedus, D.D.; Rozwadowski, K. Repression of seed maturation genes by a trihelix transcriptional repressor in Arabidopsis seedlings. Plant Cell 2009, 21, 54–71. [Google Scholar] [CrossRef]
- Osorio, M.B.; Bücker-Neto, L.; Castilhos, G.; Turchetto-Zolet, A.C.; Wiebke-Strohm, B.; Bodanese-Zanettini, M.H.; Margis-Pinheiro, M. Identification and in silico characterization of soybean trihelix-GT and bHLH transcription factors involved in stress responses. Genet. Mol. Biol. 2012, 35, 233–246. [Google Scholar] [CrossRef]
- Yu, C.; Cai, X.; Ye, Z.; Li, H. Genome-wide identification and expression profiling analysis of trihelix gene family in tomato. Biochem. Biophys. Res. Commun. 2015, 468, 653–659. [Google Scholar] [CrossRef]
- Song, A.; Wu, D.; Fan, Q.; Tian, C.; Chen, S.; Guan, Z.; Xin, J.; Zhao, K.; Chen, F. Transcriptome-wide identification and expression profiling analysis of chrysanthemum trihelix transcription factors. Int. J. Mol. Sci. 2016, 17, 198. [Google Scholar] [CrossRef]
- Kaplan-Levy, R.N.; Brewer, P.B.; Quon, T.; Smyth, D.R. The trihelix family of transcription factors—Light, stress and development. Trends Plant Sci. 2012, 17, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Maréchal, E.; Hiratsuka, K.; Delgado, J.; Nairn, A.; Qin, J.; Chait, B.T.; Chua, N.H. Modulation of GT-1 DNA-binding activity by calcium-dependent phosphorylation. Plant Mol. Biol. 1999, 40, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Yoo, C.Y.; Pence, H.E.; Jin, J.B.; Miura, K.; Gosney, M.J.; Hasegawa, P.M.; Mickelbart, M.V. The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 2010, 22, 4128–4141. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Breuer, C.; Kawamura, A.; Clark, N.M.; Rymen, B.; Braidwood, L.; Morohashi, K.; Busch, W.; Benfey, P.N.; Sozzani, R.; et al. GTL1 and DF1 regulate root hair growth through transcriptional repression of ROOT HAIR DEFECTIVE 6-LIKE 4 in Arabidopsis. Development 2018, 145, dev159707. [Google Scholar] [CrossRef]
- Xi, J.; Qiu, Y.; Du, L.; Poovaiah, B.W. Plant-specific trihelix transcription factor AtGT2L interacts with calcium/calmodulin and responds to cold and salt stresses. Plant Sci. 2012, 185–186, 274–280. [Google Scholar] [CrossRef]
- Luo, J.; Tang, S.; Mei, F.; Peng, X.; Li, X.; Yan, X.; Zeng, X.; Liu, F.; Wu, Y.; Wu, G. BnSIP1-1, a trihelix family gene, mediates abiotic stress tolerance and ABA signaling in Brassica napus. Front Plant Sci. 2017, 8, 44. [Google Scholar] [CrossRef]
- Fu, M.; Li, F.; Zhou, S.; Guo, P.; Chen, Y.; Xie, Q.; Chen, G.; Hu, Z. Trihelix transcription factor SlGT31 regulates fruit ripening mediated by ethylene in tomato. J. Exp. Bot. 2023, 74, 5709–5721. [Google Scholar] [CrossRef]
- Liu, H.F.; Zhang, T.T.; Liu, Y.Q.; Liu, R.X.; Zhnag, H.Y.; Rui, L.; Wang, D.R.; Li, C.Y.; Zhang, S.; You, C.X.; et al. The trihelix transcription factor MdSIP1-2 interacts with MdNIR1 promoter to regulate nitrate utilization in apple. Environ. Exp. Bot. 2024, 220, 105669. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Q.; Wang, H.; Zhang, H.; Xu, X.; Li, C.; Yang, C. Comprehensive analysis of trihelix genes and their expression under biotic and abiotic stresses in Populus trichocarpa. Sci. Rep. 2016, 6, 36274. [Google Scholar] [CrossRef]
- Mo, H.; Wang, L.; Ma, S.; Yu, D.; Lu, L.; Yang, Z.; Yang, Z.; Li, F. Transcriptome profiling of Gossypium arboreum during fiber initiation and the genome-wide identification of trihelix transcription factors. Gene 2019, 709, 36–47. [Google Scholar] [CrossRef]
- Xiao, J.; Hu, R.; Gu, T.; Han, J.; Qiu, D.; Su, P.; Feng, J.; Chang, J.; Yang, G.; He, G. Genome-wide identification and expression profiling of trihelix gene family under abiotic stresses in wheat. BMC Genom. 2019, 20, 287. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, M.; Sun, J.; Mao, X.; Wang, J.; Wang, J.; Liu, H.; Zheng, H.; Zhen, Z.; Zhao, H.; et al. Genome-Wide characterization and identification of trihelix transcription factor and expression profiling in response to abiotic stresses in rice (Oryza sativa L.). Int. J. Mol. Sci. 2019, 20, 251. [Google Scholar] [CrossRef] [PubMed]
- Javed, T.; Shabbir, R.; Ali, A.; Afzal, I.; Zaheer, U.; Gao, S.J. Transcription factors in plant stress responses: Challenges and potential for sugarcane improvement. Plants 2020, 9, 491. [Google Scholar] [CrossRef] [PubMed]
- Lindemose, S.; O’Shea, C.; Jensen, M.K.; Skriver, K. Structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 2013, 14, 5842–5878. [Google Scholar] [CrossRef]
- Abdullah; Faraji, S.; Mehmood, F.; Malik, H.M.T.; Ahmed, I.; Heidari, P.; Poczai, P. The GASA gene family in cacao (Theobroma cacao, malvaceae): Genome wide identification and expression analysis. Agronomy 2021, 11, 1425. [Google Scholar] [CrossRef]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Waterhouse, A.; Procter, J.; Martin, D.A.; Barton, G.J. Jalview: Visualization and analysisof molecular sequences, alignments, and structures. BMC Bioinform. 2005, 6, P28. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Sinsheimer, J.S.; Little, R.J.A.; Lake, J.A. Rooting gene trees without outgroups: EP rooting. Genome Biol. Evol. 2012, 4, 821–831. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Jiang, Z.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.Y.; Dillies, M.A. SARTools: A DESeq2- and EdgeR-based R pipeline for comprehensive differential analysis of RNA-seq data. PLoS ONE 2016, 9, e0157022. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, M.; Tan, J.; Huang, M.; Chu, X.; Li, Y.; Han, X.; Fang, T.; Tian, Y.; Jarret, R.; et al. Telomere-to-telomere Citrullus super-pangenome provides direction for watermelon breeding. Nat. Genet. 2024, 56, 1750–1761. [Google Scholar] [CrossRef]
- Li, H.; Dong, Y.; Chang, J.; He, J.; Liu, Q.; Wei, C.; Ma, J.; Zhang, Y.; Yang, J.Q.; Zhang, X. High-throughput microRNA and mRNA sequencing reveals that microRNAs may be involved in melatonin-mediated cold tolerance in Citrullus lanatus L. Front. Plant Sci. 2016, 8, 01231. [Google Scholar] [CrossRef]
- Song, Q.; Joshi, M.; Joshi, V. Transcriptomic analysis of short-term salt stress response in watermelon seedlings. Int. J. Mol. Sci. 2020, 21, 6036. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Mo, Y.; Yang, X.; Zhang, H.; Wang, Y.; Li, H.; Wei, H.; Zhang, X. Transcriptome profiling of watermelon root in response to short-term osmotic stress. PLoS ONE 2016, 11, e0166314. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Wang, Z.; Guo, Y.; Zhang, X. Comparative transcriptome profiling reveals the role of phytohormones and phenylpropanoid pathway in early-stage resistance against powdery mildew in watermelon (Citrullus lanatus L.). Front. Plant Sci. 2022, 10, 1016822. [Google Scholar] [CrossRef] [PubMed]
- Yasmeen, E.; Riaz, M.; Sultan, S.; Azeem, F.; Abbas, A.; Riaz, K.; Ali, M.A. Genome-wide analysis of trihelix transcription factor gene family in Arabidopsis thaliana. Pak. J. Agric. Sci. 2016, 53, 439–448. [Google Scholar]
- Ma, W.; Yang, D.; Qiu, M.; Gao, J.; Cui, R. Genome-wide identification and expression pattern analysis of the trihelix gene family in cucumber. Pak. J. Bot. 2024, 56, 1853–1866. [Google Scholar] [CrossRef]
- Wang, R.; Hong, G.; Han, B. Transcript abundance of rml1, encoding a putative GT1-like factor in rice, is up-regulated by Magnaporthe grisea and down-regulated by light. Gene 2004, 324, 105–115. [Google Scholar] [CrossRef]
- Breuer, C.; Kawamura, A.; Ichikawa, T.; Tominaga-Wada, R.; Wada, T.; Kondou, Y.; Muto, S.; Matsui, M.; Sugimoto, K. The trihelix transcription factor GTL1 regulates ploidy-dependent cell growth in the Arabidopsis trichome. Plant Cell 2009, 21, 2307–2322. [Google Scholar] [CrossRef]
- Fang, Y.; Xie, K.; Hou, X.; Hu, H.; Xiong, L. Systematic analysis of GT factor family of rice reveals a novel subfamily involved in stress responses. Mol. Genet. Genom. 2010, 283, 157–169. [Google Scholar] [CrossRef]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Ma, H.; dePamphilis, C.W. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef]
- Roulin, A.; Auer, P.L.; Libault, M.; Schlueter, J.; Farmer, A.; May, G.; Stacey, G.; Doerge, R.W.; Jackson, S.A. The fate of duplicated genes in a polyploid plant genome. Plant J. 2013, 73, 143–153. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, C.; Li, L.; Meng, L.; Singh, J.; Jiang, N.; Deng, X.W.; He, Z.H.; Lemaux, P.G. Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 2005, 139, 1107–1124. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Tyagi, A.K.; Khurana, J.P. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 2006, 88, 360–371. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, M.; Kaplan-Levy, R.; Tezz, Q.; Sappl, P.; Smyth, D. PETAL LOSS, a trihelix transcription factor that represses growth in Arabidopsis thaliana, binds the energysensing SnRK1 kinase AKIN10. J. Exp. Bot. 2015, 9, 2475–2485. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Qi, S.; Touqeer, A.; Li, H.; Zhang, X.; Liu, X.; Wu, S. SlGT11 controls floral organ patterning andbfloral determinacy in tomato. BMC Plant Biol. 2020, 20, 562. [Google Scholar] [CrossRef]
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A.; et al. The Sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551–556. [Google Scholar] [CrossRef]
- Liu, X.; Wu, D.; Shan, T.; Xu, S.; Qin, R.; Li, H.; Negm, M.; Wu, D.; Li, J. The trihelix transcription factor OsGTγ-2 is involved adaption to salt stress in rice. Plant Mol. Biol. 2020, 103, 545–560. [Google Scholar] [CrossRef]
- Li, F.; Chen, G.; Xie, Q.; Zhou, S.; Hu, Z. Down-regulation of SlGT-26 gene confers dwarf plants and enhances drought and salt stress resistance in tomato. Plant Physiol. Biochem. 2023, 203, 108053. [Google Scholar] [CrossRef]
- Mao, H.; Zhang, W.; Lv, J.; Yang, J.; Yang, S.; Jia, B.; Song, J.; Wu, M.; Pei, W.; Ma, J.; et al. Overexpression of cotton Trihelix transcription factor GhGT-3b_A04 enhances resistance to Verticillium dahliae and affects plant growth in Arabidopsis thaliana. J. Plant Physiol. 2023, 283, 153947. [Google Scholar] [CrossRef]
- Wang, T.; Wang, G.; Zhang, J.; Xuan, J. E3 ubiquitin ligase PUB23 in kiwifruit interacts with trihelix transcription factor GT1 and negatively regulates immune responses against Pseudomonas syringae pv. actinidiae. Int. J. Mol. Sci. 2024, 25, 1930. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef]



| Gene Name | Forward Primer (5′-3′) | Reverse Prime (5′-3′) | Size (bp) |
|---|---|---|---|
| Actin | CCATGTATGTTGCCATCCAG | GGATAGCATGGGGTAGAGCA | 135 |
| Cla97C05G083690 | GAATCTCAAGGCTACTCACT | CTCCATCTTATGACGACACT | 106 |
| Cla97C05G109140 | CCGCCATGTCTCTTCAAG | CGGAGGCAGATTCAGAAC | 110 |
| Cla97C08G147040 | CTCCACTACTTCCGTCTTG | TCATCCTCACCAGAATTGTT | 126 |
| Cla97C08G147050 | AGTGGGAAATCCGATAACAG | GGTGGTGGTGAGATTGGA | 115 |
| Cla97C09G164310 | AGACAACAAGCAGAACAGT | GCATACGAGGAGCAAGTT | 113 |
| Cla97C09G171740 | AGCATCAGCAACATACTCC | CCTCTTCTTCTTCTTCCTCTT | 103 |
| Cla97C10G197980 | GTTGGGACCCTGTATTGG | TCTCGTAATGTGGACATCC | 114 |
| Cla97C10G205470 | CCATTCTACACAGAGTTACAAG | TTCTTCGTCATCGTCAGAC | 129 |
| Gene ID | Name | CDS Size (bp) | Number of Amino Acids (aa) | Molecular Weight (kD) | pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Prediction of Subcellular Location |
|---|---|---|---|---|---|---|---|---|---|
| Cla97C01G001230 | ClGT1 | 2091 | 696 | 77.00 | 5.05 | 69.59 | 57.44 | −0.827 | Nuclear |
| Cla97C01G010480 | ClGT2 | 1473 | 490 | 55.20 | 5.86 | 60.44 | 68.88 | −0.701 | Nuclear |
| Cla97C01G023690 | ClGT3 | 1386 | 461 | 51.39 | 8.94 | 54.43 | 73.38 | −0.633 | Nuclear |
| Cla97C02G026990 | ClGT4 | 1473 | 490 | 56.13 | 5.55 | 51.33 | 62.76 | −1.025 | Nuclear |
| Cla97C02G049420 | ClGT5 | 1275 | 424 | 48.04 | 6.74 | 50.98 | 66.65 | −0.826 | Nuclear |
| Cla97C03G058990 | ClGT6 | 936 | 311 | 34.96 | 5.74 | 47.88 | 66.24 | −0.629 | Nuclear |
| Cla97C04G073600 | ClGT7 | 300 | 99 | 11.73 | 9.51 | 48.06 | 70 | −0.705 | Nuclear |
| Cla97C05G083690 | ClGT8 | 1083 | 360 | 39.75 | 9.33 | 66.93 | 52.36 | −0.919 | Nuclear |
| Cla97C05G103300 | ClGT9 | 1233 | 410 | 46.50 | 6.38 | 47.81 | 54.73 | −0.874 | Nuclear |
| Cla97C05G108590 | ClGT10 | 2712 | 903 | 101.54 | 5.79 | 42.65 | 83.93 | −0.521 | Nuclear |
| Cla97C05G109140 | ClGT11 | 1053 | 350 | 38.86 | 9.59 | 63.59 | 61.63 | −0.962 | Nuclear |
| Cla97C06G111030 | ClGT12 | 1068 | 355 | 38.67 | 5.4 | 50.41 | 75.61 | −0.642 | Nuclear |
| Cla97C06G118940 | ClGT13 | 1872 | 623 | 72.13 | 5.99 | 62.88 | 61.36 | −1.159 | Nuclear |
| Cla97C06G127520 | ClGT14 | 2709 | 902 | 99.89 | 8.71 | 46.03 | 86.78 | −0.381 | Nuclear |
| Cla97C07G131110 | ClGT15 | 858 | 285 | 32.33 | 9.56 | 48.74 | 75.61 | −0.615 | Nuclear |
| Cla97C07G138080 | ClGT16 | 978 | 325 | 38.06 | 8.67 | 57.83 | 61.05 | −1.003 | Nuclear |
| Cla97C08G147040 | ClGT17 | 1920 | 639 | 71.53 | 6.06 | 69.98 | 55.27 | −1.052 | Nuclear |
| Cla97C08G147050 | ClGT18 | 1638 | 545 | 61.81 | 5.66 | 64.55 | 51.89 | −1.096 | Nuclear |
| Cla97C08G149670 | ClGT19 | 1401 | 466 | 52.78 | 6.36 | 42.29 | 67.58 | −0.893 | Nuclear |
| Cla97C08G161040 | ClGT20 | 1224 | 407 | 46.61 | 5.83 | 53.11 | 61.84 | −0.826 | Nuclear |
| Cla97C09G163270 | ClGT21 | 1128 | 375 | 41.20 | 9.67 | 46.21 | 59.04 | −0.97 | Nuclear |
| Cla97C09G164310 | ClGT22 | 1161 | 386 | 44.45 | 4.81 | 50.15 | 50.03 | −1.26 | Nuclear |
| Cla97C09G171740 | ClGT23 | 921 | 306 | 35.41 | 5.38 | 74.28 | 64.12 | −0.925 | Nuclear |
| Cla97C10G197980 | ClGT24 | 981 | 326 | 37.76 | 6.97 | 46.64 | 66.44 | −0.839 | Nuclear |
| Cla97C10G202370 | ClGT25 | 1575 | 524 | 59.43 | 6.53 | 49.59 | 52.6 | −1.087 | Nuclear |
| Cla97C10G202580 | ClGT26 | 1815 | 604 | 66.90 | 6.43 | 44.84 | 73.44 | −0.408 | PlasmaMembrane |
| Cla97C10G202590 | ClGT27 | 1554 | 517 | 58.09 | 5.8 | 62.93 | 57.16 | −0.969 | Nuclear |
| Cla97C10G205470 | ClGT28 | 915 | 304 | 35.54 | 6.82 | 52.11 | 55.59 | −1.128 | Nuclear |
| Cla97C11G207290 | ClGT29 | 1197 | 398 | 44.88 | 9.02 | 71.04 | 61.31 | −0.951 | Nuclear |
| Homologs Genes | Ka | Ks | Ka/Ks | p-Value |
|---|---|---|---|---|
| Cla97C01G023690-Cla97C07G131110 | 0.452 | 3.845 | 0.118 | 2.912 × 10−25 |
| Cla97C03G058990-Cla97C07G131110 | 0.470 | 3.849 | 0.122 | 2.355 × 10−36 |
| Cla97C04G073600-Cla97C10G205470 | 0.392 | 3.084 | 0.127 | 1.776 × 10−15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Cheng, H.; Liang, Z.; Su, Y.; Shi, L.; Qin, N. Genome-Wide Identification of Watermelon Trihelix Genes and Their Expression Patterns Under Biotic and Abiotic Stresses. Horticulturae 2025, 11, 275. https://doi.org/10.3390/horticulturae11030275
Wang Y, Cheng H, Liang Z, Su Y, Shi L, Qin N. Genome-Wide Identification of Watermelon Trihelix Genes and Their Expression Patterns Under Biotic and Abiotic Stresses. Horticulturae. 2025; 11(3):275. https://doi.org/10.3390/horticulturae11030275
Chicago/Turabian StyleWang, Yunan, Hui Cheng, Zhonghao Liang, Yuting Su, Lijing Shi, and Nannan Qin. 2025. "Genome-Wide Identification of Watermelon Trihelix Genes and Their Expression Patterns Under Biotic and Abiotic Stresses" Horticulturae 11, no. 3: 275. https://doi.org/10.3390/horticulturae11030275
APA StyleWang, Y., Cheng, H., Liang, Z., Su, Y., Shi, L., & Qin, N. (2025). Genome-Wide Identification of Watermelon Trihelix Genes and Their Expression Patterns Under Biotic and Abiotic Stresses. Horticulturae, 11(3), 275. https://doi.org/10.3390/horticulturae11030275





