Abstract
NRL (NPH3/RPT2-Like) proteins, which are exclusive to plants, serve as critical mediators in phototropic signaling by dynamically regulating light-dependent cellular processes. We identified 24 NRL genes (VvNRL) in the Vitis vinifera L. genome, which were unevenly distributed on 11 chromosomes. Phylogenetic analysis showed that these family members were divided into six groups, and promoter analysis revealed ubiquitous light-responsive cis-elements across all 24 members, suggesting conserved regulatory mechanisms. Sequence alignment and structural similarity analysis showed that VvNRL4 and VvNRL6 were highly similar to NPH3 and NPR2. Experiments with different light qualities showed that VvNRL6 was induced by blue and red light, while VvNRL4 was not affected by light spectra, similar to NPH3 in Arabidopsis. Molecular docking prediction suggested that VvNRL4 and VvNRL6 may, respectively, interact with the LOV domain in VvPHO1 and VvPHO2, through their C-terminal coiled-coil domain and N-terminal BTB domain, to further regulate the activity of VvPHO1 and VvPHO2. In addition, 10 of the 24 VvNRLs, including VvNRL4 and VvNRL6, possessed the conserved RxS motif in their conserved C-terminal consensus sequences. This study provides a reference for further studies on the function and regulation of VvNRL family members in fruit vine plants.
1. Introduction
During the growth and morphogenesis of plants, they respond to changes in wavelength, intensity and direction of external light signals. One of these responses is phototropism, which refers to the phenomenon where plant organs exhibit bending towards or away from the specific direction of a light source [1]. In this process, the blue light receptors in plants, known as PHOT (phototropins), mediate many physiological responses such as phototropism, chloroplast movement, stomatal opening, leaf flattening, and positioning, enabling plants to adapt to different light intensities. In the model plant Arabidopsis, PHOT1 and PHOT2 regulate plant physiological responses in a light intensity-dependent manner. Specifically, PHOT1 alone mediates phototropism in hypocotyls under weak blue light conditions [2]. PHOT2 not only mediates chloroplast avoidance movement and nuclear positioning induced by strong blue light, but also regulates the positioning of chloroplasts and nuclei under dark conditions [3,4,5].
In addition, PHOT1 and PHOT2 also collectively mediate physiological responses, including the aggregation of chloroplasts, the opening of stomata, leaf flattening and positioning, as well as an increase in cytoplasmic Ca2+ [2,6,7] So far, multiple downstream genes of PHOT have been identified. Among them, NPH3 (Non-Phototropic Hypocotyl 3) from the NRL family is one of the earliest identified downstream members of PHOT [8]. The nph3 mutant does not exhibit phototropic bending under unilateral blue light illumination, indicating that it is an essential signaling component for phototropism [9].
As a plant-specific protein family, the NRL (NPH3/RPT2-Like) family is named after its two founding members, NPH3 and RPT2 (Root Phototropism 2) [10]. They were initially identified through genetic screening of Arabidopsis mutants with impaired hypocotyl and root phototropism [11,12,13]. NRL proteins exhibit a primary structure composed of three conserved regions: an N-terminal BTB (Bric-a-Brac, tramtrack, and broad complex) domain, a central NPH3 domain, and a C-terminal coiled-coil domain [10]. Furthermore, beyond NPH3 and RPT2, Arabidopsis encodes 31 additional NPH3/RPT2-like proteins (NRL1–NRL31) characterized by the presence of the NPH3 domain; among these, ten lack the C-terminal coiled-coil domain and two are missing the BTB domain [14]. The reasons for these structural variations are currently unclear, as many of these NRL proteins have not been assigned biological functions.
So far, the functions of 33 out of 10 identified Arabidopsis NRL proteins have been determined. Phylogenetic analysis indicates that in land plants, NRL proteins fall into seven distinct clades [15]. However, in Arabidopsis and other angiosperms, only six of these branches exist. NRL proteins classified into three of these branches have been shown to interact with phototropins [15]. These include NPH3 [8], RPT2 [9], NRL protein 1 (NCH1/NRL31) [15], and NRL2 [16] involved in chloroplast movement. NRL2 has been identified as a phot1 interactor via yeast two-hybrid screening, although its exact role in phototransduction remains unresolved [16]. In addition, NPH3 and RPT2 are crucial for auxin-mediated processes such as phototropism, leaf positioning, and expansion [17]. Beyond these, other NRL family members also play central roles in coordinating auxin distribution independently of phototropins. For instance, NRL23 (Defectively Organized Tributaries3) contributes to vascular development and leaf vein patterning [18], while NRL proteins including NRL6, NRL7, NRL20, NRL21, and NRL30—collectively known as naked pins in mosses (NPY)—regulate auxin transport needed for organogenesis and gravitropic root bending via AGCVIII kinases (excluding phosphorus removal) [19,20]. Moreover, NRL8 (also known as SETH6) is involved in pollen tube growth [21], a process that also depends on AGC kinase activity [22]. Collectively, these findings support the proposal that NRL proteins act as AGC kinase modules orchestrating various aspects of auxin transport and signal transduction.
Currently, the NRL gene family has been identified in model plants such as Arabidopsis, rice (Oryza sativa), tomato (Solanum lycopersicum), and maize (Zea mays) [15,23,24], but no reports have been found in the woody plant grapevine. Light plays a crucial role in the production and growth of grapes, and different light qualities also have a significant impact on the aroma components and flavor of grapes. The study of the NRL gene family in grapes holds important theoretical and practical significance. In this research, we conducted a whole-genome analysis of VvNRL and confirmed the presence of 24 VvNRL members. Then, using a series of bioinformatics methods, we analyzed the chromosomal location, phylogenetic tree, gene structure, conserved motifs, protein tertiary structure, multiple sequence comparison, collinearity, promoter cis-elements, and phosphorylation sites of these VvNRL genes. Based on the VvNRL PPI (protein–protein interaction network), we focused on the functions of VvNRL4 and VvNRL6 and examined their expression levels under different light qualities (blue, red, and green light). Additionally, utilizing the predicted 3D structures from alphafold3 and previous studies, we predicted and simulated protein–protein docking of VvNRL4, VvNRL6, VvPHO1, and VvPHO2. The aim of this research is to elucidate the potential function of VvNRL4 and provide a theoretical basis for uncovering the functional aspects of NRL genes in grapes.
2. Materials and Methods
2.1. Genome-Wide Identification and Characterization of VvNRLs in Grape
To identify all sequences related to the VvNRL family, we used the 33 AtNRL protein sequences of Arabidopsis as queries in the BLAST program (version 2.14.0) on the Ensembl Plants website (https://plants.ensembl.org/index.html, accessed on 28 August 2024) against the genomes of Vitis vinifera L. Additionally, HMMER 3.0 was utilized to perform hidden Markov model searches for candidate proteins containing NPH3 (PF03000) or BTB/POZ (IPR000210) and coiled-coil domains in two databases: UniProt (https://www.uniprot.org/, accessed on 3 October 2024) and InterPro (https://www.ebi.ac.uk/interpro/, accessed on 8 October 2024).
A total of 24 proteins were predicted as candidate grape VvNRL proteins. Their physicochemical properties—including molecular weight (MW), isoelectric point (PI), and the grand average of hydropathicity (GRAVY)—were computed using the ExPASy platform (www.expasy.org/, accessed on 14 November 2024). Additionally, the subcellular localization of these VvNRLs was predicted via the Cell-PLoc 2.0 tool (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/, accessed on 14 November 2024).
Chromosomal location and gene structure information of VvNRLs were depicted using TBtools (version 2.154) [25], based on the GFF annotation file from Ensembl Plants. The lengths of conserved domains were identified using InterPro (https://www.ebi.ac.uk/interpro/, accessed on 22 November 2024) and SMART (http://smart.embl-heidelberg.de/, accessed on 24 November 2024).
2.2. Phylogenetic Analysis of VvNRLs
Amino acid sequences for NRL proteins from Arabidopsis, a model dicot, and Oryza sativa, a representative monocot, were downloaded from TAIR (https://www.arabidopsis.org/index.jsp, accessed on 27 August 2024) and Rap-db (http://rapdb.dna.affrc.go.jp/index.html, accessed on 27 August 2024), respectively, for multiple sequence alignment. The VvNRL protein sequences were then aligned using MEGA version 11. The phylogenetic tree was constructed employing the neighbor-joining method based on amino acid p-distance, with reliability assessed through 1000 bootstrap replicates using the JTT model. Finally, the interactive Tree of Life (iTOL version 6) tool (https://itol.embl.de/, accessed on 17 January 2025) was used to generate the phylogenetic tree of the NRL proteins.
2.3. Spatiotemporal Expression Profiling During Grapevine Development
The expression profiles of the grapevine NRL genes were analyzed in the Vitis vinifera L. gene expression atlas of different organs at various developmental stages. The dataset for GSE36128 contained 54 tissues and organs of Vitis vinifera cv. ‘Corvina’ [26], which can survey the tissue-specific expression, and the heatmap was constructed using TBtools software [25], with the raw datasets standardized using min–max normalization.
To investigate the expression levels of VvNRLs under different light conditions, we utilized previously published data from Li et al. [27]. Briefly, leafy single-node cuttings (15 mm long) forming the grape cultivar “ManicureFinger” (Vitis vinifera L.) were cultured in vitro on 3/4 Murashige and Skoog medium supplemented with 0.35 mg/L indole butyric acid, 3% sucrose, and 0.55% agar. Following a 24 h dark incubation period, the cultures were exposed to normal (white light), blue light (peak at 440 nm), green light (peak at 520 nm), or red light (peak at 630 nm) fixtures for 40 days. Subsequently, leaf samples were collected for RNA extraction using TRIzol reagent (Invitrogen, Waltham, MA, USA) according to the manufacturer’s protocol. The RNA concentration and purity were measured with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and RNA integrity was verified by 1% agarose gel electrophoresis. All RNA samples were stored at −80 °C. The forward and reverse primers used were 5′-AGATGATGGAGTCCTGGGTTCAAG -3′ and 5′-TCATGACCTTCCAGGCGGACCTGAG-3′ for VvNRL4, 5′-ATGGCCGCCTCTCTCAAGGG-3′ and 5′-GGTGGAAAGGGATTAGGC-3′ for VvNRL6, 5′-AGGACGACACCCGCCAGAA-3′ and 5′-AGGAGGAACTCAGTGCGGAG-3′ for VvPHO1, and 5′-TGGAGGTGTTTGAACCGGCA-3′ and 5′-GGCATGAAGTTCTCGTCTGT-3′ for VvPHO2. To further analyze the expression patterns of VvNRLs under different light conditions, we performed multiple sequence alignment using original sequences and gene IDs extracted from a previous version of the reference genome (https://ftp.ensemblgenomes.ebi.ac.uk/pub/plants/release-60/fasta/vitis_vinifera/, accessed on 1 March 2025). We then matched these with the latest grape reference genome (https://plants.ensembl.org/index.html, accessed on 1 March 2025) to obtain RPKM (reads per kb per million reads) values for the 24 VvNRLs and to quantify their expression levels.
2.4. Identification of Cis-Acting Elements in VvNRL Promoters
To study cis-regulatory elements in the promoters of the VvNRL genes, the upstream 2000 bp regulatory regions (from the translation start site) of VvNRL genes were obtained from the grape database. The cis-regulatory elements were identified using PLACE (http://www.dna.affrc.go.jp/PLACE/?action=newplace, accessed on 1 March 2025).
2.5. Chromosomal Location and Synteny Correlation Analysis
For chromosomal locations of VvNRL genes, a Circos diagram was illustrated by annotating genes with respect to their specific chromosomal position in the genome annotation by using the TBtools software [25]. The syntenic gene relationship between the homologs of Arabidopsis and Vitis vinifera L. and within the Vitis vinifera L. was also verified. These syntentic or collinear analyses were carried out using MCScanX with gene duplication parameters [28].
2.6. Protein–Protein Interaction Network and GO Enrichment
To construct the protein–protein interaction (PPI) network between VvNRL in Vitis vinifera L., the sequences of all VvNRLs were submitted to the STRING v11.5 database (https://version-11-5.string-db.org/, accessed on 2 December 2024). The maximum number of interactors was adjusted to no more than 10 interactors for the first shell and the minimum required interaction score, with high confidence (0.700). Finally, interaction networks were illustrated using Cytoscape (v3.10.0). GO enrichment was performed with the accession numbers of significant DEGs via agriGO v2.0 (http://systemsbiology.cau.edu.cn/agriGOv2, accessed on 7 December 2024).
2.7. Three-Dimensional Structure and Protein–Protein Docking
High-confidence 3D protein structures were predicted by Alphafold3 (https://alphafold.com/, accessed on 8 December 2024). Protein–protein docking predictions were performed using the HDOCK website (http://hdock.phys.hust.edu.cn, accessed on 12 December 2024) based on Yan et al. [29], and the docking pose with the highest score was used for each interaction result. The similarity of protein structures was calculated using PyMOL software (version 2.5) and quantified using root mean square deviation (RMSD).
2.8. Prediction of Phosphorylation Sites and Disorder Regions in VvNRLs
The phosphorylation sites of each VvNRL protein in Vitis vinifera L. were predicted based on three amino acids, including serine (S), threonine (T), and tyrosine (Y), using the GPS 6.0 web server (http://gps.biocuckoo.cn/online.php, accessed on 17 December 2024) [30]. Prediction of the sites with a high percentage of confidence was based on Sp values of 98%. Intrinsic disorder regions (IDR) were predicted by an in silico analysis using flDPnn [31].
3. Results
3.1. Genome-Wide Identification of the VvNRL Family in the Grape Genome
In this study, 27 candidate NRL family genes in Vitis vinifera L. were obtained using local BLAST and HMMER analysis methods (Figure S1). Subsequently, the predicted grapevine NRL family genes were screened using InterPro (https://www.ebi.ac.uk/interpro/, accessed on 17 December 2024), SMART (http://smart.embl.de/, accessed on 1 March 2025), and Pfam (http://pfam.xfam.org/, accessed on 9 January 2025). Finally, 24 rice NRL family genes were identified and named VvNRL1~VvNRL24 (Table 1). The coding region length of rice NRL family genes ranged from 1302 to 2025 bp, while their amino acid sequence length ranged from 434 to 675. Predicted protein molecular weight ranged from 48.42 to 76.31 kD, and theoretical isoelectric point ranged from 5.12 to 9.17. Subcellular localization prediction indicated that all grapevine NRL gene family members were predicted to be located on the cell membrane, with 12 NRL gene family members predicted to be located in both the nucleus and cell membrane and 2 NRL gene family members predicted to be located in the chloroplast and cell membrane. The 24 grapevine VvNRLs are mainly distributed on the 11 chromosomes of grapevine (Figure 1a). Among them, the VvNRLs on chromosome 7 have the highest distribution, with a total of seven genes located on chromosome 7 (VvNRL8, VvNRL9, VvNRL10, VvNRL11, VvNRL12, VvNRL13, and VvNRL14), while only one NRL gene is distributed on chromosomes 4, 9, 13, and 15. We then analyzed the conserved domains of the 24 NRLs. The results showed that all NRLs contain the NPH3 domain (Figure 1b). Except for VvNRL17, all other members contain the BTB/POZ domain at the N-terminus. However, compared to other members, VvNRL17 lacks two exons at the 5′ end and only contains two exon regions, while other members contain three to six exon regions (Figure 1c). In addition to the NPH3 and BTB/POZ domains, some NRL members also contain the coiled-coil domain, which is another typical feature of the NRL family. Among the 24 members, 19 members contain the coiled-coil domain at the C-terminus, with NRL24 containing two coiled-coil domains. It is worth noting that although VvNRL12 contains all the conserved domains, its NPH3 domain is significantly shorter than other members, which indicates its independent evolution compared to other members.

Table 1.
Summary of 24 VvNRL genes identified in the genome of Vitis vinifera L.
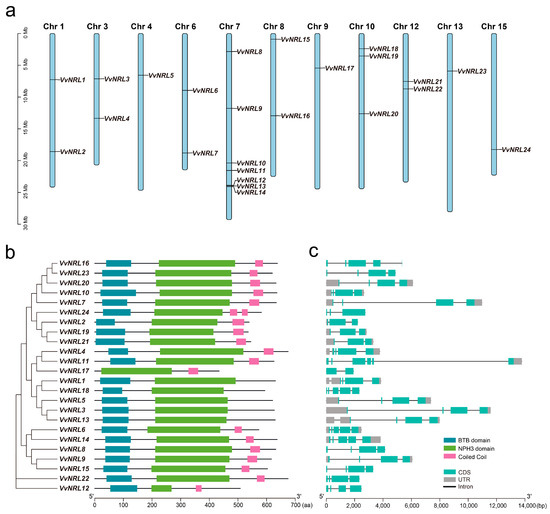
Figure 1.
Chromosome distribution, phylogenetic relationship, gene structure, and conserved domain analysis of VvNRLs. (a) Distribution of VvNRL genes on grape chromosomes. The figure displays the size of each chromosome, indicated by a scale on the left side. (b) Analysis of conserved domains in VvNRLs. Different motifs are represented by boxes in various colors. The sequence sizes (in amino acids, aa) are shown with a scale at the bottom of the figure. (c) Exon–intron structures of VvNRL genes. Untranslated regions (UTR), exons, and introns are depicted with gray boxes, green boxes, and gray lines, respectively. The sequence sizes (bp) are shown with a scale at the bottom of the figure.
3.2. Phylogenetic Analysis of the Grapevine VvNRL Gene Family’s Evolutionary Relationships
In order to further understand the functionality of the grapevine NRL gene family, NRL protein sequences from Arabidopsis (33 members) and Oryza sativa (27 members) were obtained from the online database Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html, accessed on 9 January 2025) and a systematic evolutionary relationship analysis was performed (Figure 2a). The results showed a total of 84 NRLs, which could be classified into six groups (Group I~VI) consisting of 5, 8, 21, 19, 21, and 10 members, respectively. The grapevine NRL family members were evenly distributed among these six groups. Specifically, VvNRL12 and VvNRL22 were in Group I; VvNRL6, 8, 9, 14, and 15 were in Group IV; VvNRL4, 11, and 17 were in Group VI; VvNRL1, 3, 5, 13, and 18 were in Group V; VvNRL2, 19, and 21 were in Group II; and VvNRL7, 10, 16, 20, 23, and 24 were in Group III (Figure 2a,b). It is worth noting that VvNRL4 showed a close evolutionary relationship with AtNPH3 and VvNRL6 showed a close evolutionary relationship with AtRPT2, suggesting that they may have similar functionalities.
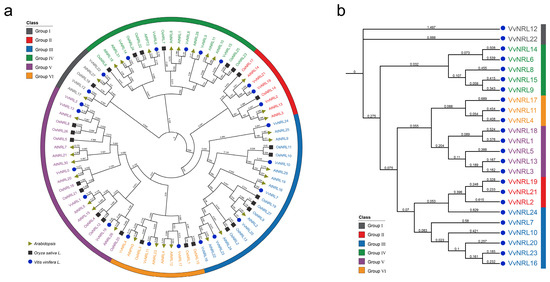
Figure 2.
Phylogenetic analysis of the NRL gene family in grapevine, rice, and Arabidopsis. (a) The phylogenetic analysis of NRL proteins between grapevine, rice, and Arabidopsis. (b) The phylogenetic analysis of NRL proteins in grapevine. A total of 84 NRL proteins were analyzed, including 27 members from Oryza sativa subsp. Japonica, 33 members from Arabidopsis, and 24 members from Vitis vinifera L. The analysis was performed using MEGA 7.0 with the neighbor-joining (NJ) method and JTT matrix-based model. Bootstrap was set to 1000. Oryza sativa is represented by black squares, Arabidopsis by triangles, and Vitis vinifera L. by circles.
3.3. Expression Pattern Analysis of VvNRL Genes
To gain insight into the potential functions of VvNRL genes during grapevine development, we conducted a gene expression atlas analysis using 54 different organs and tissues at various developmental stages (Figure 3, Table S1) based on the GEO DataSets (GSE36128). In order to compare the expression levels of VvNRL genes within the same tissues, the data were normalized. The results showed that VvNRL4, VvNRL5, VvNRL6, and VvNRL13 exhibited higher expression levels in most tissues compared to other genes. Although the expression levels of other genes were slightly lower than these four genes, some genes demonstrated significant tissue specificity. For example, VvNRL23 displayed high expression in pollen, while VvNRL23 was highly expressed in mature stems. These findings indicate the functional diversification of VvNRL genes during grapevine growth and development.
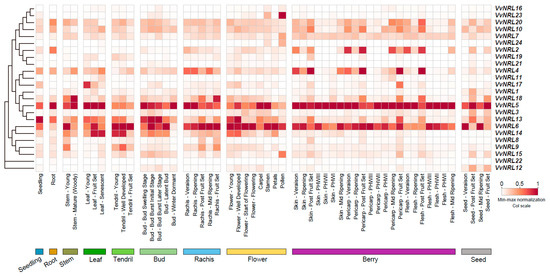
Figure 3.
Illustration of the expression patterns of the grapevine NRL gene family in different tissues at various developmental stages. The expression data were derived from the GEO DataSets (GSE36128), and subsequently, the expression levels were normalized using min–max standardization.
3.4. Analysis of Cis-Acting Elements in Grapevine NRL Gene Family Promoters
To further understand the functionality of the grapevine NRL gene family, an analysis of cis-acting elements was performed on the upstream 2000 bp promoter sequences of the grapevine NRL gene family (Figure 4a). A total of 484 responsive elements were identified in the promoter regions of the 24 VvNRL genes, which could be categorized into three groups based on their functions. Among them, 85 elements were related to light responsiveness, 168 were involved in phytohormone-responsive pathways, and 231 were associated with development and abiotic and biotic stress (Figure 4b). It is worth noting that all genes, except for VvNRL16, contained one to eight light-responsive elements.
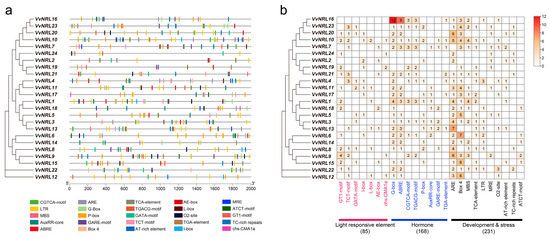
Figure 4.
Analysis of cis-acting elements in the promoter sequences of the grapevine NRL protein family members. (a) Comparison of VvNRLs’ promoter sequences: distribution of cis-elements on the promoter regions of VvNRLs. A total of 484 cis-elements were identified. Predicted putative cis-acting elements were generated using PlantCARE, and different colored bars represent different types of cis-elements. (b) Number and percentage of cis-element occurrence in the upstream sequences of 24 VvNRLs in grapevine. Depth of red represents the number of cis-acting regulatory elements.
3.5. Segmental Duplication Events Are Observed in VvNRLs
A gene family consists of a group of genes derived from a common ancestor, formed by gene duplication. Typically, these genes exhibit apparent similarities in structure and function, encoding similar protein products. Genes belonging to the same family can be closely clustered together, forming a gene cluster. However, most of the time, they are dispersed in different locations on the same chromosome or even exist on different chromosomes.
To further understand the evolutionary behavior of the grapevine NRL gene family both between species and within species, we conducted an analysis of gene collinearity in grapevine and Arabidopsis to elucidate the expansion process of NRL family members (Figure 5). In the grapevine genome, we discovered two segmental duplication events: VvNRL19 and VvNRL21, as well as VvNRL16 and VvNRL13. Additionally, several genes in the Arabidopsis genome exhibited collinear relationships with grapevine NRL genes; for example, VvNRL6 and AtRPT2, VvNRL7 and AtNRL16, VvNRL8 and AtNRL1, VvNRL8 and AtNRL8/SETH6, VvNRL9 and AtNRL28, VvNRL12 and AtNRL17, VvNRL14 and AtNRL18, VvNRL14 and AtNRL31/NCH1, VvNRL15 and AtNRL11, VvNRL16 and AtNRL22, VvNRL17 and AtNRL4, VvNRL17 and AtNRL12, VvNRL20 and AtNRL2, and VvNRL21 and AtNRL1.
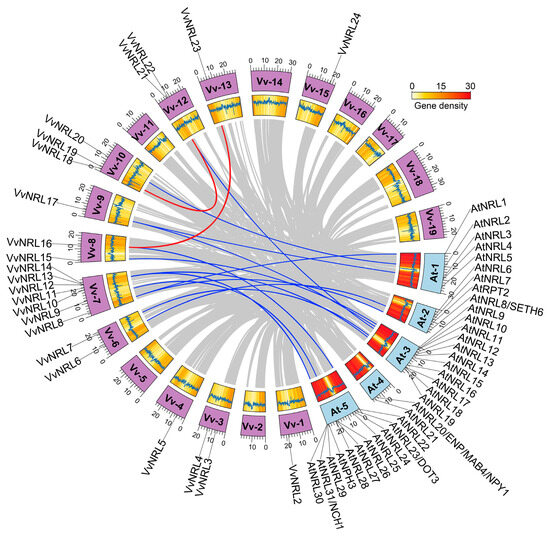
Figure 5.
Illustration of the distribution and synteny analysis of VvNRLs and AtNRLs with respect to the chromosomes of grapevine and Arabidopsis. Synteny relationships between VvNRLs themselves, as well as between AtNRLs and VvNRLs, are depicted by connecting lines in red and blue, respectively. The syntenic relationships of other genes are shown using gray lines. Chromosome lengths are indicated to scale, and gene density is represented using a heatmap, with a window size of 100 kb. The line graph represents CG content, with a window size of 10 kb. Vv1–Vv19 corresponds to the 19 grapevine chromosomes, while At1–At5 represents the 5 Arabidopsis chromosomes.
3.6. Protein Docking of VvNRL4, VvNRL6 and VvPHO1, and VvPHO2
Two genes, VvNRL4 and VvNRL6, among all 24 VvNRLs, are of particular interest. The NJ evolutionary tree based on amino acid sequences reveals a close relationship between VvNRL4 and AtNPH3, while VvNRL6 shows a close affinity with AtRPT2 (Figure 2a and Figure 6a). AI structure prediction indicates a high similarity between VvNRL4 and AtNPH3, as well as between VvNRL6 and AtRPT2, with RMSD values of 1.001 and 1.118, respectively (Figure 6b,c). These findings suggest that VvNRL4, VvNRL6, AtNPH3, and AtRPT2 may have highly similar functions. Previous studies have shown that AtNPH3 is not light-induced at the transcriptional level [8,32], while both blue and red light can induce significant expression of AtRPT2 [9]. Based on the sequence and structural similarities between VvNRL4 and AtNPH3, as well as VvNRL6 and AtRPT2, it can be inferred that VvNRL6 may also be induced by blue and red light.
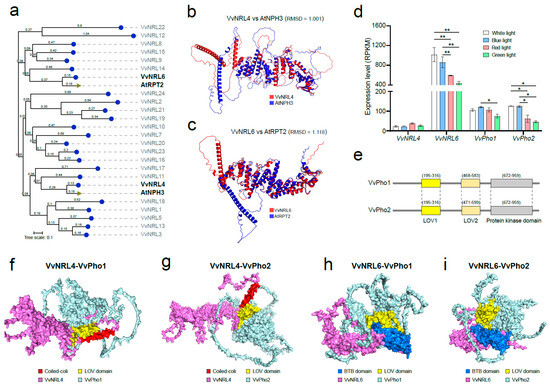
Figure 6.
Phylogenetic relationship and structure analysis of NRL proteins from different plant species. (a) Phylogenetic tree of the VvNRL protein family (grape) and AtNPH3 and AtRPT2 (Arabidopsis). The NRL gene family was subjected to a systematic evolutionary analysis using the neighbor-joining (NJ) method and the JTT matrix-based model in MEGA 7.0. Bootstrap was set to 1000. Circles represent grape; triangles represent Arabidopsis. (b) Structural comparison of VvNRL4 (red) and AtNPH3 (blue). (c) Structural comparison of VvNRL6 (red) and AtRPT2 (blue). (d) Expression levels of VvNRL4, VvNRL6, VvPho1, and VvPho2 under different light conditions. (e) Conserved domains in VvPho1 (1004 aa) and VvPho2 (1001 aa), with LOV1 and LOV2 indicating the LOV domain. Gray dashed lines represent collinearity within the same domain. (f,g) Docking modes of VvNRL4 (violet) with VvPHO1 and VvPHO2 (wathet) proteins. The coiled-coil domain of VvNRL4 is highlighted in red, while the LOV domain of VvPHO1 and VvPHO2 is highlighted in yellow. (h,i) Docking modes of VvNRL6 (violet) with VvPHO1 and VvPHO2 (wathet) proteins. The coiled-coil domain of VvNRL6 is highlighted in red, while the LOV domain of VvPHO1 and VvPHO2 is highlighted in yellow. * and ** indicate significant differences at p < 0.05 and p < 0.01 respectively (Student’s t-test).
We subsequently examined the transcription levels of VvNRL4, VvNRL6, VvPHO1, and VvPHO2 under different light conditions: white light from a fluorescent lamp (wavelengths from 350 to 750 nm), blue light (peak at 440 nm), red light (peak at 630 nm), and green light (peak at 520 nm). The results showed that, like AtRPT2, transcriptional expression of VvNRL6 was induced by blue and red light (Figure 6d). The expression of VvNRL4 did not show any differences under different spectra, similar to NPH3 in Arabidopsis. Although the expression of NPH3 is not regulated by light, the electrophoretic mobility of the NPH3 protein undergoes a significant increase following blue light stimulation. This phenomenon has been attributed to potential changes in the phosphorylation states of NPH3, which may explain the observed alterations in its electrophoretic mobility [9,32,33,34]. Meanwhile, the two phototropins in grape, VvPHOT1 and VvPHOT2, were significantly induced by blue light stimulation. We subsequently examined the expression of VvNRLs and NRL under different light conditions. The results showed that the expression of VvNRL6, VvPHOT1, and VvPHOT2 was induced by white and blue light.
Phototropins are plasma membrane-associated kinases containing two light, oxygen, or voltage-sensing domains (LOV1 and LOV2) at their N-terminus, which bind oxidized flavin mononucleotide (FMN) as a UV/blue light-absorbing cofactor [33,34]. In Arabidopsis, it has been demonstrated that the C-terminal portion of AtNPH3, which includes the coiled-coil domain, is believed to play a role in localizing NPH3 to the plasma membrane [35] and mediating direct interaction with phot1 [8]. In contrast to AtNPH3, the N-terminal region of AtRPT2 contains the BTB domain, which interacts with the LOV1 domain at the N-terminus of PHOT1. RPT2 affects phot1 photosensitivity through this LOV1 domain [9,36]. Similar to the PHO1 and PHO2 proteins in Arabidopsis, VvPHO1 and VvPHO2 in grapes also possess a C-terminal protein kinase domain and N-terminal LOV1 and LOV2 domains, showing high sequence and structural similarity (Figure 6e and Figure S2), suggesting their potential functional redundancy. Subcellular localization prediction using Cell-PLoc 2.0 revealed that VvPHO1 and VvPHO2 are localized in the cell membrane, similar to VvNRL4 and VvNRL6. To further investigate the molecular structure and interaction models of VvNRL4 and VvNRL6 with VvPHO1 and VvPHO2, we employed alphafold3 to predict their 3D structures. The interactions between VvNRL4 and VvNRL6 with VvPHO1 and VvPHO2 proteins were simulated using the HDOCK website [29], and the docking pose with the highest score was selected for each interaction. The results revealed a high coincidence between the docking sites of the coiled-coil domain of VvNRL4 and the LOV domain of VvPHO1 and VvPHO2 (Figure 6f,g), as well as between the docking sites of the BTB domain of VvNRL6 and the LOV domain of VvPHO1 and VvPHO2 (Figure 6h,i). These findings are consistent with those observed in Arabidopsis. The results of molecular docking suggest that VvNRL4 and VvNRL6 may interact with the LOV domain of VvPHO1 and VvPHO2, respectively, through their C-terminal coiled-coil domain and N-terminal BTB domain to further regulate the activity of VvPHO1 and VvPHO2.
3.7. Network Analysis of VvNRL-Mediated Genes for Development and Light Responsiveness
To predict the regulatory pathway mediated by VvNRLs, we utilized all VvNRL proteins and constructed a network of proteins associated with VvNRL-mediated biological processes using the STRING database (Figure 7a). To further understand the functions of these predicted interacted genes, we performed a Gene Ontology (GO) enrichment analysis. The results demonstrated significant enrichment of these genes in various pathways within the protein–protein interaction (PPI) network, including response to blue light (GO:0009637), auxin-activated signaling pathway (GO:0009734; Figure 7b), membrane (GO:0016020), photoreceptor activity (GO:0009881), blue light photoreceptor activity (GO:0009882), and protein ubiquitination (GO:0016567).
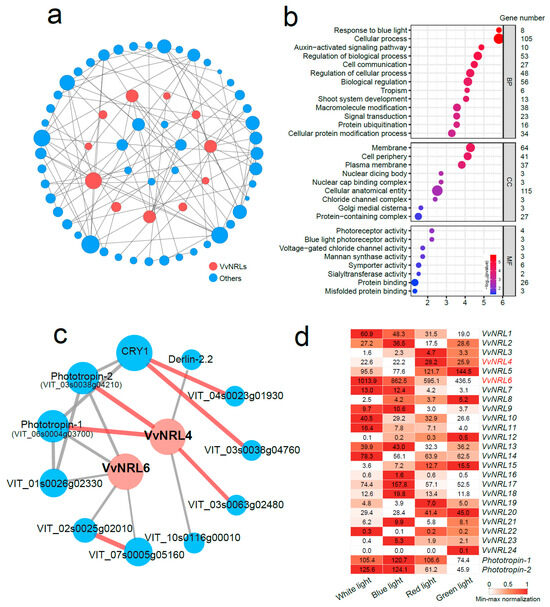
Figure 7.
VvNRL protein–protein interaction (PPI) network and biological function annotation. (a) Network showing the interactions between VvNRLs and their co-expressed proteins. Gray lines represent minimum required interaction scores with high confidence (>0.700 and <0.900), while red lines represent minimum required interaction scores with the highest confidence (>0.900). (b) Gene Ontology (GO) enrichment analysis of the genes in the PPI network, categorized into biological process (BP), cellular component (CC), and molecular function (MF). (c) Co-expression of genes with VvNRL4 and VvNRL6. Expression levels are shown on a scale (min–max normalization, row scale). (d) Expression levels of VvNRLs, Phototropin-1, and Phototropin-2 under different light conditions. RNA-seq data from Li et al. [27]. Expression levels were normalized using the min–max method (row scale).
Previous studies have indicated the interaction between NPH3 and CUL3 (CULLIN3), forming part of the CRLNPH3 ubiquitin ligase complex. Together, they contribute to the ubiquitination of PHOT1 under blue light induction, highlighting the involvement of NPH3 as a key factor in the blue light-induced phototropic response [37]. Along with NPH3, two other NRL family members also have known roles in phot signaling pathways. RPT2 interacts with both phot1 and NPH3 [9,16,38], and it is proposed to influence NPH3 phosphorylation status and promote the reconstitution of the phot1–NPH3 complex to sustain signaling under higher light intensities [38].
Due to the close evolutionary relationship between VvNRL4 and AtNPH3, as well as between VvNRL6 and AtRPT2, these genes exhibit highly similar phenotypes. Additionally, genes within the VvNRL protein-protein interaction (PPI) network are enriched in the blue light response pathway (Figure 7b). Subsequently, we conducted protein interaction predictions for VvNRL4 and VvNRL6 individually using the STRING database. It is worth noting that among all 11 PPI network genes, three genes (Phototropin-1, Phototropin-2, VIT_03s0063g02480) showed the highest confidence (0.900) with either VvNRL4 or VvNRL6 (Figure 7c). We further examined the expression patterns of VvNRLs, VvPHO1, and VvPHO2 under different light conditions. The results revealed that the expression of VvNRL6, VvNRL17, VvPHO1, and VvPHO2 is induced by both white light and blue light (Figure 7d).
3.8. Prediction of the Phosphorylation Site of VvNRL Proteins
Blue light receptors, phototropins (PHOT), are members of the AGC kinase family. Arabidopsis nph3 mutants exhibit a loss of hypocotyl phototropism under unilateral blue light of any intensity [10,39,40], indicating that NPH3 regulates the blue light-induced bending response of the hypocotyl mediated by PHOT1 and PHOT2. Under dark conditions, phosphorylated NPH3 protein binds to PHOT1 and localizes to the cell membrane. Upon weak unilateral blue light irradiation, NPH3 rapidly undergoes PHOT1-dependent dephosphorylation, leading to its detachment from the cell membrane [38,41]. Altogether, this has led to the current model, which states that the phosphorylation status of NPH3 determines its subcellular localization and function: phosphorylation of NPH3 promotes its action in mediating phototropic signaling from the plasma membrane, whereas NPH3 dephosphorylation reduces it by internalizing NPH3 into aggregates.
In addition, NRL6/NPY2, NRL7/NPY4, NRL20/NPY1, NRL21/NPY5, and NRL30/NPY3 function redundantly to regulate organ development and auxin transport in gravitropic responses in association with AGCVIII kinases [20,41]. This evidence suggests that phosphorylation plays a crucial role in the functioning of NRL genes.
In order to further investigate the distribution of phosphorylation sites in VvNRLs, phosphorylation sites within VvNRLs were predicted based on three amino acids: serine (S), threonine (T), and tyrosine (Y) (Figure 8a). The results showed that the number of phosphorylation sites in VvNRLs ranged from 9 to 33. Among them, VvNRL1 had the highest number of phosphorylation sites (33), while VvNRL22 had the fewest (9) (Figure 8b). Among all the phosphorylation sites, 45.7% (247/540) were located within the conserved domains (NPH3, BTB3, and coiled-coil domain) of the genes. Specifically, VvNRL24 had 70% (7/10) of its phosphorylation sites within conserved domains, while VvNRL18 had 15.8% (3/19) (Figure S3). As NPH3 is a key determinant of photophilic growth regulated by the phototropin AGC kinase, we further examined the number of phosphorylation sites associated with AGC kinases within VvNRLs. Among all the phosphorylation sites identified in VvNRLs, only 10.7% (58/540) were found to be phosphorylation sites associated with AGC kinases (Figure 8c). Each VvNRL contained one to five phosphorylation sites associated with AGC kinases.
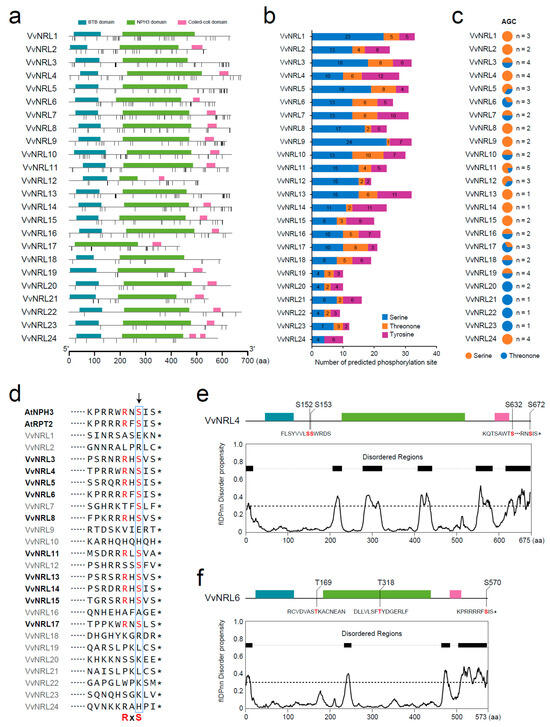
Figure 8.
Prediction of phosphorylation sites in VvNRL proteins, and conservation of the RxS phosphorylation sites of VvNRLs. (a) Distribution of predicted phosphorylation sites (Serine/threonine kinase) in VvNRL1 to 24 using the GPS 6.0 web server with a high threshold. (b) Number of serine (S), threonine (T), and tyrosine (Y) phosphorylation sites in VvNRL proteins. (c) Types and quantities of phosphorylation sites for the AGC kinase class in VvNRL proteins. (d) Conservation of the PHOT1 phosphorylation sequence of AtNPH3, with amino acid alignment of the last 10 residues in the AtRPT2 and VvNRL protein family. The position of S744 is indicated by an arrow, and sequences containing an RxS phosphorylation motif are denoted with an asterisk. (e,f) Distribution of phosphorylation sites for the AGC kinase class and prediction of disorder regions in VvNRL4 and VvNRL6. Disorder regions were predicted using flDPnn [31]. * indicate stop codon.
Recent studies have demonstrated that NPH3 undergoes direct phosphorylation by phot1 within a conserved C-terminal consensus sequence (RxS) in Arabidopsis. This phosphorylation is crucial for promoting phototropism and petiole positioning. Additionally, RxS phosphorylation leads to 14-3-3 binding and modulates NPH3 phosphorylation and localization. Furthermore, the conservation of RxS phosphorylation extends to other members of the NRL family, suggesting a common mechanism for regulating plant growth based on light conditions [42].
To investigate whether the conserved C-terminal consensus sequence (RxS) exists in VvNRLs, we examined the last ten amino acids of 24 VvNRLs. The results showed that 41.6% (10/24), including VvNRL4 and VvNRL6, possessed the conserved RxS motif. Moreover, in these genes with the conserved RxS motif, the last amino acid was predominantly serine (S) in approximately 90% of cases (Figure 8d). It is noteworthy that both VvNRL4 and VvNRL6 with the conserved RxS motif are located in disorder regions and belong to AGC kinase phosphorylation sites (Figure 8e,f), indicating their potential roles in signal transduction and regulation processes of VvNRL4 and VvNRL6.
4. Discussion
Plant response to external light signals is a crucial factor in plant growth and morphogenesis. Among them, phototropism is an important response mechanism, which refers to the bending or movement of plant organs towards or away from specific light sources [43]. The blue light receptor phototropin (PHOT) plays a significant role in regulating plant physiological responses [2,3,4]. Among the downstream genes of PHOT, NPH3 is one of the earliest identified genes. NPH3 belongs to the NRL (Nonphototropic Hypocotyl and Root Phototropism) family, which is a plant-specific protein family named after its two founding members, NPH3 and root phototropism 2 (RPT) [8,9,14,42]. There are 31 other NRL-like proteins in Arabidopsis, which show structural variations, but most of their biological functions remain unclear. To date, 33 functions have been identified in 10 Arabidopsis NRL proteins, which play important roles in light signal transduction, auxin-mediated processes, vascular development, vein formation, gravity bending, pollen tube growth, and more [14].
In this study, we conducted a whole-genome analysis of NRL genes in grapevines and identified 24 VvNRL members. Most of these genes contain both the NPH3 domain and the BTB/POZ domain, while some also contain a coiled-coil domain. Among the 24 VvNRLs, we focused on two genes, VvNRL4 and VvNRL6, because the NJ phylogenetic tree based on amino acid sequences shows a close relationship between VvNRL4 and AtNPH3, as well as between VvNRL6 and AtRPT2. Additionally, based on AI-based structural prediction, VvNRL4 and VvNRL6 exhibit high similarity to AtNPH3 and AtRPT2, respectively. These findings suggest that VvNRL4, VvNRL6, AtNPH3, and AtRPT2 may have highly similar functions. The transcriptional regulation of NRL genes appears to be tightly linked to light signaling pathways, as evidenced by our promoter analysis. The identification of light-responsive cis-elements in 23 out of 24 VvNRL promoters (Figure 4a) provides important genomic context for understanding their light-dependent expression patterns. Previous studies have shown that AtNPH3 is not light-induced at the transcriptional level [8,32], while the expression of AtRPT2 can be significantly induced by blue and red light [9]. Based on the sequence and structural similarities between VvNRL4 and AtNPH3, as well as between VvNRL6 and AtRPT2, it can be inferred that VvNRL6 may also be induced by blue and red light. To verify this inference, we examined the transcription levels of VvNRL4, VvNRL6, VvPHO1, and VvPHO2 under different light conditions (Figure 6d and Figure 7d). The results showed that, similar to AtRPT2, the transcriptional expression of VvNRL6 was induced by blue and red light. However, VvNRL4 expression did not show any differences under different spectra. In addition, the study also found that two phototropin proteins (VvPHOT1 and VvPHOT2) in grapes were significantly induced after blue light stimulation. Further testing showed that the expression of VvNRL6, Phototropin-1, and Phototropin-2 was induced by white and blue light. Intriguingly, although VvNRL4’s promoter contains more light-responsive elements than VvNRL6 (Figure 4b), its transcriptional levels remained unchanged under different light conditions (Figure 6d). This apparent discrepancy suggests that the presence of cis-elements alone may not guarantee transcriptional activation, and additional regulatory mechanisms (e.g., epigenetic modifications or protein co-factor requirements) might determine functional light responsiveness.
Phototropins are membrane-associated kinases with two light, oxygen, or voltage-sensing domains (LOV1 and LOV2) at the N-terminus, which bind oxidized flavin mononucleotide (FMN) as UV/blue-light-absorbing chromophores [33,34]. In Arabidopsis, it has been demonstrated that the C-terminal domain of AtNPH3 plays a role in localizing NPH3 to the plasma membrane and mediates its direct interaction with PHOT1 [8,35]. Unlike AtNPH3, the N-terminal region of AtRPT2 contains a BTB domain, which interacts with the N-terminal LOV1 domain of PHOT1. RPT2 influences the photosensitivity of phot1 through the LOV1 domain [9,36]. Similarly, VvPHO1 and VvPHO2 in grapevines also possess C-terminal protein kinase domains, as well as N-terminal LOV1 and LOV2 domains, showing high sequence and structural similarity. Based on the predicted structure models using Alphafold2 and molecular docking results, the coiled-coil domain of VvNRL4 is highly compatible with the LOV domains of VvPHO1 and VvPHO2, while the BTB domain of VvNRL6 highly matches the LOV domains of VvPHO1 and VvPHO2. These results are consistent with the observations in Arabidopsis. The molecular docking results indicate that VvNRL4 and VvNRL6 may interact with the LOV domains of VvPHO1 and VvPHO2, respectively, through their C-terminal coiled-coil domain and N-terminal BTB domain, further regulating the activity of VvPHO1 and VvPHO2 (Figure 6f–i).
Phosphorylation plays a crucial role in the blue light activation of PHOT1-mediated NPH3 dephosphorylation and CRL3NPH3-dependent PHOT1 ubiquitination, which are essential physiological and biochemical processes for phototropic responses induced by PHOT1 [10,27,38,40,41]. The blue light receptors, phototropins, belong to the AGC kinase family [27]. Studies on Arabidopsis nph3 mutants have shown that under dark conditions, phosphorylated NPH3 protein binds to PHOT1 and localizes to the plasma membrane. Under weak unilateral blue light irradiation, NPH3 rapidly undergoes PHOT1-dependent dephosphorylation, leading to dissociation from the plasma membrane [38,41]. These findings support the current model that the phosphorylation status of NPH3 determines its subcellular localization and function. Phosphorylation of NPH3 promotes its role in mediating light-induced signal transduction from the plasma membrane, while dephosphorylation reduces its activity by internalizing NPH3 into aggregates. The NRL gene family is functionally associated with AGCVIII kinases and participates in gravity response by cooperatively regulating organ development and auxin transport. Research has shown that NRL6/NPY2, NRL7/NPY4, NRL20/NPY1, NRL21/NPY5, and NRL30/NPY3 redundantly regulate organ development and auxin transport in association with AGCVIII kinases, indicating the important role of phosphorylation in NRL gene function [20,41].
Our investigation revealed multiple phosphorylation sites in VvNRLs (Figure 8). Among these phosphorylation sites, 45.7% are located within the conserved domains of the genes, and different genes have varying numbers of phosphorylation sites in the conserved domains. Moreover, only 10.7% of the phosphorylation sites are associated with AGC kinases. Recent studies have shown that in Arabidopsis, NPH3 is phosphorylated by phot1 within the conserved C-terminal consensus sequence (RxS), which is crucial for promoting phototropic responses and stem positioning. Furthermore, RxS phosphorylation leads to binding with 14-3-3 proteins, regulating the phosphorylation and localization of NPH3. The conservation of RxS phosphorylation extends to other members of the NRL family, indicating a common mechanism for light-regulated plant growth modulation based on subcellular localization. Here we show that the conserved C-terminal consensus sequence (RxS) exists in VvNRLs. Moreover, in genes with the conserved RxS motif, the last amino acid was predominantly serine (S) in approximately 90% of cases (Figure 8d).
Protein intrinsic disorder regions (IDRs) are increasingly recognized as important determinants of cellular signal transduction [44] and can provide accessibility for assembling protein complexes. IDRs are often target regions for post-translational modifications [45], and are involved in the dynamic self-assembly of membraneless organelles or cytoplasmic structures [46]. It is noteworthy that both VvNRL4 and VvNRL6 with the conserved RxS motif are located in disorder regions and belong to AGC kinase phosphorylation sites (Figure 8e,f), indicating their potential roles in signal transduction and regulation processes of VvNRL4 and VvNRL6.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae11030274/s1: Table S1: Expression pattern analysis of VvNRL genes. Figure S1: Venn diagram illustrating the 24 VvNRL gene family members identified by BLASTP and HMMER analyses. Figure S2: Structural comparison of VvPHO1 and VvPHO2. Figure S3: Distribution of phosphorylation sites within conserved domains among the 24 VvNRL proteins.
Author Contributions
Conceptualization, H.Z. (Hongxia Zhang) and N.Q.; methodology, S.L.; software, H.G. and H.Z. (Hang Zhao); validation, S.L., H.G. and X.L.; formal analysis, S.L. and Y.L.; investigation, Y.L.; resources, X.L.; data curation, H.G.; writing—original draft preparation, S.L.; writing—review and editing, N.Q. and H.Z. (Hongxia Zhang); visualization, S.L.; supervision, N.Q.; funding acquisition, H.Z. (Hang Zhao) and H.Z. (Hongxia Zhang). All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science Foundation of China (32071733, 32371915, 32201762) and the Natural Science Foundation of Shandong Province (ZR2020MC144).
Data Availability Statement
All relevant data are presented in the article and its Supplementary Materials. Any further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Holland, J.J.; Roberts, D.; Liscum, E. Understanding Phototropism: From Darwin to Today. J. Exp. Bot. 2009, 60, 1969–1978. [Google Scholar] [CrossRef] [PubMed]
- Sakai, T.; Kagawa, T.; Kasahara, M.; Swartz, T.E.; Christie, J.M.; Briggs, W.R.; Wada, M.; Okada, K. Arabidopsis nph1 and npl1: Blue Light Receptors that Mediate both Phototropism and Chloroplast Relocation. Proc. Natl. Acad. Sci. USA 2001, 98, 6969–6974. [Google Scholar] [CrossRef] [PubMed]
- Iwabuchi, K.; Sakai, T.; Takagi, S. Blue Light-Dependent Nuclear Positioning in Arabidopsis thaliana Leaf Cells. Plant Cell Physiol. 2007, 48, 1291–1298. [Google Scholar] [CrossRef] [PubMed]
- Kagawa, T.; Sakai, T.; Suetsugu, N.; Oikawa, K.; Ishiguro, S.; Kato, T.; Tabata, S.; Okada, K.; Wada, M. Arabidopsis NPL1: A Phototropin Homolog Controlling the Chloroplast High-Light Avoidance Response. Science 2001, 291, 2138–2141. [Google Scholar] [CrossRef]
- Suetsugu, N.; Kagawa, T.; Wada, M. An Auxilin-Like J-Domain Protein, JAC1, Regulates Phototropin-Mediated Chloroplast Movement in Arabidopsis. Plant Physiol. 2005, 139, 151–162. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, Y.L.; Qiao, X.R.; Wang, J.; Wang, L.D.; Xu, C.S.; Zhang, X. Phototropins Function in High-Intensity Blue Light-Induced Hypocotyl Phototropism in Arabidopsis by Altering Cytosolic Calcium. Plant Physiol. 2013, 162, 1539–1551. [Google Scholar] [CrossRef]
- Christie, J.M. Phototropin Blue-Light Receptors. Annu. Rev. Plant Biol. 2007, 58, 21–45. [Google Scholar] [CrossRef]
- Motchoulski, A.; Liscum, E. Arabidopsis NPH3: A NPH1 Photoreceptor-Interacting Protein Essential for Phototropism. Science 1999, 286, 961–964. [Google Scholar] [CrossRef]
- Inada, S.; Ohgishi, M.; Mayama, T.; Okada, K.; Sakai, T. RPT2 Is a Signal Transducer Involved in Phototropic Response and Stomatal Opening by Association with Phototropin 1 in Arabidopsis thaliana. Plant Cell 2004, 16, 887–896. [Google Scholar] [CrossRef]
- Liscum, E.; Askinosie, S.K.; Leuchtman, D.L.; Morrow, J.; Willenburg, K.T.; Coats, D.R. Phototropism: Growing towards an Understanding of Plant Movement. Plant Cell 2014, 26, 38–55. [Google Scholar] [CrossRef]
- Okada, K.; Shimura, Y. Mutational Analysis of Root Gravitropism and Phototropism of Arabidopsis thaliana Seedlings. Aust. J. Plant Physiol. 1992, 19, 439–448. [Google Scholar] [CrossRef]
- Liscum, E.; Briggs, W.R. Mutations in the nph1 Locus of Arabidopsis Disrupt the Perception of Phototropic Stimuli. Plant Cell 1995, 7, 473–485. [Google Scholar] [CrossRef] [PubMed]
- Sakai, T.; Wada, T.; Ishiguro, S.; Okada, K. RPT2: A Signal Transducer of the Phototropic Response in Arabidopsis. Plant Cell 2000, 12, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Pedmale, U.V.; Celaya, R.B.; Liscum, E. Phototropism: Mechanism and Outcomes. Arab. Book 2010, 2010, e0125. [Google Scholar] [CrossRef] [PubMed]
- Suetsugu, N.; Takemiya, A.; Kong, S.G.; Higa, T.; Komatsu, A.; Shimazaki, K.; Kohchi, T.; Wada, M. RPT2/NCH1 Subfamily of NPH3-Like Proteins Is Essential for the Chloroplast Accumulation Response in Land Plants. Proc. Natl. Acad. Sci. USA 2016, 113, 10424–10429. [Google Scholar] [CrossRef]
- Sullivan, S.; Thomson, C.E.; Kaiserli, E.; Christie, J.M. Interaction Specificity of Arabidopsis 14-3-3 Proteins with Phototropin Receptor Kinases. Febs Lett. 2009, 583, 2187–2193. [Google Scholar] [CrossRef]
- Christie, J.M.; Blackwood, L.; Petersen, J.; Sullivan, S. Plant Flavoprotein Photoreceptors. Plant Cell Physiol. 2015, 56, 401–413. [Google Scholar] [CrossRef]
- Petricka, J.J.; Clay, N.K.; Nelson, T.M. Vein Patterning Screens and the Defectively Organized Tributaries Mutants in Arabidopsis thaliana. Plant J. 2008, 56, 251–263. [Google Scholar] [CrossRef]
- Cheng, Y.F.; Qin, G.J.; Dai, X.H.; Zhao, Y.D. NPY Genes and AGC Kinases Define Two Key Steps in Auxin-Mediated Organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 21017–21022. [Google Scholar] [CrossRef]
- Li, Y.T.; Dai, X.H.; Cheng, Y.F.; Zhao, Y.D. NPY Genes Play an Essential Role in Root Gravitropic Responses in Arabidopsis. Mol. Plant 2011, 4, 171–179. [Google Scholar] [CrossRef]
- Lalanne, E.; Honys, D.; Johnson, A.; Borner, G.H.H.; Lilley, K.S.; Dupree, P.; Grossniklaus, U.; Twell, D. SETH1 and SETH2, Two Components of the Glycosylphosphatidylinositol Anchor Biosynthetic Pathway, Are Required for Pollen Germination and Tube Growth in Arabidopsis. Plant Cell 2004, 16, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wengier, D.; Shuai, B.; Gui, C.P.; Muschietti, J.; McCormick, S.; Tang, W.H. The Pollen Receptor Kinase LePRK2 Mediates Growth-Promoting Signals and Positively Regulates Pollen Germination and Tube Growth. Plant Physiol. 2008, 148, 1368–1379. [Google Scholar] [CrossRef] [PubMed]
- Christie, J.M.; Suetsugu, N.; Sullivan, S.; Wada, M. Shining Light on the Function of NPH3/RPT2-Like Proteins in Phototropin Signaling. Plant Physiol. 2018, 176, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.T.; Meng, X.C.; Zhang, L.; Zhang, Y.Y.; Cai, Z.R.; Huang, Z.Q.; Su, M.; Wang, Y.; Li, M.Z.; Li, F.Y.; et al. A Mechanically Robust Conducting Polymer Network Electrode for Efficient Flexible Perovskite Solar Cells. Joule 2019, 3, 2205–2218. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Fasoli, M.; Dal Santo, S.; Zenoni, S.; Tornielli, G.B.; Farina, L.; Zamboni, A.; Porceddu, A.; Venturini, L.; Bicego, M.; Murino, V.; et al. The Grapevine Expression Atlas Reveals a Deep Transcriptome Shift Driving the Entire Plant into a Maturation Program. Plant Cell 2012, 24, 3489–3505. [Google Scholar] [CrossRef]
- Li, C.X.; Xu, Z.G.; Dong, R.Q.; Chang, S.X.; Wang, L.Z.; Khalil-Ur-Rehman, M.; Tao, J.M. An RNA-Seq Analysis of Grape Plantlets Grown in Vitro Reveals Different Responses to Blue, Green, Red LED Light, and White Fluorescent Light. Front. Plant Sci. 2017, 8, 78. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Yan, Y.M.; Tao, H.Y.; He, J.H.; Huang, S.Y. The HDOCK Server for Integrated Protein–Protein Docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Chen, M.M.; Zhang, W.Z.; Gou, Y.J.; Xu, D.Y.; Wei, Y.X.; Liu, D.; Han, C.; Huang, X.H.; Li, C.Z.; Ning, W.S.; et al. GPS 6.0: An Updated Server for Prediction of Kinase-Specific Phosphorylation Sites in Proteins. Nucleic Acids Res. 2023, 51, W243–W250. [Google Scholar] [CrossRef]
- Hu, G.; Katuwawala, A.; Wang, K.; Wu, Z.H.; Ghadermarzi, S.; Gao, J.Z.; Kurgan, L. flDPnn: Accurate Intrinsic Disorder Prediction with Putative Propensities of Disorder Functions. Nat. Commun. 2021, 12, 4438. [Google Scholar] [CrossRef] [PubMed]
- Pedmale, U.V.; Liscum, E. Regulation of Phototropic Signaling in Arabidopsis Via Phosphorylation State Changes in the Phototropin 1-Interacting Protein NPH3. J. Biol. Chem. 2007, 282, 19992–20001. [Google Scholar] [CrossRef] [PubMed]
- Christie, J.M.; Salomon, M.; Nozue, K.; Wada, M.; Briggs, W.R. LOV (Light, Oxygen, or Voltage) Domains of the Dlue-Light Photoreceptor Phototropin (nph1): Binding Sites for the Chromophore Flavin Mononucleotide. Proc. Natl. Acad. Sci. USA 1999, 96, 8779–8783. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, S.; Thomson, C.E.; Lamont, D.J.; Jones, M.A.; Christie, J.M. In Vivo Phosphorylation Site Mapping and Functional Characterization of Arabidopsis Phototropin 1. Mol. Plant 2008, 1, 178–194. [Google Scholar] [CrossRef]
- Inoue, S.; Kinoshita, T.; Takemiya, A.; Doi, M.; Shimazaki, K. Leaf Positioning of Arabidopsis in Response to Blue Light. Mol. Plant 2008, 1, 15–26. [Google Scholar] [CrossRef]
- Kimura, T.; Tsuchida-Mayama, T.; Imai, H.; Okajima, K.; Ito, K.; Sakaia, T. Arabidopsis ROOT PHOTOTROPISM2 Is a Light-Dependent Dynamic Modulator of Phototropin1. Plant Cell 2020, 32, 2004–2019. [Google Scholar] [CrossRef]
- Roberts, D.; Pedmale, U.V.; Morrow, J.; Sachdev, S.; Lechner, E.; Tang, X.B.; Zheng, N.; Hannink, M.; Genschik, P.; Liscum, E. Modulation of Phototropic Responsiveness in Arabidopsis through Ubiquitination of Phototropin 1 by the CUL3-Ring E3 Ubiquitin Ligase CRL3NPH3. Plant Cell 2011, 23, 3627–3640. [Google Scholar] [CrossRef]
- Haga, K.; Tsuchida-Mayama, T.; Yamada, M.; Sakai, T. Arabidopsis ROOT PHOTOTROPISM2 Contributes to the Adaptation to High-Intensity Light in Phototropic Responses. Plant Cell 2015, 27, 1098–1112. [Google Scholar] [CrossRef]
- Liscum, E.; Briggs, W.R. Mutations of Arabidopsis in Potential Transduction and Response Components of the Phototropic Signaling Pathway. Plant Physiol. 1996, 112, 291–296. [Google Scholar] [CrossRef]
- Fankhauser, C.; Christie, J.M. Plant Phototropic Growth. Curr. Biol. 2015, 25, R384–R389. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, Q.P.; Xu, C.Y.; Wang, J.; Zhu, J.D.; Shang, B.S.; Zhang, X. Phot2-Regulated Relocation of NPH3 Mediates Phototropic Response to High-Intensity Blue Light in Arabidopsis thaliana. J. Integr. Plant Biol. 2018, 60, 562–577. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, S.; Waksman, T.; Henderson, L.; Paliogianni, D.; Lütkemeyer, M.; Suetsugu, N.; Christie, J.M. Regulation of Plant Phototropic Growth by NPH3/RPT2-Like Substrate Phosphorylation and 14-3-3 Binding. Nat. Commun. 2021, 12, 6129. [Google Scholar] [CrossRef] [PubMed]
- Fankhauser, C.; Chory, J. Light Control of Plant Development. Annu. Rev. Cell Dev. Biol. 1997, 13, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically Disordered Proteins in Cellular Signalling and Regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef]
- Bah, A.; Forman-Kay, J.D. Modulation of Intrinsically Disordered Protein Function by Post-Translational Modifications. J. Biol. Chem. 2016, 291, 6696–6705. [Google Scholar] [CrossRef]
- Cuevas-Velazquez, C.L.; Dinneny, J.R. Organization out of Disorder: Liquid–Liquid Phase Separation in Plants. Curr. Opin. Plant Biol. 2018, 45, 68–74. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).