Genetic Diversity and Population Structure of Saffron (Crocus sativus L.) in Morocco Revealed by Sequence-Related Amplified Polymorphism Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Extraction
2.3. SRAP-PCR Analysis
2.4. Statistical Analyses
- PICi is the polymorphic information content of marker ‘i’,
- fi is the frequency of the amplified allele (band present),
- 1 − fi is the frequency of the null allele.
3. Results
3.1. Variation for SRAP Markers
3.2. Genetic Diversity of Moroccan C. sativus
3.3. Genetic Structure of Global C. sativus Accessions Based on Geographic Origin
3.4. Cluster Analysis
3.5. Principal Coordinates Analysis (PCoA)
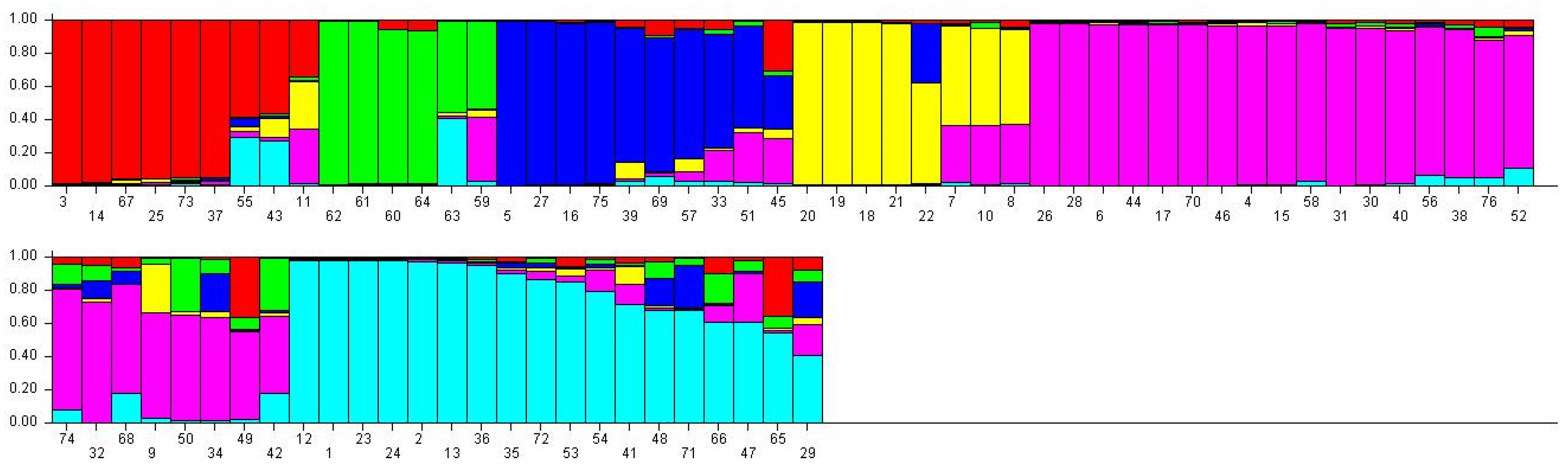
3.6. Population Genetic Structure
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gresta, F.; Lombardo, G.; Siracusa, L.; Ruberto, G. Saffron, an alternative crop for sustainable agricultural systems. A review. Agron. Sustain. Dev. 2008, 28, 95–112. [Google Scholar] [CrossRef]
- Petersen, G.; Seberg, O.; Thorsøe, S.; Jørgensen, T.; Mathew, B. A phylogeny of the genus Crocus (Iridaceae) based on sequence data from five plastid regions. Taxon 2008, 57, 487–499. [Google Scholar]
- Harpke, D.; Carta, A.; Tomović, G.; Ranđelović, V.; Ranđelović, N.; Blattner, F.R.; Peruzzi, L. Phylogeny, karyotype evolution and taxonomy of Crocus series Verni (Iridaceae). Plant Syst. Evol. 2015, 301, 309–325. [Google Scholar] [CrossRef]
- Dolatyari, A.; Tolyat Abolhasani, M.; Ardalani, F.; Rukšāns, J. A taxonomic revision of the genus Crocus (Iridaceae) in Iran. Nord. J. Bot. 2024, 2024, e04270. [Google Scholar] [CrossRef]
- Yadav, P.; Yadav, K.; Mishra, A.; Singh, K. An assessment and analysis of diseases of economically important plant members of family Iridaceae. J. Plant Dis. Prot. 2024, 131, 329–346. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, S.; Wang, Y.; Tian, Y.; Wang, X.; Xin, T.; Li, Z.; Hua, X.; Tan, S.; Sun, W. Crocus genome reveals the evolutionary origin of crocin biosynthesis. Acta Pharm. Sin. B 2024, 14, 1878–1891. [Google Scholar] [CrossRef]
- Kothari, D.; Thakur, M.; Joshi, R.; Kumar, A.; Kumar, R. Agro-climatic suitability evaluation for saffron production in areas of western Himalaya. Front. Plant Sci. 2021, 12, 657819. [Google Scholar] [CrossRef] [PubMed]
- Babaei, S.; Talebi, M.; Bahar, M.; Zeinali, H. Analysis of genetic diversity among saffron (Crocus sativus) accessions from different regions of Iran as revealed by SRAP markers. Sci. Hortic. 2014, 171, 27–31. [Google Scholar] [CrossRef]
- Caiola, M.G.; Caputo, P.; Zanier, R. RAPD analysis in Crocus sativus L. accessions and related Crocus species. Biol. Plant. 2004, 48, 375–380. [Google Scholar] [CrossRef]
- Alsayied, N.F.; Fernández, J.A.; Schwarzacher, T.; Heslop-Harrison, J. Diversity and relationships of Crocus sativus and its relatives analysed by inter-retroelement amplified polymorphism (IRAP). Ann. Bot. 2015, 116, 359–368. [Google Scholar] [CrossRef]
- Nemati, Z.; Harpke, D.; Gemicioglu, A.; Kerndorff, H.; Blattner, F.R. Saffron (Crocus sativus) is an autotriploid that evolved in Attica (Greece) from wild Crocus cartwrightianus. Mol. Phylogenet. Evol. 2019, 136, 14–20. [Google Scholar] [CrossRef]
- Kazemi-Shahandashti, S.-S.; Mann, L.; El-Nagish, A.; Harpke, D.; Nemati, Z.; Usadel, B.; Heitkam, T. Ancient artworks and Crocus genetics both support saffron’s origin in early Greece. Front. Plant Sci. 2022, 13, 834416. [Google Scholar] [CrossRef]
- Koocheki, A.; Moghaddam, P.R.; Aghhavani-Shajari, M.; Fallahi, H.-R. Corm weight or number per unit of land: Which one is more effective when planting corm, based on the age of the field from which corms were selected? Ind. Crops Prod. 2019, 131, 78–84. [Google Scholar] [CrossRef]
- Branca, F.; Argento, S. Evaluation of saffron pluriannual growing cycle in central Sicily. In Proceedings of the III International Symposium on Saffron: Forthcoming Challenges in Cultivation, Research and Economics 850, Kozani, Greece, 20–23 May 2009; pp. 153–158. [Google Scholar]
- Mansotra, R.; Vakhlu, J. Crocus sativus saffron: A 360-degree overview. In The Saffron Genome; Springer International Publishing: Cham, Swizerland, 2022; pp. 3–25. [Google Scholar]
- Abdusamat, B.; Samir, M.A.A.; Farid, M.A. Bioecology And Introduction of Saffron (Crocus sativus L.). Tex. J. Multidiscip. Stud. 2021, 2, 162–167. [Google Scholar]
- Moraga, Á.R.; Rambla, J.L.; Ahrazem, O.; Granell, A.; Gómez-Gómez, L. Metabolite and target transcript analyses during Crocus sativus stigma development. Phytochemistry 2009, 70, 1009–1016. [Google Scholar] [CrossRef] [PubMed]
- Farrokhi, H.; Asgharzadeh, A.; Samadi, M.K. Yield and qualitative and biochemical characteristics of saffron (Crocus sativus L.) cultivated in different soil, water, and climate conditions. Ital. J. Agrometeorol. 2021, 2014, 43–55. [Google Scholar] [CrossRef]
- Salas, M.d.C.; Montero, J.L.; Diaz, J.G.; Berti, F.; Quintero, M.F.; Guzmán, M.; Orsini, F. Defining optimal strength of the nutrient solution for soilless cultivation of saffron in the Mediterranean. Agronomy 2020, 10, 1311. [Google Scholar] [CrossRef]
- Rubio-Moraga, A.; Castillo-López, R.; Gómez-Gómez, L.; Ahrazem, O. Saffron is a monomorphic species as revealed by RAPD, ISSR and microsatellite analyses. BMC Res. Notes 2009, 2, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, M. The economics of saffron in Iran. In Proceedings of the II International Symposium on Saffron Biology and Technology 739, Masshad, Iran, 28 October 2006; pp. 321–331. [Google Scholar]
- Abolhassani, L.; Khorramdel, S.; Reed, M.; Saghaian, S. Environmental economic analysis of saffron production. In Saffron; Woodhead Publishing: Sawston, UK, 2020; pp. 367–390. [Google Scholar]
- Lambarraa-Lehnhardt, F.; Lmouden, A. Marketing Prospects for Saffron in Domestic Market: The Case of Moroccan PDO “Saffron of Taliouine”. In The Saffron Genome; Springer: Berlin/Heidelberg, Germany, 2022; pp. 289–300. [Google Scholar]
- Annemer, S.; Ez zoubi, Y.; Ramzi, A.; El Hadrami, E.M.; El Ouali Lalami, A.; Satrani, B.; Farah, A. Variations in saffron quality in Morocco (Taliouine and Taznakht) according to altitude and provenance: Chemometric investigation. J. Food Process. Preserv. 2022, 46, e16292. [Google Scholar] [CrossRef]
- Ait-Oubahou, A.; El-Otmani, M. Saffron cultivation in Morocco. In Saffron: Crocus sativus L.; CRC Press: London, UK, 1999; pp. 87–94. [Google Scholar]
- Khan, R.; Farooq, M.S.; Khelifi, A.; Ahmad, U.; Ahmad, F.; Riaz, S. Internet of things (IoT) based saffron cultivation system in greenhouse. Sci. Rep. 2024, 14, 22589. [Google Scholar] [CrossRef] [PubMed]
- Mzabri, I.; Charif, K.; Rimani, M.; Kouddane, N.; Boukroute, A.; Berrichi, A. History, biology, and culture of Crocus sativus: Overview and perspectives. Arab. J. Chem. Environ. Res. 2021, 8, 1–28. [Google Scholar]
- Maggi, M.A.; Bisti, S.; Picco, C. Saffron: Chemical composition and neuroprotective activity. Molecules 2020, 25, 5618. [Google Scholar] [CrossRef] [PubMed]
- Spence, C. Saffron: The colourful spice. Int. J. Gastron. Food Sci. 2023, 34, 100821. [Google Scholar] [CrossRef]
- Semeniuc, C.A.; Mandrioli, M.; Urs, M.J.; Toschi, T.G. Quality and authenticity of saffron and sensory aspects. Int. J. Gastron. Food Sci. 2024, 38, 101067. [Google Scholar] [CrossRef]
- José Bagur, M.; Alonso Salinas, G.L.; Jiménez-Monreal, A.M.; Chaouqi, S.; Llorens, S.; Martínez-Tomé, M.; Alonso, G.L. Saffron: An old medicinal plant and a potential novel functional food. Molecules 2018, 23, 30. [Google Scholar] [CrossRef]
- Pandita, D. Saffron (Crocus sativus L.): Phytochemistry, therapeutic significance and omics-based biology. In Medicinal and Aromatic Plants; Elsevier: Amsterdam, The Netherlands, 2021; pp. 325–396. [Google Scholar]
- Srivastava, R.; Ahmed, H.; Dixit, R.K.; Saraf, S. Crocus sativus L.: A comprehensive review. Pharmacogn. Rev. 2010, 4, 200. [Google Scholar] [CrossRef] [PubMed]
- De-Los-Mozos-Pascual, M.; Fernández, J.-A.; Roldán, M. Preserving biodiversity in saffron: The Crocusbank Project and the world saffron and Crocus collection. In Proceedings of the III International Symposium on Saffron: Forthcoming Challenges in Cultivation, Research and Economics 850, Kozani, Greece, 20–23 May 2009; pp. 23–28. [Google Scholar]
- Husaini, A.M. Challenges of climate change: Omics-based biology of saffron plants and organic agricultural biotechnology for sustainable saffron production. GM Crops Food 2014, 5, 97–105. [Google Scholar] [CrossRef]
- Kouzegaran, S.; Mousavi Baygi, M.; Babaeian, I.; Khashei-Siuki, A. Future projection of the effects of climate change on saffron yield and spatial-temporal distribution of cultivation by incorporating the effect of extreme climate indices. Theor. Appl. Climatol. 2020, 141, 1109–1118. [Google Scholar] [CrossRef]
- Cardone, L.; Castronuovo, D.; Perniola, M.; Cicco, N.; Candido, V. Saffron (Crocus sativus L.), the king of spices: An overview. Sci. Hortic. 2020, 272, 109560. [Google Scholar] [CrossRef]
- Kour, K.; Gupta, D.; Gupta, K.; Juneja, S.; Kaur, M.; Alharbi, A.H.; Lee, H.-N. Controlling agronomic variables of saffron crop using IoT for sustainable agriculture. Sustainability 2022, 14, 5607. [Google Scholar] [CrossRef]
- Salehi, A.; Shariatifar, N.; Pirhadi, M.; Zeinali, T. An overview on different detection methods of saffron (Crocus sativus L.) adulterants. J. Food Meas. Charact. 2022, 16, 4996–5006. [Google Scholar] [CrossRef]
- Malavi, D.; Nikkhah, A.; Alighaleh, P.; Einafshar, S.; Raes, K.; Van Haute, S. Detection of saffron adulteration with Crocus sativus style using NIR-hyperspectral imaging and chemometrics. Food Control 2024, 157, 110189. [Google Scholar] [CrossRef]
- Raina, A.; Kaul, S.; Dhar, M.K. Sniffing out adulteration in saffron: Detection methods and health risks. Food Control 2024, 155, 110042. [Google Scholar] [CrossRef]
- Fernández, J.-A.; Santana, O.; Guardiola, J.-L.; Molina, R.-V.; Heslop-Harrison, P.; Borbely, G.; Branca, F.; Argento, S.; Maloupa, E.; Talou, T. The world saffron and Crocus collection: Strategies for establishment, management, characterisation and utilisation. Genet. Resour. Crop Evol. 2011, 58, 125–137. [Google Scholar] [CrossRef]
- Treccarichi, S.; Infurna, G.; Ciulla, A.; Rossitto, A.; Argento, S.; Fallahi, H.; Branca, F. Evaluation of innovative growing techniques for organic saffron production in the Mediterranean countries. In Proceedings of the III International Organic Fruit Symposium and I International Organic Vegetable Symposium 1354, Catania, Italy, 14–17 December 2021; pp. 57–62. [Google Scholar]
- Alavi-Siney, S.M.; Saba, J.; Nasiri, J. Genetic variability and population genetic structure in autotriploid saffron using allelic phenotypes of microsatellite markers. Sci. Hortic. 2022, 299, 111043. [Google Scholar] [CrossRef]
- Alavi-Siney, S.M.; Saba, J.; Siahpirani, A.F.; Nasiri, J. ISSR-assisted spatial genetic structure, population admixture, and biodiversity estimates across locally adopted saffron ecotypes from 18 different provenances of Iran. J. Appl. Res. Med. Aromat. Plants 2023, 35, 100467. [Google Scholar] [CrossRef]
- Iqbal, A.M.; Lone, A.A.; Khan, M.H.; uddin Sofi, M.; Alie, B.A.; Hassan, M.G.; Dar, N.A.; Khan, A.; Fayaz, U.; Dar, S.A. Genetic variability assessment of indigenous and exotic saffron germplasm through morpho-agronomic characterization at Jammu and Kashmir, India. Plant Genet. Resour. 2023, 21, 389–398. [Google Scholar] [CrossRef]
- Javadi, B.; Sahebkar, A.; Emami, S.A. A survey on saffron in major Islamic traditional medicine books. Iran. J. Basic Med. Sci. 2013, 16, 1. [Google Scholar]
- Salgotra, R.K.; Chauhan, B.S. Genetic diversity, conservation, and utilization of plant genetic resources. Genes 2023, 14, 174. [Google Scholar] [CrossRef]
- Ayoub, I.B.; Ara, S.; Lone, S.A. Evaluating the Sensitivity of Saffron Yield to Climate Change in Western Himalaya, India. A Study from Kashmir Valley. In Climate Crisis, Social Responses and Sustainability: Socio-Ecological Study on Global Perspectives; Springer: Berlin/Heidelberg, Germany, 2024; pp. 159–173. [Google Scholar]
- El Caid, M.B.; Lachheb, M.; Lagram, K.; Wang, X.; Serghini, M.A. Ecotypic variation and environmental influence on saffron (Crocus sativus L.) vegetative growth: A multivariate performance analysis. J. Appl. Res. Med. Aromat. Plants 2024, 43, 100601. [Google Scholar] [CrossRef]
- Cardone, L.; Castronuovo, D.; Perniola, M.; Cicco, N.; Molina, R.V.; Renau-Morata, B.; Nebauer, S.G.; Candido, V. Crocus sativus L. Ecotypes from Mediterranean Countries: Phenological, Morpho-Productive, Qualitative and Genetic Traits. Agronomy 2021, 11, 551. [Google Scholar] [CrossRef]
- Boudadi, I.; Lachheb, M.; El Merzougui, S.; Lachguer, K.; Serghini, M.A. Exploring genetic variation in saffron (Crocus sativus L.) accessions through two-locus DNA barcoding. Ind. Crops Prod. 2024, 220, 119177. [Google Scholar] [CrossRef]
- Izadpanah, F.; Kalantari, S.; Hassani, M.; Naghavi, M.; Shokrpour, M. Variation in Saffron (Crocus sativus L.) accessions and Crocus wild species by RAPD analysis. Plant Syst. Evol. 2014, 300, 1941–1944. [Google Scholar] [CrossRef]
- Anabat, M.M.; Riahi, H.; Sheidai, M.; Koohdar, F. Population genetic study and barcoding in Iran saffron (Crocus sativus L.). Ind. Crops Prod. 2020, 143, 111915. [Google Scholar] [CrossRef]
- Busconi, M.; Colli, L.; Sánchez, R.A.; Santaella, M.; Pascual, M.D.-L.-M.; Santana, O.; Roldán, M.; Fernández, J.-A. AFLP and MS-AFLP analysis of the variation within saffron Crocus (Crocus sativus L.) germplasm. PLoS ONE 2015, 10, e0123434. [Google Scholar] [CrossRef]
- Siracusa, L.; Gresta, F.; Avola, G.; Albertini, E.; Raggi, L.; Marconi, G.; Lombardo, G.M.; Ruberto, G. Agronomic, chemical and genetic variability of saffron (Crocus sativus L.) of different origin by LC-UV–vis-DAD and AFLP analyses. Genet. Resour. Crop Evol. 2013, 60, 711–721. [Google Scholar] [CrossRef]
- Alavi-Siney, S.M.; Saba, J.; Alavikia, S.S.; Azimi, M.R. Identification of ISSR markers related to quantitative traits of Crocus sativus L. ecotypes. Saffron Agron. Technol. 2020, 8, fa598–fa608. [Google Scholar]
- Mir, M.A.; Mansoor, S.; Sugapriya, M.; Alyemeni, M.N.; Wijaya, L.; Ahmad, P. Deciphering genetic diversity analysis of saffron (Crocus sativus L.) using RAPD and ISSR markers. Saudi J. Biol. Sci. 2021, 28, 1308–1317. [Google Scholar] [CrossRef]
- Lachheb, M.; Ben El Caid, M.; El Merzougui, S.; Boudadi, I.; Serghini, M. Genetic diversity analysis of Moroccan saffron (Crocus sativus L.) of different origins based on intermicrosatellite markers. J. Appl. Hortic. 2021, 23, 242. [Google Scholar] [CrossRef]
- Lachheb, M.; El Merzougui, S.; Boudadi, I.; El Caid, M.B.; El Mousadik, A.; Serghini, M.A. Assessing genetic diversity using the first polymorphic set of EST-SSRs markers and barcoding of Moroccan saffron. J. Appl. Res. Med. Aromat. Plants 2022, 29, 100376. [Google Scholar] [CrossRef]
- Haq, S.A.; Salami, S.A.; Husaini, A.M. Omics in saffron (Crocus sativus L.): A spice of immense medicinal value. In Omics in Horticultural Crops; Elsevie: Amsterdam, The Netherlands, 2022; pp. 573–587. [Google Scholar]
- Li, G.; Quiros, C.F. Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: Its application to mapping and gene tagging in Brassica. Theor. Appl. Genet. 2001, 103, 455–461. [Google Scholar] [CrossRef]
- Salami, S.A.; Husaini, A.M. Genetic Mapping and Molecular Markers in Saffron. In The Saffron Genome; Springer: Berlin/Heidelberg, Germany, 2022; pp. 83–94. [Google Scholar]
- Gautam, N.; Bhattacharya, A. Molecular marker based assessment of genetic homogeneity within the in vitro regenerated plants of Crocus sativus L.—A globally important high value spice crop. S. Afr. J. Bot. 2021, 140, 461–467. [Google Scholar] [CrossRef]
- Bhagat, N.; Sharma, S.; Ambardar, S.; Raj, S.; Trakroo, D.; Horacek, M.; Zouagui, R.; Sbabou, L.; Vakhlu, J. Microbiome fingerprint as biomarker for geographical origin and heredity in Crocus sativus: A Feasibility Study. Front. Sustain. Food Syst. 2021, 5, 688393. [Google Scholar] [CrossRef]
- Naim, N.; Ennahli, N.; Hanine, H.; Lahlali, R.; Tahiri, A.; Fauconnier, M.-L.; Madani, I.; Ennahli, S. ATR-FTIR spectroscopy combined with DNA barcoding and GC-MS to assess the quality and purity of saffron (Crocus sativus L.). Vib. Spectrosc. 2022, 123, 103446. [Google Scholar] [CrossRef]
- Krstić, N.; Jaćimović, G.; Ljevnaić-Mašić, B.; Petrović, S.; Prijić, Ž.; Krstić, Đ.; Banjac, B. Morphological Trait Variations and Flower Color Differences in Wild Crocus Species. Horticulturae 2024, 10, 1214. [Google Scholar] [CrossRef]
- Chaachouay, N.; Azeroual, A.; Ansari, M.K.A.; Zidane, L. Use of plants as medicines and aromatics by indigenous communities of Morocco: Pharmacognosy, ecology and conservation. In Plants as Medicine and Aromatics; CRC Press: Boca Raton, FL, USA, 2023; pp. 33–44. [Google Scholar]
- Dessein, J. Territorialisation in practice: The case of saffron cultivation in Morocco. In Cultural Sustainability and Regional Development; Routledge: London, UK, 2015; pp. 108–124. [Google Scholar]
- Lachheb, M.; El Merzougui, S.; Boudadi, I.; El Caid, M.B.; El Mousadik, A.; Serghini, M.A. Deciphering phylogenetic relationships and genetic diversity of Moroccan saffron (Crocus sativus L.) using SSRg markers and chloroplast DNA SNP markers. S. Afr. J. Bot. 2023, 162, 1–9. [Google Scholar] [CrossRef]
- Wang, S.-M.; Zhang, Z.-W. The State of the World’s Plant Genetic Resources gor Food and Agriculture. J. Plant Genet. Resour. 2011, 12, 325–338. [Google Scholar]
- Busconi, M.; Wischnitzki, E.; Del Corvo, M.; Colli, L.; Soffritti, G.; Stagnati, L.; Fluch, S.; Sehr, E.M.; de los Mozos Pascual, M.; Fernández, J.A. Epigenetic variability among saffron Crocus (Crocus sativus L.) accessions characterized by different phenotypes. Front. Plant Sci. 2021, 12, 642631. [Google Scholar] [CrossRef]
- Liu, B.; Ding, P.; Ye, R.; Li, Y.; Ou, S.; Gentile, A.; Ma, X.; Deng, Z. Genetic Diversity Analysis of Guangxi Kumquat (Fortunella Swing) Germplasm Using SRAP Markers. Horticulturae 2023, 9, 689. [Google Scholar] [CrossRef]
- Sun, L.; Peng, L.; Li, Z.; Hou, R.; Mou, Y. Construction of genetic linkage map of plum (Prunus salicina L.) with ISSR and SRAP markers. Guangdong Agric. Sci 2022, 49, 40–48. [Google Scholar]
- Al-Ghamedi, K.; Alaraidh, I.; Afzal, M.; Mahdhi, M.; Al-Faifi, Z.; Oteef, M.D.; Tounekti, T.; Alghamdi, S.S.; Khemira, H. Assessment of genetic diversity of local coffee populations in southwestern Saudi Arabia using SRAP markers. Agronomy 2023, 13, 302. [Google Scholar] [CrossRef]
- Alzahib, R.H.; Migdadi, H.M.; Ghamdi, A.A.A.; Alwahibi, M.S.; Afzal, M.; Elharty, E.H.; Alghamdi, S.S. Exploring genetic variability among and within hail tomato landraces based on sequence-related amplified polymorphism markers. Diversity 2021, 13, 135. [Google Scholar] [CrossRef]
- Mousa, M.A.; Abo-Elyousr, K.A.; Ibrahim, O.H. Evaluation of Genetic Variability within a Collection of Cumin Genotypes Using RAPD, ISSR, SRAP and SCoT Markers and Variability of In Vitro Callus Induced Therefrom. Horticulturae 2023, 9, 742. [Google Scholar] [CrossRef]
- Rebrean, F.A.; Fustos, A.; Szabo, K.; Lisandru, T.-T.; Rebrean, M.S.; Varga, M.I.; Pamfil, D. Genetic Diversity and Structure of Quercus petraea (Matt.) Liebl. Populations in Central and Northern Romania Revealed by SRAP Markers. Diversity 2023, 15, 1093. [Google Scholar] [CrossRef]
- Lassner, M.W.; Peterson, P.; Yoder, J.I. Simultaneous amplification of multiple DNA fragments by polymerase chain reaction in the analysis of transgenic plants and their progeny. Plant Mol. Biol. Report. 1989, 7, 116–128. [Google Scholar] [CrossRef]
- Roldàn-Ruiz, I.; Dendauw, J.; Van Bockstaele, E.; Depicker, A.; De Loose, M. AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol. Breed. 2000, 6, 125–134. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Nei, M.; Li, W.-H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Hajyzadeh, M.; Olmez, F.; Khawar, K.M. Molecular approaches to determine phylogeny in saffron. In Saffron; Elsevier: Amsterdam, The Netherlands, 2020; pp. 57–68. [Google Scholar]
- Amiteye, S. Basic concepts and methodologies of DNA marker systems in plant molecular breeding. Heliyon 2021, 7, e08093. [Google Scholar] [CrossRef]
- Budak, H.; Shearman, R.; Parmaksiz, I.; Gaussoin, R.; Riordan, T.; Dweikat, I. Molecular characterization of Buffalograss germplasm using sequence-related amplified polymorphism markers. Theor. Appl. Genet. 2004, 108, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Ennami, M.; Briache, F.Z.; Mansi, J.M.; Gaboun, F.; Ghaouti, L.; Belqadi, L.; Mentag, R. Genetic Diversity of Moroccan Orobanche crenata Populations Revealed by Sequence-Related Amplified Polymorphism Markers. J. Agric. Sci. 2017, 9, 164. [Google Scholar] [CrossRef][Green Version]
- Yi, L.; Dong, Z.; Lei, Y.; Zhao, J.; Xiong, Y.; Yang, J.; Xiong, Y.; Gou, W.; Ma, X. Genetic diversity and molecular characterization of worldwide prairie grass (Bromus catharticus Vahl) accessions using SRAP markers. Agronomy 2021, 11, 2054. [Google Scholar] [CrossRef]
- Bidyananda, N.; Jamir, I.; Nowakowska, K.; Varte, V.; Vendrame, W.A.; Devi, R.S.; Nongdam, P. Plant Genetic Diversity Studies: Insights from DNA Marker Analyses. Int. J. Plant Biol. 2024, 15, 607–640. [Google Scholar] [CrossRef]
- Guo, L.; Liu, X.; Liu, X.; Yang, Z.; Kong, D.; He, Y.; Feng, Z. Construction of genetic map in barley using sequence-related amplified polymorphism markers, a new molecular marker technique. Afr. J. Biotechnol. 2012, 11, 13858–13862. [Google Scholar]
- Almarri, N.B.; Alghamdi, S.S.; ElShal, M.H.; Afzal, M. Estimating genetic diversity among durum wheat (Triticum durum desf.) landraces using morphological and SRAP markers. J. Saudi Soc. Agric. Sci. 2023, 22, 273–282. [Google Scholar] [CrossRef]
- Keify, F.; Beiki, A.H. Exploitation of random amplified polymorphic DNA (RAPD) and sequence-related amplified polymorphism (SRAP) markers for genetic diversity of saffron collection. J. Med. Plants Res. 2012, 6, 2761–2768. [Google Scholar]
- Zhou, L.; Yarra, R.; Cao, H.; Zhao, Z. Sequence-related amplified polymorphism (SRAP) markers based genetic diversity and population structure analysis of oil palm (Elaeis guineensis Jacq.). Trop. Plant Biol. 2021, 14, 63–71. [Google Scholar] [CrossRef]
- Choudhury, D.R.; Sharma, L.; Suma, A.; Singh, G.; Singh, R. Exploring the genetic diversity and population structure of Bacopa monnieri (L.) using random amplified (RAPD and ISSR) and gene-targeted (SCoT and CBDP) markers. In Genetic Resources and Crop Evolution; Springer: Berlin/Heidelberg, Germany, 2024; pp. 1–21. [Google Scholar] [CrossRef]
- Nemli, S.; Kianoosh, T.; Tanyolac, M.B. Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) accessions through retrotransposon-based interprimer binding sites (iPBSs) markers. Turk. J. Agric. For. 2015, 39, 940–948. [Google Scholar] [CrossRef]
- Rasul, K.S.; Majeed, H.O.; Faraj, J.M.; Lateef, D.D.; Tahir, N.A.-r. Genetic diversity and relationships among Iris aucheri genotypes determined via ISSR and CDDP markers. In Genetic Resources and Crop Evolution; Springer: Berlin/Heidelberg, Germany, 2024; pp. 1–14. [Google Scholar] [CrossRef]
- Fernández, J. Biology, biotechnology and biomedicine of saffron. In Research Developments in Plant Science; Research Signpost: Trivandrum, India, 2004; pp. 127–159. [Google Scholar]
- Brighton, C.A. Cytology of Crocus sativus and its allies (Iridaceae). Plant Syst. Evol. 1977, 128, 137–157. [Google Scholar] [CrossRef]
- Choudhary, V.; Choudhary, A.; Gahlaut, V.; Jaiswal, V. Genetic and Molecular Advancements in Saffron (Crocus sativus L.). In Genetics and Genomics of High-Altitude Crops; Springer: Berlin/Heidelberg, Germany, 2024; pp. 65–88. [Google Scholar]
- Mir, J.; Ahmed, N.; Singh, D.; Khan, M.; Zaffer, S.; Shafi, W. Breeding and biotechnological opportunities in saffron crop improvement. Afr. J. Agric. Res. 2015, 10, 970–974. [Google Scholar]
- Lawson, D.J.; Van Dorp, L.; Falush, D. A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots. Nat. Commun. 2018, 9, 3258. [Google Scholar] [CrossRef] [PubMed]
- Dong, A.; Huang, J.; Lan, S.; Wang, T.; Xiao, L.; Wang, W.; Zhao, T.; Zheng, X.; Liu, F.; Gao, G. Synthesis of N-halamine-functionalized silica–polymer core–shell nanoparticles and their enhanced antibacterial activity. Nanotechnology 2011, 22, 295602. [Google Scholar] [CrossRef] [PubMed]
- Jarocki, P.; Podleśny, M.; Komoń-Janczara, E.; Kucharska, J.; Glibowska, A.; Targoński, Z. Comparison of various molecular methods for rapid differentiation of intestinal bifidobacteria at the species, subspecies and strain level. BMC Microbiol. 2016, 16, 159. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Athe, R.; Naha, B.; Neerasa, G.; Parthasarathi, B.; Nukala, R.; Devara, D. Molecular markers-characteristics and applications in animal breeding. Int. J. Livest. Res. 2018, 8, 1–7. [Google Scholar] [CrossRef]
- Gedik, A.; Duygu, A.; Erdogmus, S.; Comertpay, G.; Tanyolac, M.B.; Ozkan, H. Genetic diversity of Crocus sativus and its close relative species analyzed by iPBS-retrotransposons. Turk. J. Field Crops 2017, 22, 243–252. [Google Scholar] [CrossRef]
- Salami, S.A.; Husaini, A.M. SaffronOMICS: Novel Approaches Toward Putting Saffron Data at Work. In The Saffron Genome; Springer: Berlin/Heidelberg, Germany, 2022; pp. 43–62. [Google Scholar]



| Localities | Number of Samples | Geographical Locality (Town) | Code | Latitude | Longitude | Altitude |
|---|---|---|---|---|---|---|
| 1 | 6 | Imin tlafoumlil | IMI-1 | 30.568818 | −7.85346 | 1876 |
| 2 | 5 | Taoulouzte | TAO-2 | 30.738507 | −7.779522 | 1964 |
| 3 | 6 | Aglimhakki | AGL-3 | 30.808924 | −7.786903 | 2035 |
| 4 | 5 | Tighboula | TIG-4 | 30.80522 | −7.772141 | 2121 |
| 5 | 6 | Dar Ifrane | DAR-5 | 30.232073 | −7.796592 | 1832 |
| 6 | 6 | Izourzen | IZO-6 | 30.233111 | −7.794296 | 1780 |
| 7 | 6 | Talaght | TAL-7 | 30.148356 | −8.266525 | 1615 |
| 8 | 6 | Azghar Nizri | AZG-8 | 30.147929 | −8.289528 | 1650 |
| 9 | 6 | Agzoule | AGR-9 | 30.154140 | −8.266954 | 1620 |
| 10 | 6 | Issoura | ISS-10 | 30.571637 | −7.611294 | 1676 |
| 11 | 6 | Guersafen | GUE-11 | 30.572882 | −7.61065 | 1680 |
| 12 | 6 | Idawtala | IDA-12 | 30.610221 | −7.6117 | 1700 |
| 13 | 6 | Bettale | BET-13 | 30.757580 | −7.374315 | 1604 |
| Forward | Reverse | ||
|---|---|---|---|
| Name | Sequence (5′-3′) | Name | Sequence (5′-3′) |
| ME1 | TGAGTCCAAACCGGATA | EM1 | GACTGCGTACGAATTAAT |
| ME2 | TGAGTCCAAACCGGAGC | EM2 | GACTGCGTACGAATTTGC |
| ME3 | TGAGTCCAAACCGGAAT | EM3 | GACTGCGTACGAATTGAC |
| ME4 | TGAGTCCAAACCGGACC | EM4 | GACTGCGTACGAATTTGA |
| ME5 | TGAGTCCAAACCGGAAG | EM5 | GACTGCGTACGAATTAAC |
| EM6 | GACTGCGTACGAATTGCA | ||
| SRAP Primers Combinations | Total Number of Bands | Number of Polymorphic Bands | Polymorphism Rate (%) | PICv |
|---|---|---|---|---|
| ME2/EM1 | 13 | 13 | 100 | 0.28 |
| ME1/EM3 | 10 | 10 | 100 | 0.36 |
| ME2/EM4 | 10 | 9 | 90 | 0.29 |
| ME1/EM2 | 13 | 13 | 100 | 0.30 |
| ME1/EM5 | 10 | 9 | 90 | 0.34 |
| ME3/EM3 | 7 | 7 | 100 | 0.26 |
| Average | 10.5 | 10.6 | 96.66 | 0.30 |
| Total | 63 | 61 | - | - |
| Localities | Na (±SE) | Ne (±SE) | I (±SE) | He (±SE) | PPL (%) |
|---|---|---|---|---|---|
| IMI-1 | 1.921 (0.041) | 1.663 (0.042) | 0.538 (0.025) | 0.369 (0.019) | 93.65 |
| TAO-2 | 1.159 (0.085) | 1.216 (0.048) | 0.174 (0.035) | 0.118 (0.024) | 31.75 |
| AGL-3 | 1.810 (0.059) | 1.589 (0.047) | 0.481 (0.031) | 0.329 (0.023) | 84.13 |
| TIG-4 | 0.968 (0.099) | 1.252 (0.053) | 0.186 (0.038) | 0.131 (0.027) | 28.57 |
| DAR-5 | 1.810 (0.063) | 1.620 (0.045) | 0.501 (0.030) | 0.345 (0.022) | 85.71 |
| IZO-6 | 1.635 (0.089) | 1.630 (0.049) | 0.485 (0.035) | 0.340 (0.025) | 76.19 |
| TAL-7 | 1.540 (0.087) | 1.472 (0.053) | 0.379 (0.038) | 0.261 (0.027) | 65.08 |
| AZG-8 | 1.429 (0.084) | 1.403 (0.053) | 0.319 (0.040) | 0.222 (0.028) | 52.38 |
| AGR-9 | 1.730 (0.076) | 1.639 (0.050) | 0.494 (0.034) | 0.344 (0.025) | 80.95 |
| ISS-10 | 1.619 (0.083) | 1.513 (0.050) | 0.419 (0.036) | 0.288 (0.026) | 71.43 |
| GUE-11 | 1.190 (0.084) | 1.246 (0.049) | 0.197 (0.037) | 0.135 (0.026) | 33.33 |
| IDA-12 | 1.460 (0.098) | 1.447 (0.050) | 0.368 (0.038) | 0.252 (0.027) | 63.49 |
| BET-13 | 1.810 (0.063) | 1.652 (0.046) | 0.515 (0.030) | 0.356 (0.022) | 85.71 |
| Average | 1.545 (0.024) | 1.488 (0.015) | 0.389 (0.011) | 0.268 (0.008) | 65.57 |
| IMI-1 | TAO-2 | AGL-3 | TIG-4 | DAR-5 | IZO-6 | TAL-7 | AZG-8 | AGR-9 | ISS-10 | GUE-11 | IDA-12 | BET-13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| **** | 0.771 | 0.945 | 0.784 | 0.938 | 0.869 | 0.885 | 0.806 | 0.896 | 0.875 | 0.647 | 0.844 | 0.938 | IMI-1 |
| 0.260 | **** | 0.748 | 0.782 | 0.737 | 0.708 | 0.757 | 0.834 | 0.768 | 0.738 | 0.701 | 0.708 | 0.742 | TAO-2 |
| 0.056 | 0.291 | **** | 0.760 | 0.886 | 0.821 | 0.874 | 0.803 | 0.881 | 0.856 | 0.682 | 0.854 | 0.907 | AGL-3 |
| 0.244 | 0.245 | 0.275 | **** | 0.767 | 0.707 | 0.781 | 0.732 | 0.769 | 0.735 | 0.583 | 0.714 | 0.755 | TIG-4 |
| 0.064 | 0.305 | 0.122 | 0.265 | **** | 0.910 | 0.887 | 0.836 | 0.848 | 0.909 | 0.611 | 0.863 | 0.870 | DAR-5 |
| 0.141 | 0.345 | 0.197 | 0.347 | 0.095 | **** | 0.797 | 0.812 | 0.810 | 0.839 | 0.690 | 0.804 | 0.784 | IZO-6 |
| 0.122 | 0.278 | 0.135 | 0.247 | 0.120 | 0.227 | **** | 0.804 | 0.829 | 0.813 | 0.661 | 0.809 | 0.873 | TAL-7 |
| 0.215 | 0.181 | 0.219 | 0.312 | 0.180 | 0.209 | 0.218 | **** | 0.763 | 0.833 | 0.739 | 0.840 | 0.767 | AZG-8 |
| 0.109 | 0.264 | 0.127 | 0.262 | 0.165 | 0.211 | 0.187 | 0.270 | **** | 0.835 | 0.732 | 0.809 | 0.902 | AGR-9 |
| 0.134 | 0.303 | 0.156 | 0.307 | 0.096 | 0.176 | 0.207 | 0.183 | 0.180 | **** | 0.642 | 0.869 | 0.827 | ISS-10 |
| 0.436 | 0.356 | 0.383 | 0.540 | 0.493 | 0.371 | 0.414 | 0.302 | 0.313 | 0.442 | **** | 0.718 | 0.693 | GUE-11 |
| 0.169 | 0.345 | 0.158 | 0.337 | 0.148 | 0.218 | 0.213 | 0.174 | 0.212 | 0.141 | 0.331 | **** | 0.838 | IDA-12 |
| 0.064 | 0.298 | 0.097 | 0.281 | 0.139 | 0.243 | 0.136 | 0.266 | 0.104 | 0.190 | 0.367 | 0.176 | **** | BET-13 |
| Source of Variation | Df | SS | MS | Est. Var. | % | PhiPT | p |
|---|---|---|---|---|---|---|---|
| Among Accessions | 12 | 284.773 | 23.731 | 2.372 | 19 | 0.194 | 0.000 |
| Within Accessions | 63 | 621.767 | 9.869 | 9.869 | 81 | ||
| Total | 75 | 906.539 | 12.241 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ennami, M.; Khouya, K.; Taimourya, H.; Benbya, A.; Kaddi, M.; Khayi, S.; Diria, G.; Abdelwahd, R.; Gaboun, F.; Mentag, R. Genetic Diversity and Population Structure of Saffron (Crocus sativus L.) in Morocco Revealed by Sequence-Related Amplified Polymorphism Markers. Horticulturae 2025, 11, 174. https://doi.org/10.3390/horticulturae11020174
Ennami M, Khouya K, Taimourya H, Benbya A, Kaddi M, Khayi S, Diria G, Abdelwahd R, Gaboun F, Mentag R. Genetic Diversity and Population Structure of Saffron (Crocus sativus L.) in Morocco Revealed by Sequence-Related Amplified Polymorphism Markers. Horticulturae. 2025; 11(2):174. https://doi.org/10.3390/horticulturae11020174
Chicago/Turabian StyleEnnami, Mounia, Khadija Khouya, Houda Taimourya, Abdellah Benbya, Mohamed Kaddi, Slimane Khayi, Ghizlan Diria, Rabha Abdelwahd, Fatima Gaboun, and Rachid Mentag. 2025. "Genetic Diversity and Population Structure of Saffron (Crocus sativus L.) in Morocco Revealed by Sequence-Related Amplified Polymorphism Markers" Horticulturae 11, no. 2: 174. https://doi.org/10.3390/horticulturae11020174
APA StyleEnnami, M., Khouya, K., Taimourya, H., Benbya, A., Kaddi, M., Khayi, S., Diria, G., Abdelwahd, R., Gaboun, F., & Mentag, R. (2025). Genetic Diversity and Population Structure of Saffron (Crocus sativus L.) in Morocco Revealed by Sequence-Related Amplified Polymorphism Markers. Horticulturae, 11(2), 174. https://doi.org/10.3390/horticulturae11020174






