Effective Tomato Spotted Wilt Virus Resistance Assessment Using Non-Destructive Imaging and Machine Learning
Abstract
1. Introduction
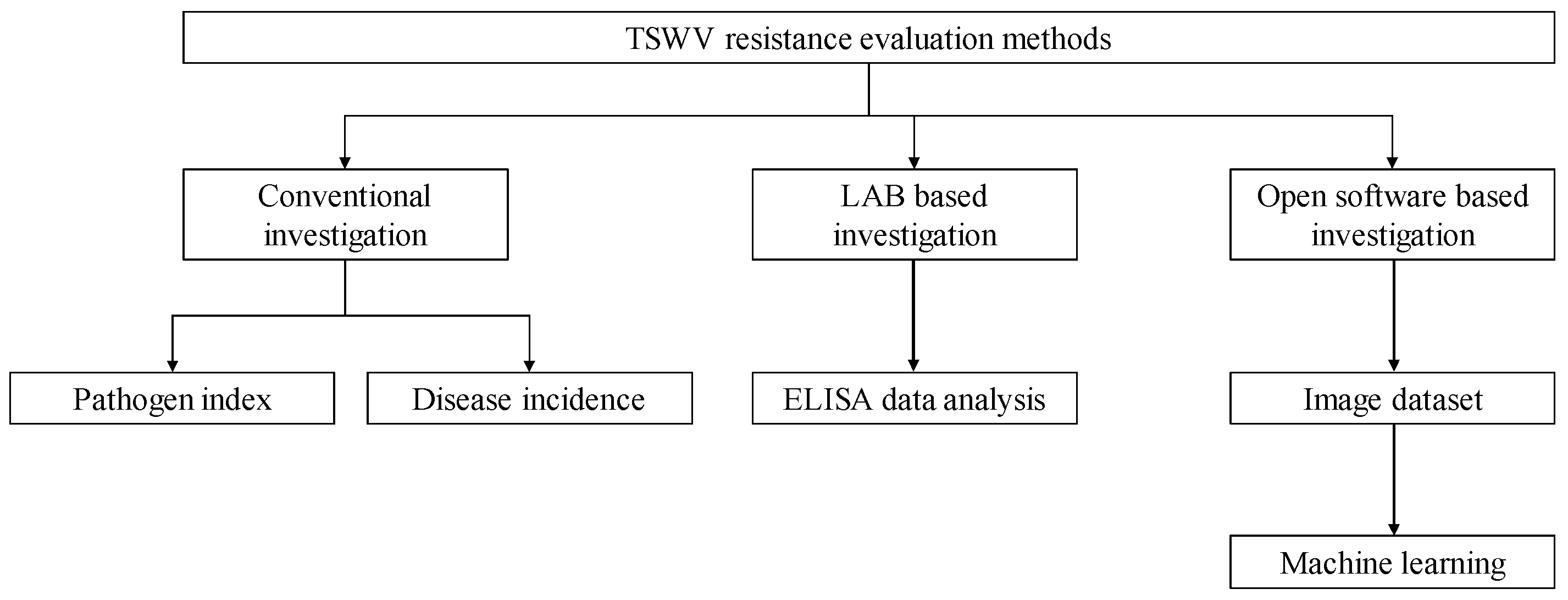
2. Materials and Methods
2.1. Samples and Virus
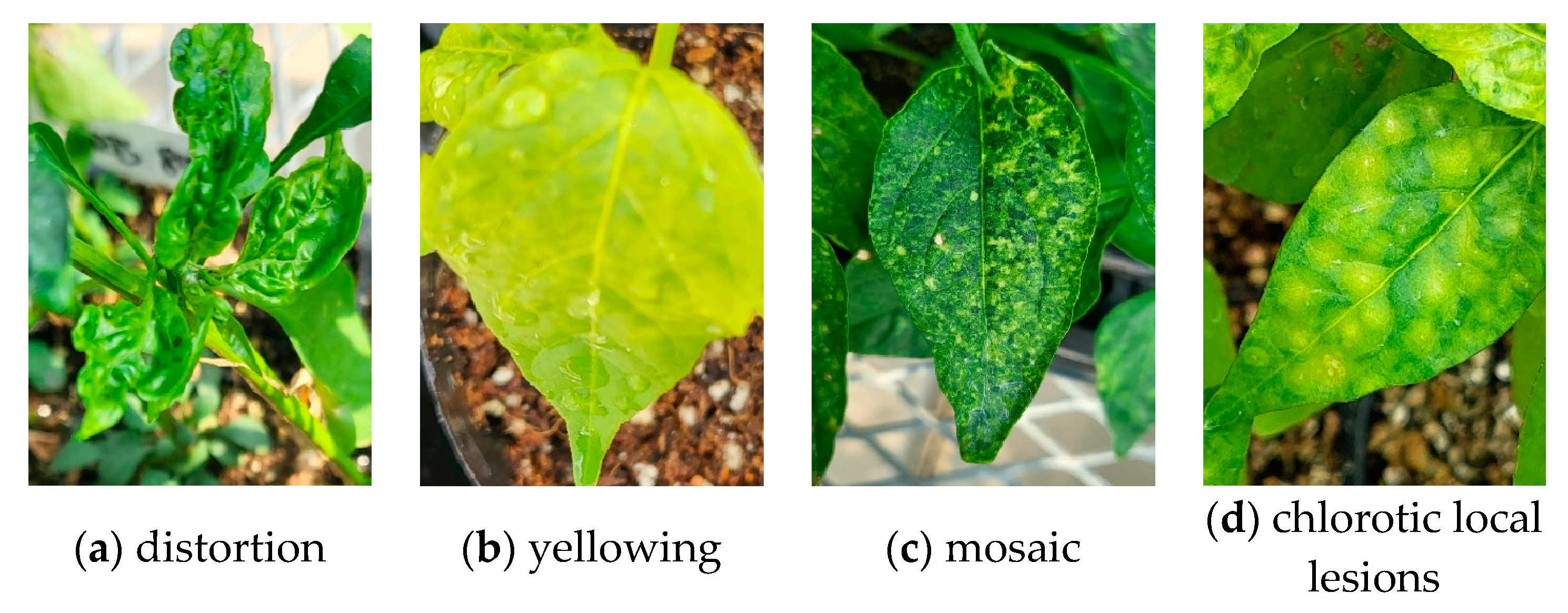
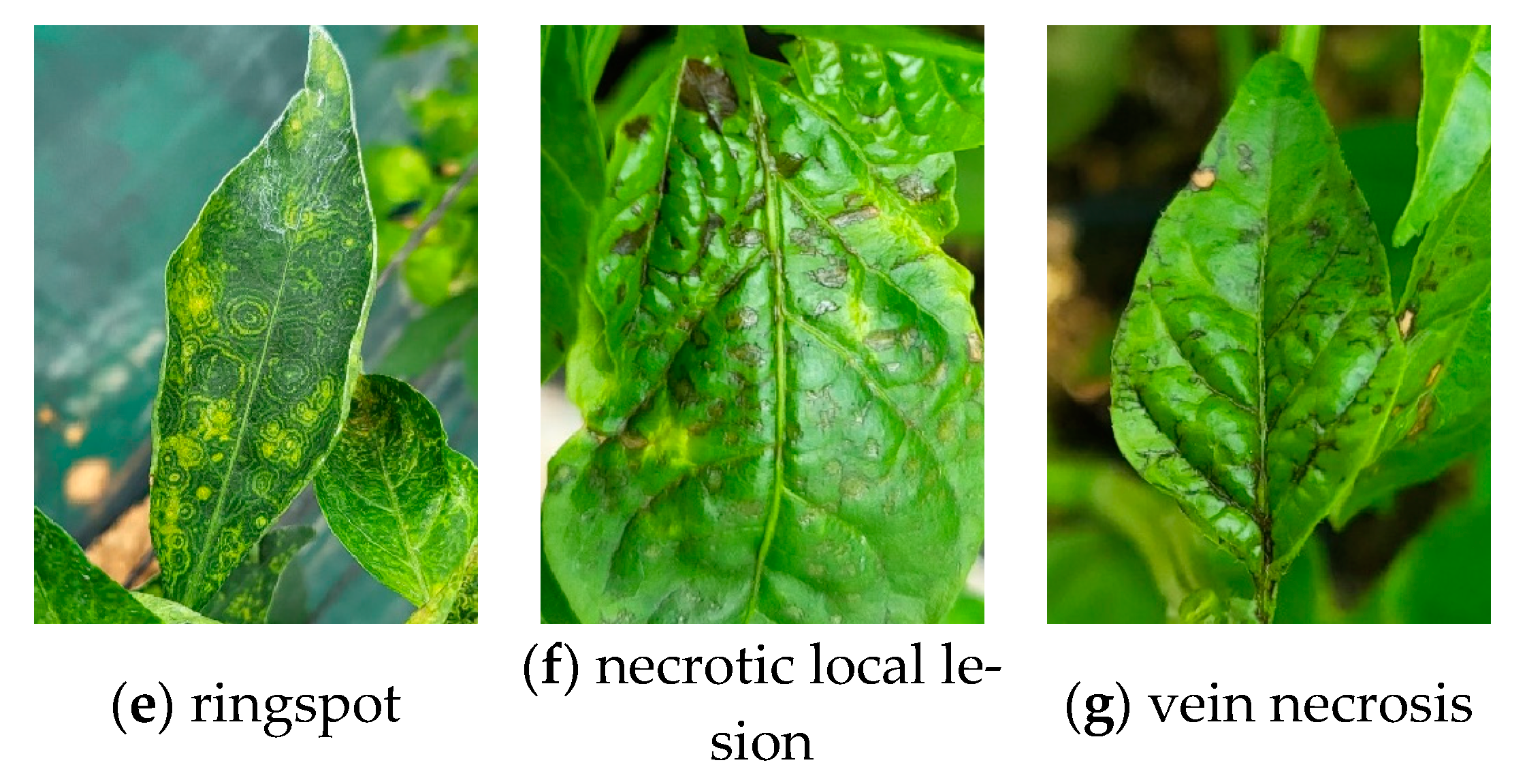
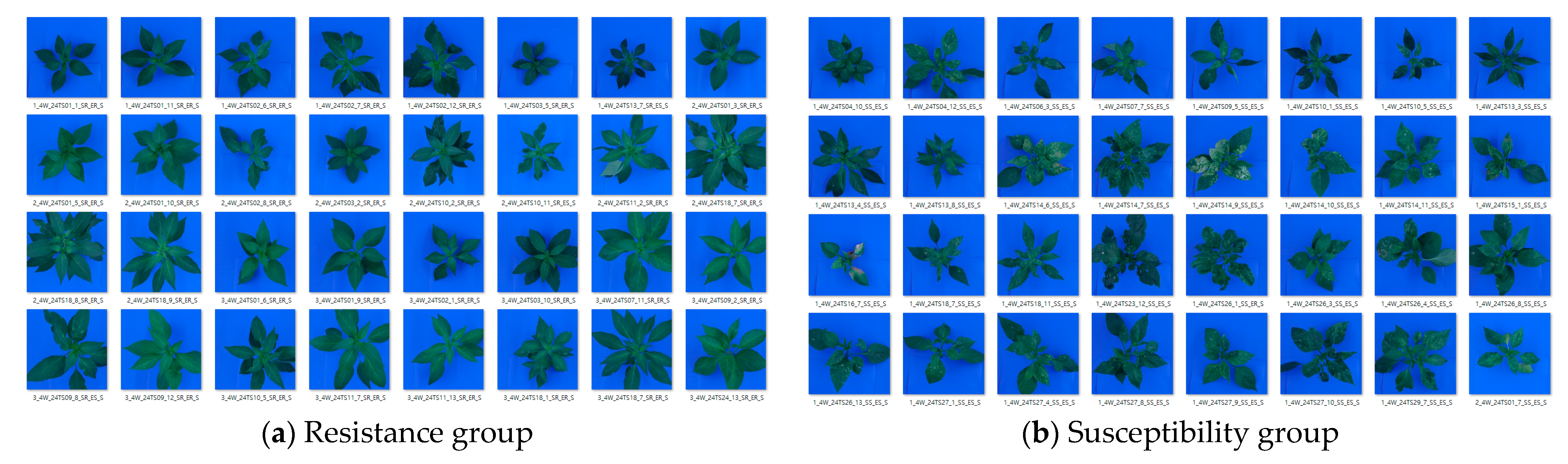
2.2. Tomato Spotted Wilt Virus (TSWV) Symptoms
2.3. Conventional Investigation
2.4. LAB Based Investigation (ELISA)
2.5. Open Software Based Investigation
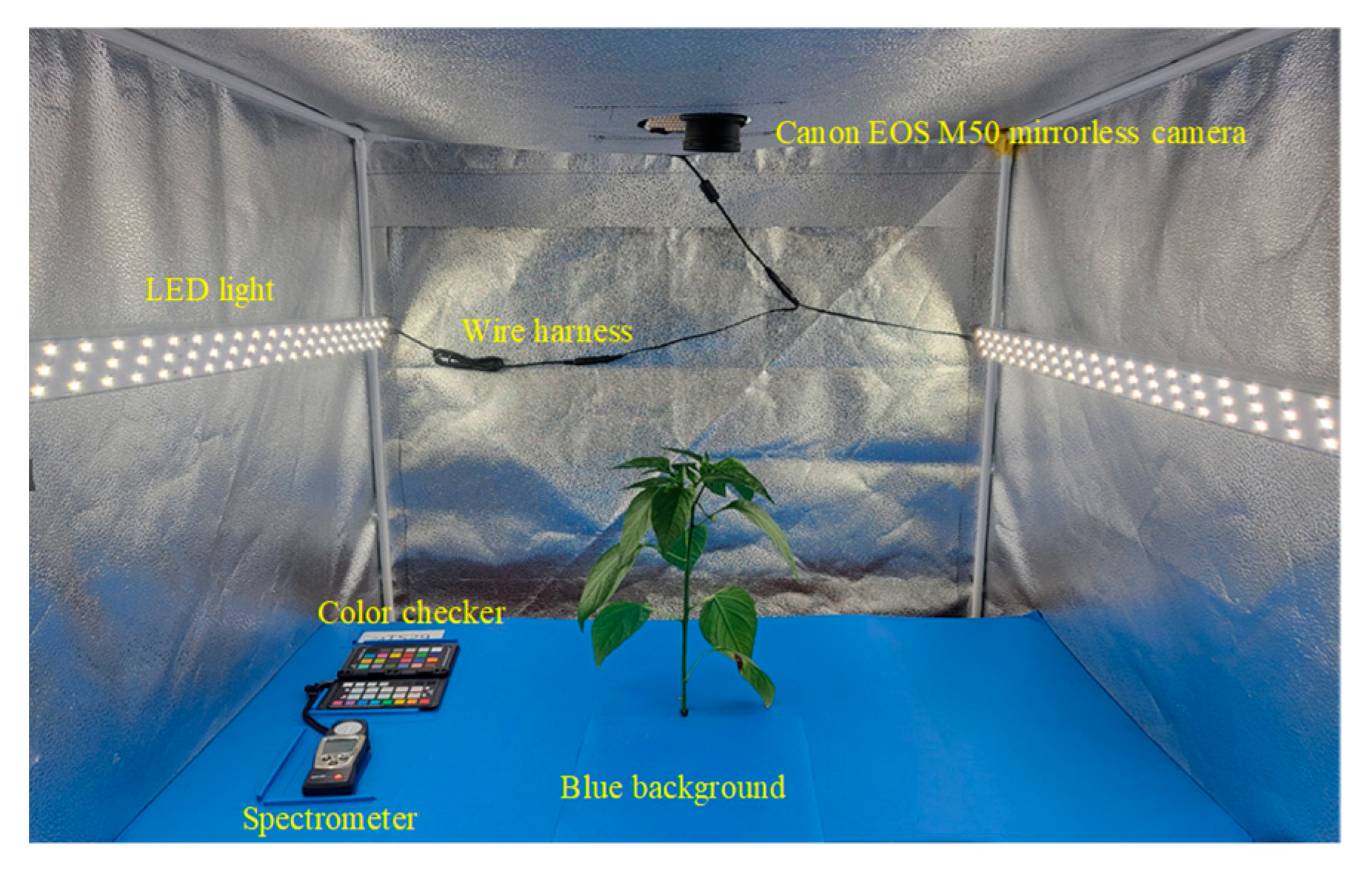
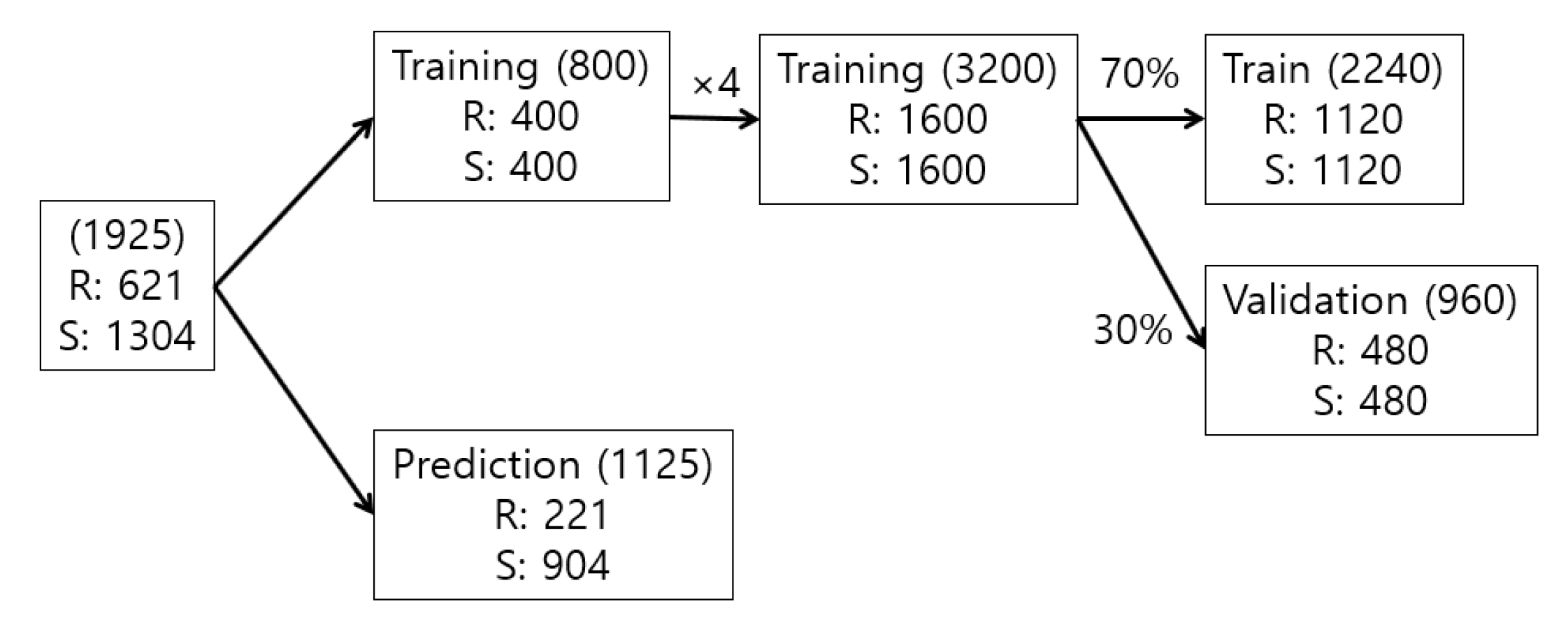
2.5.1. Image Dataset Collection
2.5.2. Image Data Usage Scheme
2.5.3. Machine Learning
2.5.4. Metrics
3. Results
3.1. Resistance Determination
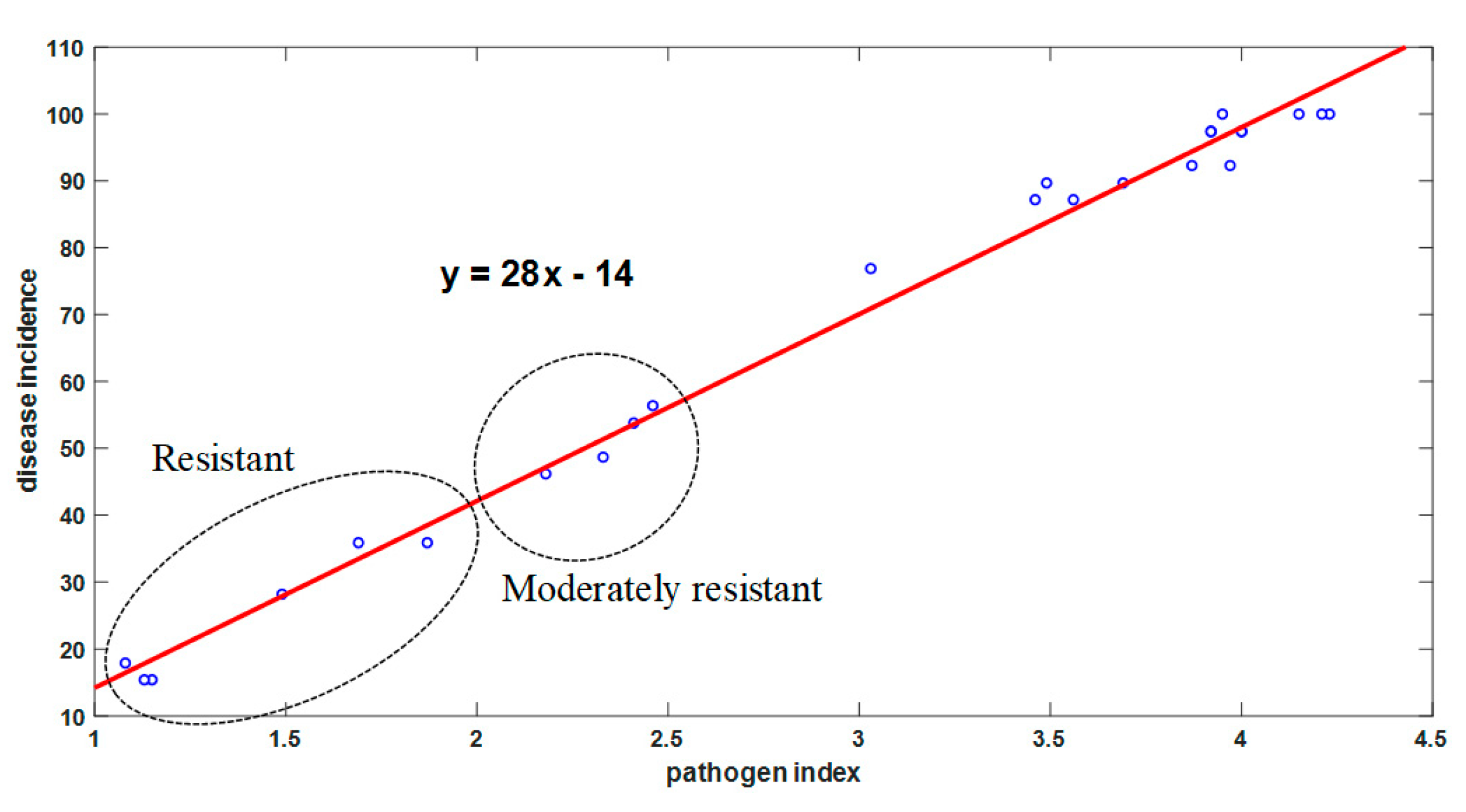
3.1.1. Conventional Investigation Results
3.1.2. LAB-Based Investigation Results
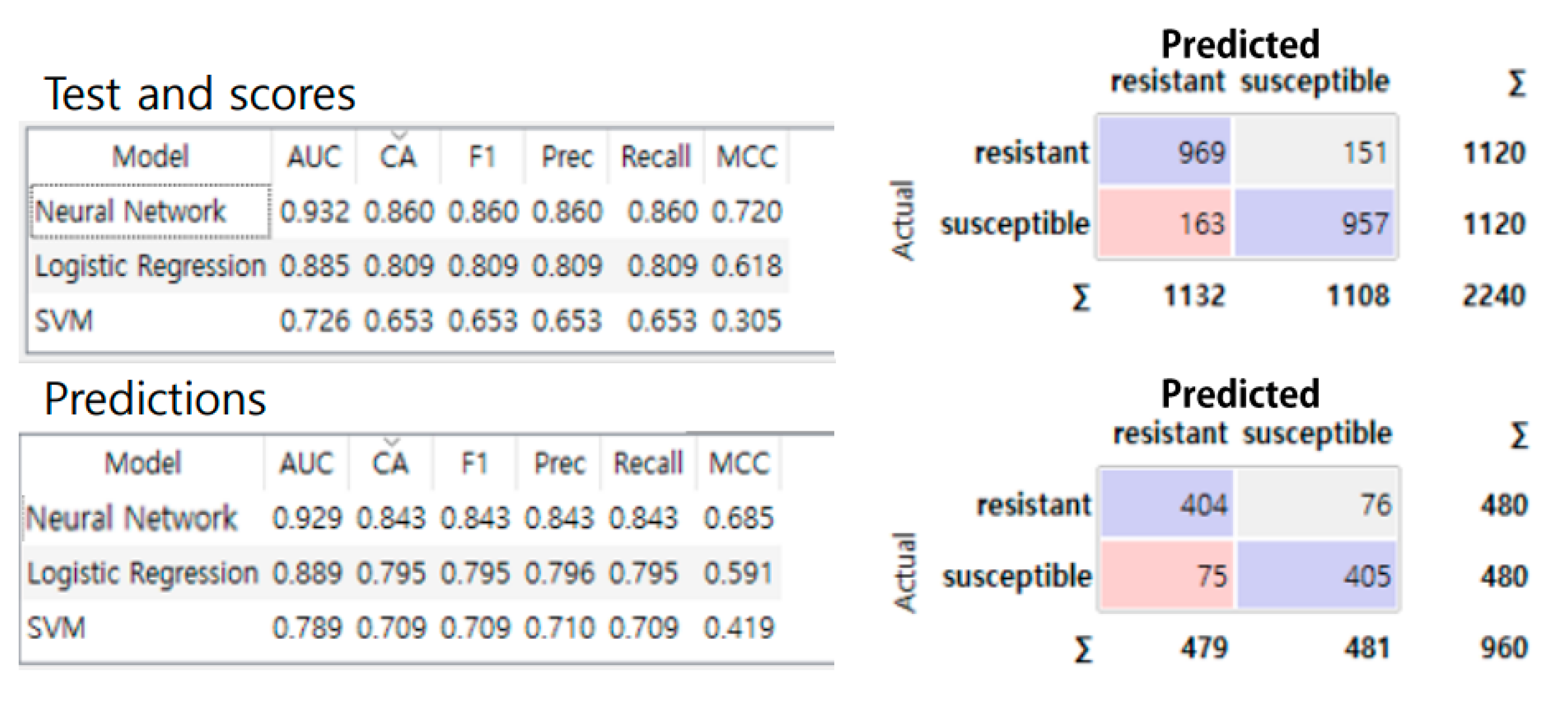
3.1.3. Machine Learning
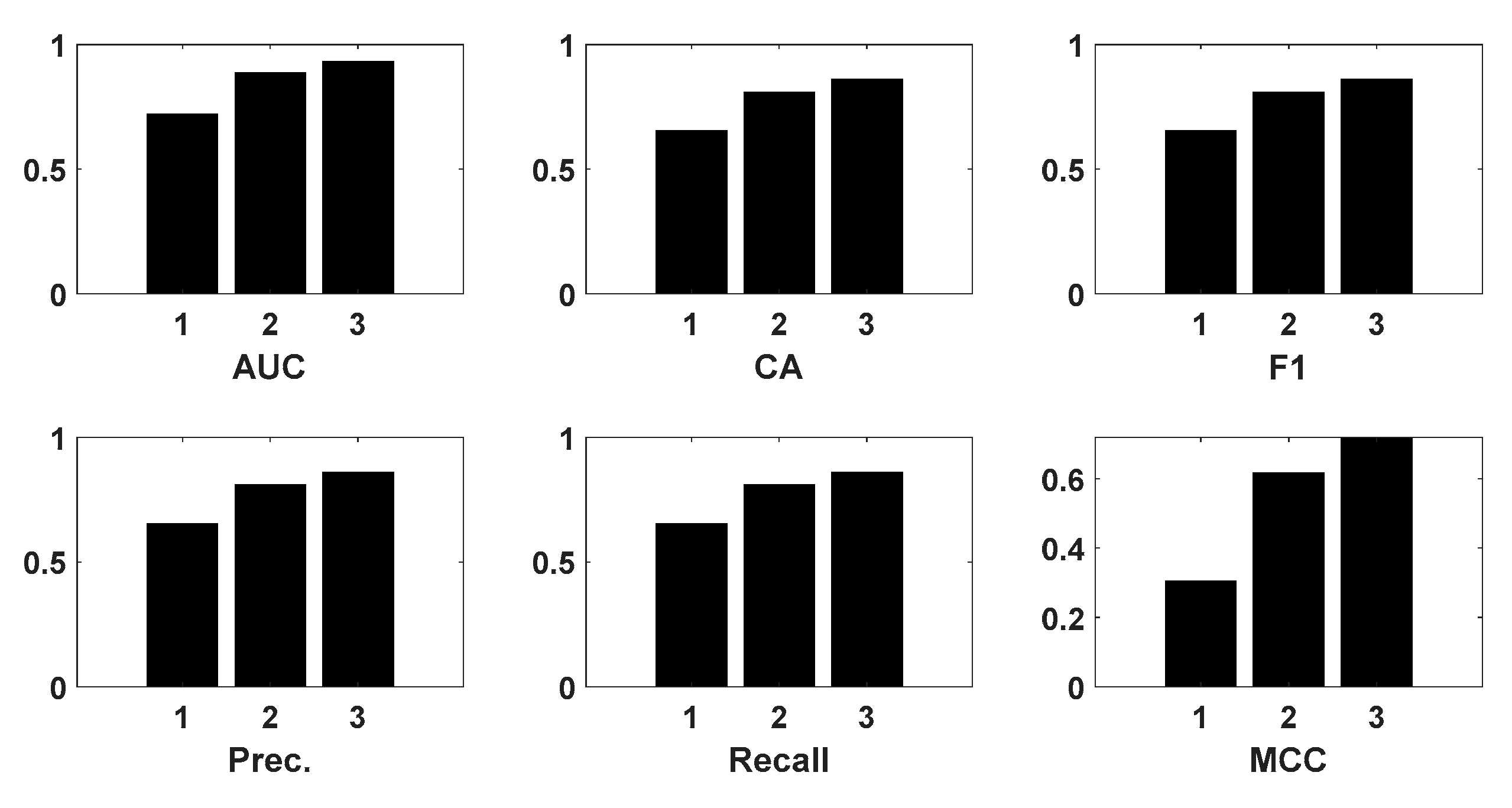
3.2. Causality of Analytical Methods
3.2.1. Average Causality
3.2.2. STD Causality
4. Discussion
4.1. Interpretation of Causality Analysis Results
4.2. Practical Scenarios
- Signs of mutation occur (macroscopic evaluation by visual inspection);
- Variety selection process based on initial mutation occurrence (micro-evaluation by ELISA);
- Continuous monitoring according to the spread of mutations (evaluation of mutation trends by open software).
4.3. Performance Comparison
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Altizani-Júnior, J.C.; Cicero, S.M.; de Lima, C.B.; Alves, R.M.; Gomes-Junior, F.G. Optimizing basil seed vigor evaluations: An automatic approach using computer vision-based technique. Horticulturae 2024, 10, 1220. [Google Scholar] [CrossRef]
- Fuentes-Peñailillo, F.; Carrasco Silva, G.; Pérez Guzmán, R.; Burgos, I.; Ewertz, F. Automating seedling counts in horticulture using computer vision and AI. Horticulturae 2023, 9, 1134. [Google Scholar] [CrossRef]
- Zhang, W.; Dang, L.M.; Nguyen, L.Q.; Alam, N.; Bui, N.D.; Park, H.Y.; Moon, H. Adapting the segment anything model for plant recognition and automated phenotypic parameter measurement. Horticulturae 2024, 10, 398. [Google Scholar] [CrossRef]
- García-Amaro, E.; Cervantes-Canales, J.; García-Lamont, F.; Lara-Viveros, F.M.; Ruiz-Castilla, J.S.; Espejel Cabrera, J. Use of computer vision techniques for recognition of diseases and pasts in tomato plants. Comput. Sist. 2024, 28, 709–723. [Google Scholar] [CrossRef]
- Clohessy, J.W.; Sanjel, S.; O’Brien, G.K.; Barocco, R.; Kumar, S.; Adkins, S.; Tillman, B.; Wright, D.L.; Small, I.M. Development of a high-throughput plant disease symptom severity assessment tool using machine learning image analysis and integrated geolocation. Comput. Electron. Agric. 2021, 184, 106089. [Google Scholar] [CrossRef]
- García-Vera, Y.E.; Polochè-Arango, A.; Mendivelso-Fajardo, C.A.; Gutiérrez-Bernal, F.J. Hyperspectral image analysis and machine learning techniques for crop disease detection and identification: A review. Sustainability 2024, 16, 6064. [Google Scholar] [CrossRef]
- Ahmad, A.; Saraswat, D.; El Gamal, A. A survey on using deep learning techniques for plant disease diagnosis and recommendations for development of appropriate tools. Smart Agric. Technol. 2023, 3, 100083. [Google Scholar] [CrossRef]
- Braga, P.; Crusiol, L.G.T.; Nanni, M.R.; Caranhato, A.L.H.; Fuhrmann, M.B.; Nepomuceno, A.L.; Neumaier, N.; Farias, J.R.B.; Koltun, A.; Gonçalves, L.S.A.; et al. Vegetation indices and NIR-SWIR spectral bands as a phenotyping tool for water status determination in soybean. Precis. Agric. 2021, 22, 249–266. [Google Scholar] [CrossRef]
- Lee, S.-D.; Gil, C.-S.; Lee, J.-H.; Jeong, H.-B.; Kim, J.-H.; Jang, Y.-A.; Kim, D.-Y.; Lee, W.-M.; Moon, J.-H. Internal quality prediction technology for ‘Sulhyang’ strawberry fruit using organic analysis and hyperspectral imaging. Spectrochim. Acta Part A 2024, 323, 124912. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-D.; Lee, J.-H.; Kim, J.-H.; Jang, Y.-a.; Moon, J.-H. Evaluation technologies for assessing drought tolerance of Kimchi cabbage seedlings using hyperspectral imaging and principal component analysis. Microchem. J. 2024, 206, 111499. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Lafrance, R.; Villicaña, C.; Valdéz-Torres, J.B.; García-Estrada, R.S.; Báez Sañudo, M.A.; Esparza-Araiza, M.J.; León-Félix, J. Selection of tomato (Solanum lycopersicum) Hybrids Resistant to Fol, TYLCV, and TSWV with early maturity and good fruit quality. Horticulturae 2024, 10, 839. [Google Scholar] [CrossRef]
- Oh, B.G.; Chung, J.-S.; Ju, H.-J.; Yoon, J.Y.; Baek, E. Arctium lappa is a new natural host of Tomato spotted wilt virus in Korea. Plant Dis. 2024, 108, 3206. [Google Scholar] [CrossRef] [PubMed]
- Abdisa, E.; Kwon, J.; Jin, G.; Kim, Y. Thrips and TSWV occurrence in geographically different open fields cultivating hot peppers. Korean J. Appl. Entomol. 2024, 63, 43–51. [Google Scholar] [CrossRef]
- Shymanovich, T.; Saville, A.C.; Mohammad, N.; Wei, Q.; Rasmussen, D.; Lahre, K.A.; Rotenberg, D.; Whitfield, A.E.; Ristaino, J.B. Disease progress and detection of a california resistance-breaking strain of Tomato spotted wilt virus in tomato with LAMP and CRISPR-Cas12a assays. PhytoFrontiers 2024, 4, 50–60. [Google Scholar] [CrossRef]
- Juárez, I.D.; Steczkowski, M.X.; Chinnaiah, S.; Rodriguez, A.; Gadhave, K.R.; Kurouski, D. Using Raman spectroscopy for early detection of resistance-breaking strains of tomato spotted wilt orthotospovirus in tomatoes. Front. Plant Sci. 2024, 14, 1283399. [Google Scholar] [CrossRef]
- Caruso, A.G.; Ragona, A.; Agrò, G.; Bertacca, S.; Yahyaoui, E.; Galipienso, L.; Rubio, L.; Panno, S.; Davino, S. Rapid detection of tomato spotted wilt virus by real-time RT-LAMP and in-field application. J. Plant Pathol. 2024, 106, 697–712. [Google Scholar] [CrossRef]
- Jahn, M.; Paran, I.; Hoffmann, K.; Radwanski, E.R.; Livingstone, K.D.; Grube, R.C.; Aftergoot, E.; Lapidot, M.; Moyer, J. Genetic mapping of the Tsw locus for resistance to the Tospovirus Tomato spotted wilt virus in Capsicum spp. and its relationship to the Sw-5 gene for resistance to the same pathogen in tomato. Mol. Plant-Microbe Interact. 2000, 13, 673–682. [Google Scholar] [CrossRef]
- Parisi, M.; Alioto, D.; Tripodi, P. Overview of botic stresses in pepper (Capsicum spp.): Sources of genetic resistance, molecular breeding and genomics. Int. J. Mol. Sci. 2020, 21, 2587. [Google Scholar] [CrossRef]
- Roggero, P.; Masenga, V.; Tavella, L. Field isolates of Tomato spotted wilt virus overcoming resistance in pepper and their spread to other hosts in Italy. Plant Dis. 2002, 86, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.Y.; Her, N.H.; Cho, I.S.; Chung, B.N.; Choi, S.K. First report of a resistance-breaking strain of Tomato spotted wilt orthotospovirus infecting Capsicum annuum carrying the Tsw resistance gene in South Korea. Plant Dis. 2021, 105, 2259. [Google Scholar] [CrossRef]
- Han, J.-H.; Lee, W.-P.; Lee, J.-D.; Kim, M.-K.; Choi, H.-S.; Yoon, J.-B. Symptom and resistance of cultivated and wild Capsicum accessions to Tomato spotted wilt virus. Res. Plant Dis. 2011, 17, 59–65. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X. Plant diseases and pests detection based on deep learning: A review. Plant Methods 2021, 17, 22. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L.; Nitschke, G. Improving deep learning with generic data augmentation. In Proceedings of the 2018 IEEE Symposium Series on Computational Intelligence (SSCI), Bangalore, India, 18–21 November 2018; pp. 1542–1547. [Google Scholar]
- Duffy, S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018, 16, e3000003. [Google Scholar] [CrossRef] [PubMed]
- Ciuffo, M.; Finetti-Sialer, M.M.; Gallitelli, D.; Turina, M. First report in Italy of a resistance-breaking strain of Tomato spotted wilt virus infecting tomato cultivars carrying the Sw5 resistance gene. Plant Pathol. 2005, 54, 564. [Google Scholar] [CrossRef]
- Deligoz, I.; Sokmen, M.A.; Sari, S. First report of resistance breaking strain of Tomato spotted wilt virus (Tospovirus; Bunyaviridae) on resistant sweetpepper cultivars in Turkey. New Dis. Rep. 2014, 30, 26. [Google Scholar] [CrossRef]
- Orecchio, C.; Sacco Botto, C.; Alladio, E.; D’Errico, C.; Vincenti, M.; Noris, E. Non-invasive and early detection of tomato spotted wilt virus infection in tomato plants using a hand-held Raman spectrometer and machine learning modelling. Plant Stress 2025, 15, 100732. [Google Scholar] [CrossRef]
- Macedo, M.A.; Rojas, M.R.; Gilbertson, R.L. First report of a resistance-breaking strain of tomato spotted wilt orthotospovirus infecting sweet pepper with the Tsw resistance gene in California, U.S.A. Plant Dis. 2019, 103, 1048. [Google Scholar] [CrossRef]
- Ferrand, L.; Almeida, M.M.S.; Orílio, A.F.; Dal Bó, E.; Resende, R.O.; García, M.L. Biological and molecular characterization of tomato spotted wilt virus (TSWV) resistance-breaking isolates from Argentina. Plant Pathol. 2019, 68, 1587–1601. [Google Scholar] [CrossRef]




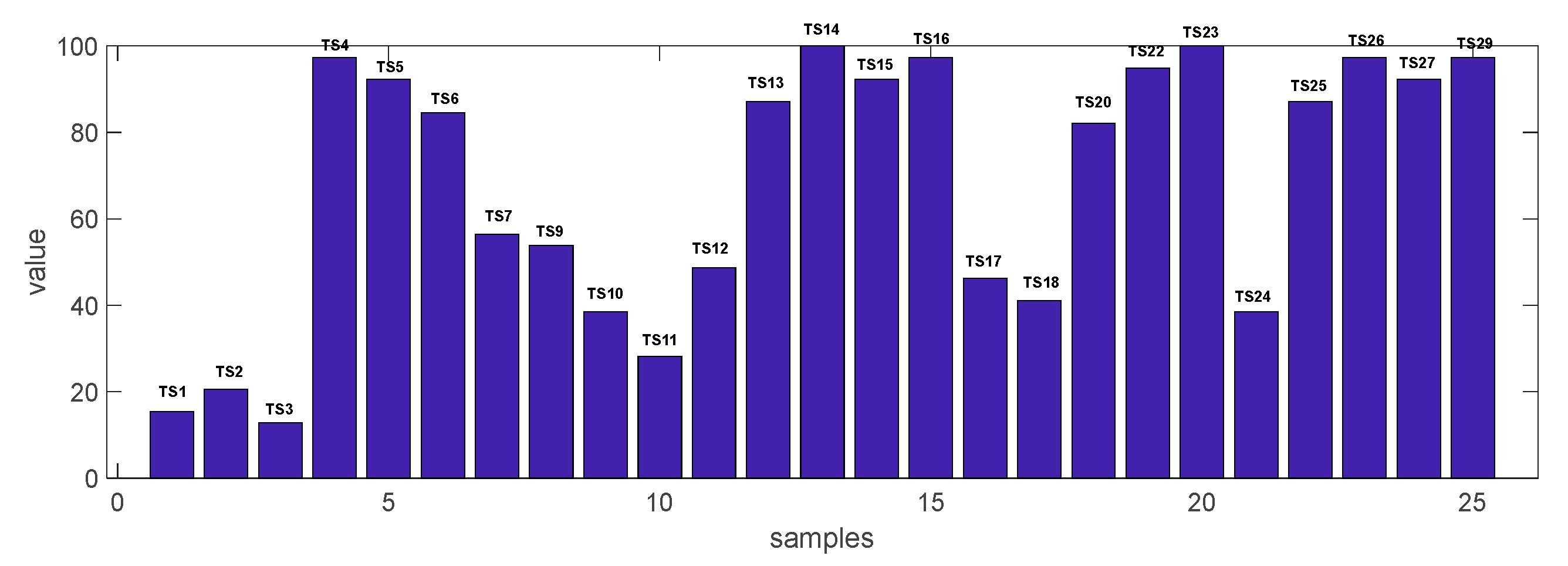
| Germplasm | Pathogen Index z | Disease Incidence y | ELISA x | Machine Learning w | ||||
|---|---|---|---|---|---|---|---|---|
| TS1 | 1.15 ± 0.61 | a | 15.4 ± 6.3 | a | 15.4 ± 6.3 | ab | 20.51 ± 7.3 | a |
| TS2 | 1.13 ± 0.64 | a | 15.4 ± 6.3 | a | 20.5 ± 9.6 | a–c | 15.4 ± 10.9 | a |
| TS3 | 1.08 ± 0.91 | a | 17.9 ± 25.4 | ab | 12.8 ± 18.1 | a | 28.2 ± 13.1 | a |
| TS4 | 3.92 ± 0.11 | fe | 97.4 ± 3.6 | g | 97.4 ± 3.6 | f | 94.9 ± 7.3 | d |
| TS5 | 3.87 ± 0.24 | ef | 92.3 ± 10.9 | fg | 92.3 ± 10.9 | f | 82.4 ± 15.4 | d |
| TS6 | 3.56 ± 0.26 | ef | 87.2 ± 9.6 | e–g | 84.6 ± 10.9 | ef | 76.9 ± 18.8 | cd |
| TS7 | 2.46 ± 0.38 | b–d | 56.4 ± 18.1 | c–f | 56.4 ± 18.1 | ed | 46.2 ± 21.8 | a–c |
| TS9 | 2.33 ± 0.67 | bc | 48.7 ± 25.4 | a–d | 53.8 ± 25.1 | c–e | 23.1 ± 21.8 | a |
| TS10 | 1.69 ± 0.35 | ab | 35.9 ± 22.1 | a–c | 38.5 ± 21.8 | a–d | 15.4 ± 16.6 | a |
| TS11 | 1.87 ± 0.31 | ab | 35.9 ± 18.1 | a–c | 28.2 ± 18.1 | a–d | 28.2 ± 7.3 | a |
| TS12 | 2.41 ± 0.89 | c–d | 53.8 ± 33.2 | b–e | 48.7 ± 28.3 | b–d | 46.2 ± 28.8 | a–c |
| TS13 | 3.46 ± 0.51 | d–f | 87.2 ± 13.1 | e–g | 87.2 ± 9.6 | ef | 66.7 ± 15.8 | b–d |
| TS14 | 4.15 ± 0.11 | ef | 100 ± 0.0 | g | 100 ± 0.0 | f | 94.9 ± 7.3 | d |
| TS15 | 4.23 ± 0.19 | fe | 100 ± 0.0 | g | 92.3 ± 6.3 | f | 97.4 ± 3.6 | d |
| TS16 | 4.00 ± 0.11 | ef | 97.4 ± 3.6 | g | 97.4 ± 3.6 | f | 94.7 ± 3.8 | d |
| TS17 | 3.03 ± 0.73 | c–d | 76.9 ± 22.6 | d–g | 46.2 ± 28.8 | a–d | 69.2 ± 22.6 | b–d |
| TS18 | 2.18 ± 0.67 | a–c | 46.2 ± 28.8 | a–d | 41.0 ± 26.1 | a–d | 43.6 ± 23.8 | ab |
| TS20 | 3.97 ± 0.35 | ef | 92.3 ± 6.3 | fg | 82.1 ± 9.6 | ef | 82.2 ± 13.7 | d |
| TS22 | 4.21 ± 0.29 | fe | 100 ± 0.0 | g | 94.9 ± 7.3 | f | 94.9 ± 3.6 | d |
| TS23 | 4.00 ± 0.13 | ef | 97.4 ± 3.6 | g | 100 ± 0.0 | f | 85.9 ± 14.1 | d |
| TS24 | 1.49 ± 0.80 | ab | 28.2 ± 25.4 | a–c | 38.5 ± 22.6 | a–d | 17.9 ± 13.1 | a |
| TS25 | 3.49 ± 0.44 | d–f | 89.7 ± 14.5 | e–g | 87.2 ± 9.6 | ef | 79.5 ± 3.6 | d |
| TS26 | 3.95 ± 0.07 | ef | 100 ± 0.0 | g | 97.4 ± 3.6 | f | 94.9 ± 3.6 | d |
| TS27 | 3.69 ± 0.44 | ef | 89.7 ± 14.5 | e–g | 92.3 ± 10.9 | f | 92.3 ± 10.9 | d |
| TS29 | 3.92 ± 0.17 | ef | 97.4 ± 3.6 | g | 97.4 ± 3.6 | f | 89.7 ± 7.3 | d |
| Metrics | Pathogen Index | Disease Incidence | NN Model |
|---|---|---|---|
| mean | significant | insignificant | significant |
| standard deviation | insignificant | insignificant | insignificant |
| Visual | ELISA | Neural Network | |
|---|---|---|---|
| advantages | simple | sensitive | accurate |
| disadvantages | inaccurate | complex and expensive | difficult |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.G.; Lee, S.-D.; Lee, W.-M.; Jeong, H.-B.; Yu, N.; Lee, O.-J.; Lee, H.-E. Effective Tomato Spotted Wilt Virus Resistance Assessment Using Non-Destructive Imaging and Machine Learning. Horticulturae 2025, 11, 132. https://doi.org/10.3390/horticulturae11020132
Kim SG, Lee S-D, Lee W-M, Jeong H-B, Yu N, Lee O-J, Lee H-E. Effective Tomato Spotted Wilt Virus Resistance Assessment Using Non-Destructive Imaging and Machine Learning. Horticulturae. 2025; 11(2):132. https://doi.org/10.3390/horticulturae11020132
Chicago/Turabian StyleKim, Sang Gyu, Sang-Deok Lee, Woo-Moon Lee, Hyo-Bong Jeong, Nari Yu, Oak-Jin Lee, and Hye-Eun Lee. 2025. "Effective Tomato Spotted Wilt Virus Resistance Assessment Using Non-Destructive Imaging and Machine Learning" Horticulturae 11, no. 2: 132. https://doi.org/10.3390/horticulturae11020132
APA StyleKim, S. G., Lee, S.-D., Lee, W.-M., Jeong, H.-B., Yu, N., Lee, O.-J., & Lee, H.-E. (2025). Effective Tomato Spotted Wilt Virus Resistance Assessment Using Non-Destructive Imaging and Machine Learning. Horticulturae, 11(2), 132. https://doi.org/10.3390/horticulturae11020132





