AP2/ERF Gene Family in Mango: Genome-Wide Identification and Transcription Analysis During Anthocyanin Biosynthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification and Analysis of AP2/ERF Genes in Mango
2.2. Physicochemical Property Analysis and Subcellular Localization Prediction of MiAP2/ERF Proteins
2.3. Construction of the MiAP2/ERF Phylogenetic Tree and Analysis of Conserved Motifs and Gene Structures
2.4. Chromosomal Localization and Collinearity Analysis
2.5. RNA-Seq Expression Pattern Analysis
2.6. Plant Material and Treatment
2.7. RNA Extraction and qPCR Analysis
2.8. Primer Design and Synthesis
2.9. Statistical Analysis
3. Results
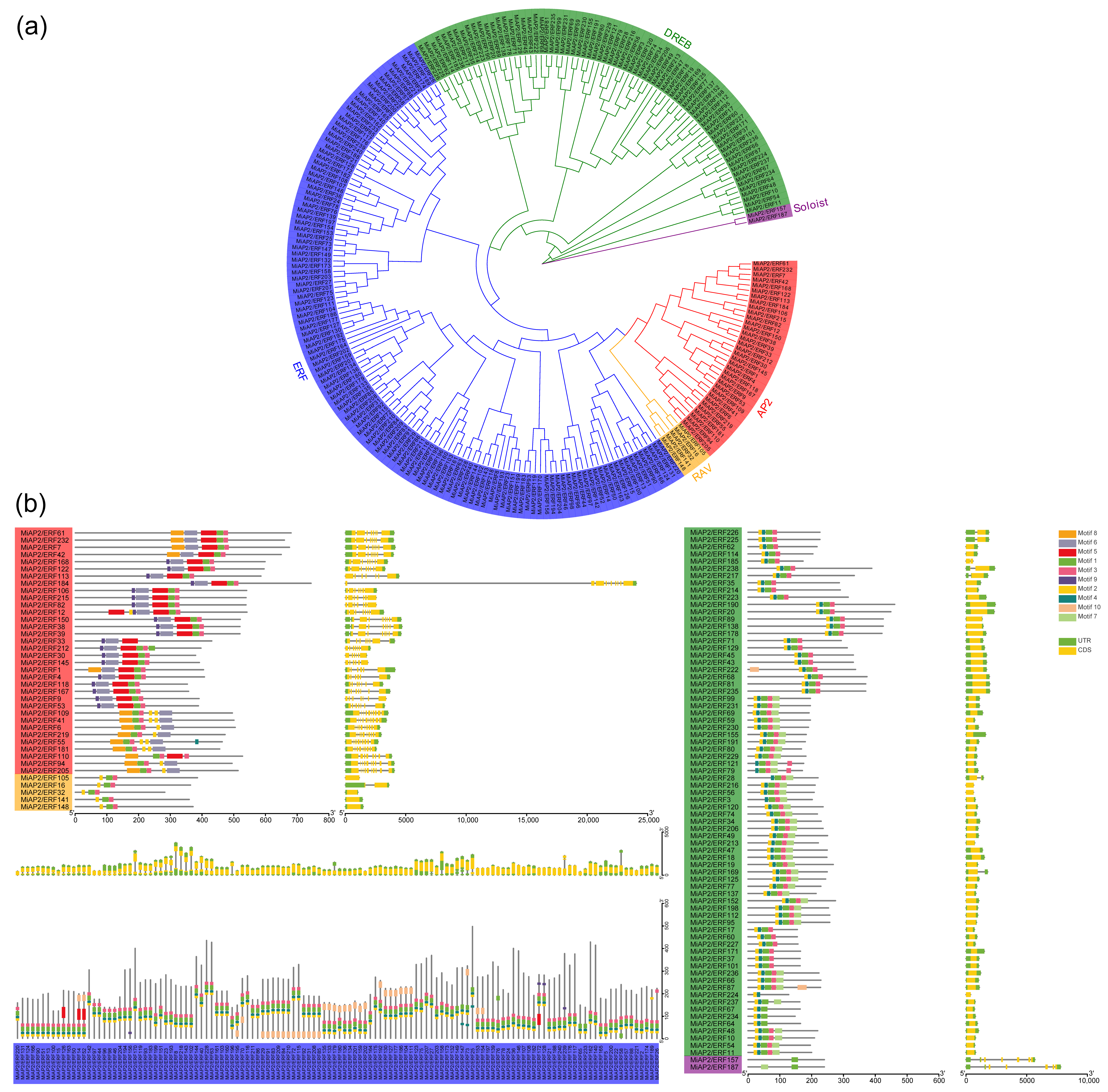
3.1. Identification and Phylogenetic Analysis of MiAP2/ERF Proteins
3.2. Gene Structure and Conserved Motif Analysis of MiAP2/ERF Proteins
3.3. Chromosomal Distribution and Syntenic Analysis of MiAP2/ERF Genes
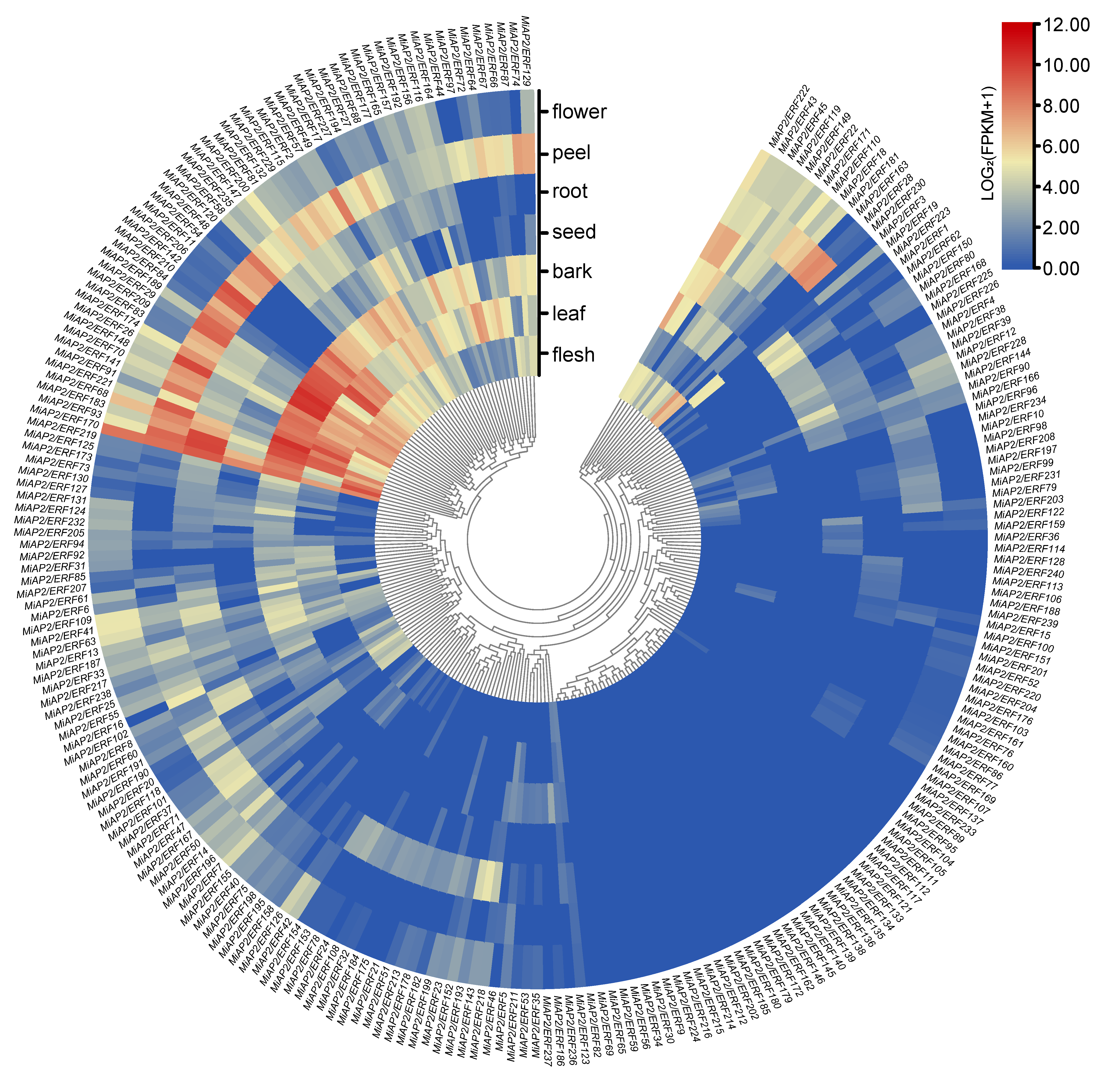
3.4. Organ-Specific Expression Profiling of MiAP2/ERF Genes
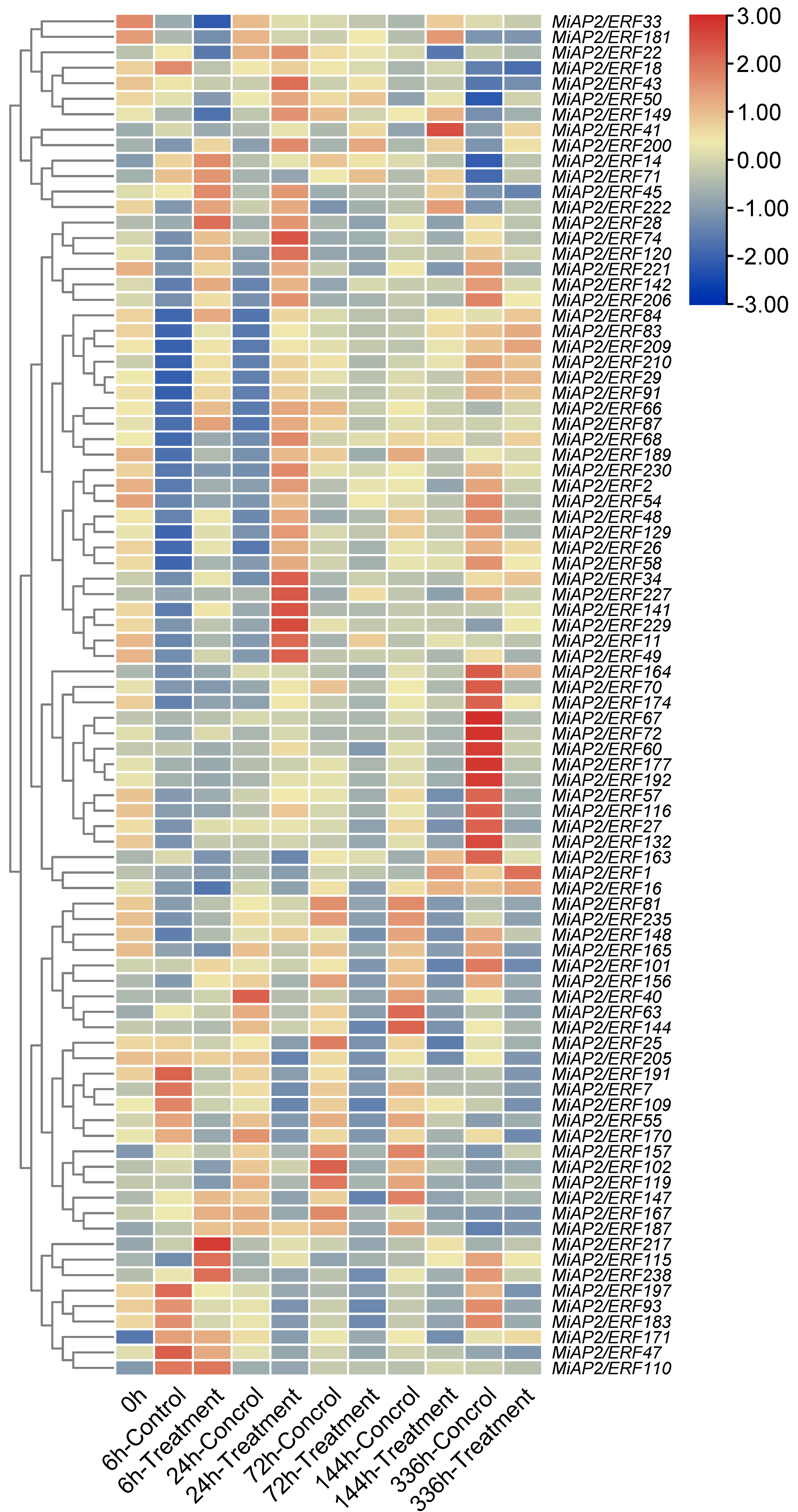
3.5. Analysis of MiAP2/ERF Gene Expression Patterns in Mango Peel Subjected to Preharvest “Bagging–Debagging” Treatment
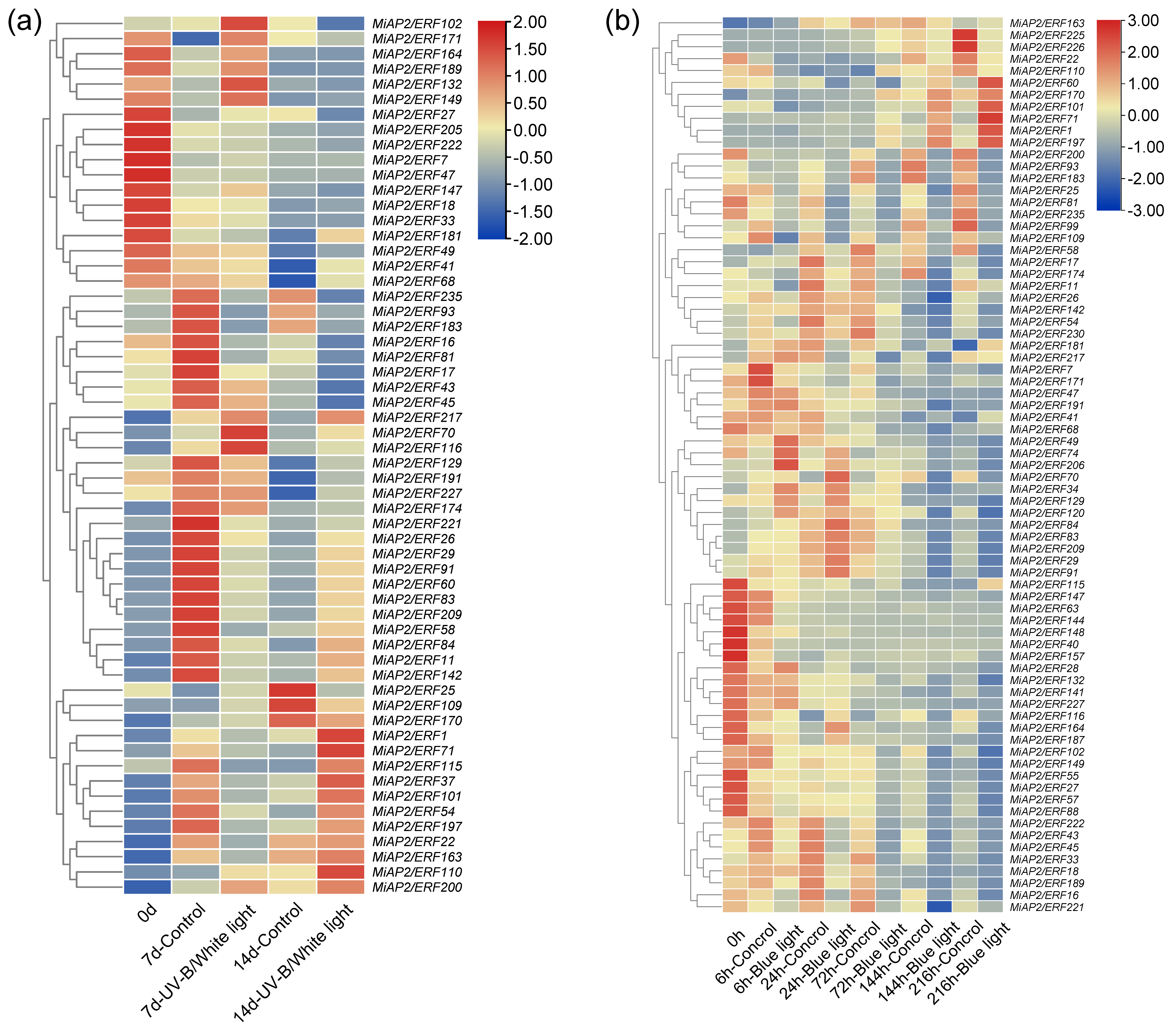
3.6. Analysis of MiAP2/ERF Gene Expression Patterns Under UV-B/White Light and Blue Light Treatments
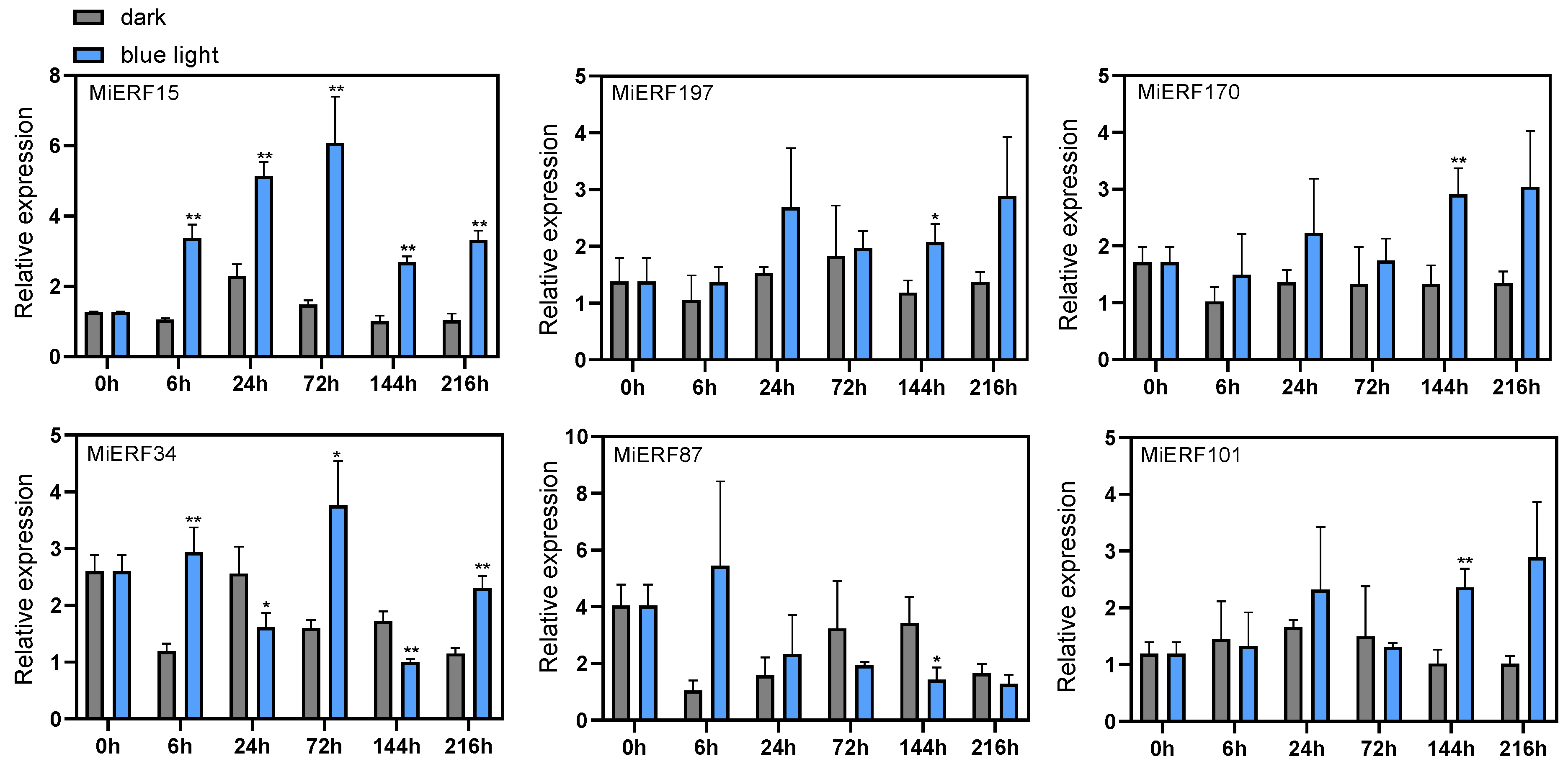
3.7. qPCR Analysis of MiAP2/ERF Expression Patterns During Blue-Light-Induced Anthocyanin Accumulation in Mango
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ledesma, N.; Campbell, R.J. The Status of Mango Cultivars, Market Perspectives and Mango Cultivar Improvement for the Future. Acta Hortic. 2019, 1244, 23–28. [Google Scholar] [CrossRef]
- Zhu, W.; Wu, H.; Yang, C.; Shi, B.; Zheng, B.; Ma, X.; Zhou, K.; Qian, M. Postharvest Light-Induced Flavonoids Accumulation in Mango (Mangifera indica L.) Peel Is Associated with the up-Regulation of Flavonoids-Related and Light Signal Pathway Genes. Front. Plant Sci. 2023, 14, 1136281. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Wu, H.; Zheng, B.; Qian, M.; Gao, A.; Zhou, K. Analysis of Light-Independent Anthocyanin Accumulation in Mango (Mangifera indica L.). Horticulturae 2021, 7, 423. [Google Scholar] [CrossRef]
- Jaakola, L. New Insights into the Regulation of Anthocyanin Biosynthesis in Fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Araguirang, G.E.; Richter, A.S. Activation of Anthocyanin Biosynthesis in High Light—What Is the Initial Signal? New Phytol. 2022, 236, 2037–2043. [Google Scholar] [CrossRef]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 Synergistically Specify the Expression of BANYULS and Proanthocyanidin Biosynthesis in Arabidopsis thaliana. Plant J. 2004, 39, 366–380. [Google Scholar] [CrossRef]
- Schaart, J.G.; Dubos, C.; Romero De La Fuente, I.; van Houwelingen, A.M.M.L.; de Vos, R.C.H.; Jonker, H.H.; Xu, W.; Routaboul, J.-M.; Lepiniec, L.; Bovy, A.G. Identification and Characterization of MYB-bHLH-WD40 Regulatory Complexes Controlling Proanthocyanidin Biosynthesis in Strawberry (Fragaria × Ananassa) Fruits. New Phytol. 2013, 197, 454–467. [Google Scholar] [CrossRef]
- Lu, N.; Rao, X.; Li, Y.; Jun, J.H.; Dixon, R.A. Dissecting the Transcriptional Regulation of Proanthocyanidin and Anthocyanin Biosynthesis in Soybean (Glycine max). Plant Biotechnol. J. 2021, 19, 1429–1442. [Google Scholar] [CrossRef]
- Ji, X.; Li, Z.; Zhang, M.; Lang, S.; Song, X. ChMYB1-ChbHLH42-ChTTG1 Module Regulates Abscisic Acid-Induced Anthocyanin Biosynthesis in Cerasus humilis. Hortic. Plant J. 2024, 10, 51–65. [Google Scholar] [CrossRef]
- Shoji, T.; Yuan, L. ERF Gene Clusters: Working Together to Regulate Metabolism. Trends Plant Sci. 2021, 26, 23–32. [Google Scholar] [CrossRef]
- Tang, Q.; Wei, S.; Zheng, X.; Tu, P.; Tao, F. APETALA2/Ethylene-Responsive Factors in Higher Plant and Their Roles in Regulation of Plant Stress Response. Crit. Rev. Biotechnol. 2024, 44, 1533–1551. [Google Scholar] [CrossRef]
- Wu, Y.; Li, X.; Zhang, J.; Zhao, H.; Tan, S.; Xu, W.; Pan, J.; Yang, F.; Pi, E. ERF Subfamily Transcription Factors and Their Function in Plant Responses to Abiotic Stresses. Front. Plant Sci. 2022, 13, 1042084. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Haxim, Y.; Liang, Y.; Qiao, S.; Gao, B.; Zhang, D.; Li, X. Genome-Wide Investigation of AP2/ERF Gene Family in the Desert Legume Eremosparton Songoricum: Identification, Classification, Evolution, and Expression Profiling under Drought Stress. Front. Plant Sci. 2022, 13, 885694. [Google Scholar] [CrossRef] [PubMed]
- Jofuku, K.D.; den Boer, B.G.; Van Montagu, M.; Okamuro, J.K. Control of Arabidopsis Flower and Seed Development by the Homeotic Gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-Wide Analysis of the ERF Gene Family in Arabidopsis and Rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Zhuang, J.; Chen, J.-M.; Yao, Q.-H.; Xiong, F.; Sun, C.-C.; Zhou, X.-R.; Zhang, J.; Xiong, A.-S. Discovery and Expression Profile Analysis of AP2/ERF Family Genes from Triticum aestivum. Mol. Biol. Rep. 2011, 38, 745–753. [Google Scholar] [CrossRef]
- Zhuang, J.; Cai, B.; Peng, R.-H.; Zhu, B.; Jin, X.-F.; Xue, Y.; Gao, F.; Fu, X.-Y.; Tian, Y.-S.; Zhao, W.; et al. Genome-Wide Analysis of the AP2/ERF Gene Family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 2008, 371, 468–474. [Google Scholar] [CrossRef]
- Licausi, F.; Giorgi, F.M.; Zenoni, S.; Osti, F.; Pezzotti, M.; Perata, P. Genomic and Transcriptomic Analysis of the AP2/ERF Superfamily in Vitis vinifera. BMC Genom. 2010, 11, 719. [Google Scholar] [CrossRef]
- Girardi, C.L.; Rombaldi, C.V.; Dal Cero, J.; Nobile, P.M.; Laurens, F.; Bouzayen, M.; Quecini, V. Genome-Wide Analysis of the AP2/ERF Superfamily in Apple and Transcriptional Evidence of ERF Involvement in Scab Pathogenesis. Sci. Hortic. 2013, 151, 112–121. [Google Scholar] [CrossRef]
- Xie, X.; Shen, S.; Yin, X.; Xu, Q.; Sun, C.; Grierson, D.; Ferguson, I.; Chen, K. Isolation, Classification and Transcription Profiles of the AP2/ERF Transcription Factor Superfamily in Citrus. Mol. Biol. Rep. 2014, 41, 4261–4271. [Google Scholar] [CrossRef]
- Canher, B.; Lanssens, F.; Zhang, A.; Bisht, A.; Mazumdar, S.; Heyman, J.; Wolf, S.; Melnyk, C.W.; De Veylder, L. The Regeneration Factors ERF114 and ERF115 Regulate Auxin-Mediated Lateral Root Development in Response to Mechanical Cues. Mol. Plant 2022, 15, 1543–1557. [Google Scholar] [CrossRef]
- Guyomarc’h, S.; Boutté, Y.; Laplaze, L. AP2/ERF Transcription Factors Orchestrate Very Long Chain Fatty Acid Biosynthesis during Arabidopsis Lateral Root Development. Mol. Plant 2021, 14, 205–207. [Google Scholar] [CrossRef] [PubMed]
- Gambhir, P.; Singh, V.; Parida, A.; Raghuvanshi, U.; Kumar, R.; Sharma, A.K. Ethylene Response Factor ERF.D7 Activates Auxin Response Factor 2 Paralogs to Regulate Tomato Fruit Ripening. Plant Physiol. 2022, 190, 2775–2796. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Geng, M.; Ma, L.; Yao, G.; Zhang, M.; Zhang, H. The H2S-Responsive Transcription Factor ERF.D3 Regulates Tomato Abscisic Acid Metabolism, Leaf Senescence, and Fruit Ripening. Plant Physiol. 2025, 197, kiae560. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Cui, Y.; Fan, Z.; Huang, H.; Wang, Z.; Chen, S.; Ma, H. Ficus carica ERF12 Improves Fruit Firmness at Ripening. Hortic. Plant J. 2024. [Google Scholar] [CrossRef]
- Cai, X.; Zeng, X.; Wang, X.; Pan, D.; Zhang, J.; Li, Z.; Yang, J.; Zhang, Y.; Zeng, J.; Zhang, Q.; et al. Hormone Metabolic Profiling and Transcriptome Analysis Reveal Phytohormone Crosstalk and the Role of OfERF017 in the Flowering and Senescence of Sweet Osmanthus. Hortic. Plant J. 2025. [Google Scholar] [CrossRef]
- Kong, L.; Song, Q.; Wei, H.; Wang, Y.; Lin, M.; Sun, K.; Zhang, Y.; Yang, J.; Li, C.; Luo, K. The AP2/ERF Transcription Factor PtoERF15 Confers Drought Tolerance via JA-Mediated Signaling in Populus. New Phytol. 2023, 240, 1848–1867. [Google Scholar] [CrossRef]
- Xu, L.; Yang, L.; Li, A.; Guo, J.; Wang, H.; Qi, H.; Li, M.; Yang, P.; Song, S. An AP2/ERF Transcription Factor Confers Chilling Tolerance in Rice. Sci. Adv. 2024, 10, eado4788. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Bürger, M.; Wang, Y.; Chory, J. Two Interacting Ethylene Response Factors Regulate Heat Stress Response. Plant Cell 2021, 33, 338–357. [Google Scholar] [CrossRef]
- Ni, J.; Bai, S.; Zhao, Y.; Qian, M.; Tao, R.; Yin, L.; Gao, L.; Teng, Y. Ethylene Response Factors Pp4ERF24 and Pp12ERF96 Regulate Blue Light-Induced Anthocyanin Biosynthesis in ‘Red Zaosu’ Pear Fruits by Interacting with MYB114. Plant Mol. Biol. 2019, 99, 67–78. [Google Scholar] [CrossRef]
- Ni, J.; Premathilake, A.T.; Gao, Y.; Yu, W.; Tao, R.; Teng, Y.; Bai, S. Ethylene-Activated PpERF105 Induces the Expression of the Repressor-Type R2R3-MYB Gene PpMYB140 to Inhibit Anthocyanin Biosynthesis in Red Pear Fruit. Plant J. 2021, 105, 167–181. [Google Scholar] [CrossRef]
- Ni, J.; Wang, S.; Yu, W.; Liao, Y.; Pan, C.; Zhang, M.; Tao, R.; Wei, J.; Gao, Y.; Wang, D.; et al. The Ethylene-Responsive Transcription Factor PpERF9 Represses PpRAP2.4 and PpMYB114 via Histone Deacetylation to Inhibit Anthocyanin Biosynthesis in Pear. Plant Cell 2023, 35, 2271–2292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Shi, Y.; Ma, Y.; Yang, X.; Yin, X.; Zhang, Y.; Xiao, Y.; Liu, W.; Li, Y.; Li, S.; et al. The Strawberry Transcription Factor FaRAV1 Positively Regulates Anthocyanin Accumulation by Activation of FaMYB10 and Anthocyanin Pathway Genes. Plant Biotechnol. J. 2020, 18, 2267–2279. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Tomes, S.; Gleave, A.P.; Zhang, H.; Dare, A.P.; Plunkett, B.; Espley, R.V.; Luo, Z.; Zhang, R.; Allan, A.C.; et al. microRNA172 Targets APETALA2 to Regulate Flavonoid Biosynthesis in Apple (Malus domestica). Hortic. Res. 2022, 9, uhab007. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, Z.; Zhan, X.; Lin, M.; Li, X.; Wang, R.; Sun, L.; Gu, H.; Wei, F.; Fang, J.; et al. AaEIL2 and AaERF059 Are Involved in Fruit Coloration and Ripening by Crossly Regulating Ethylene and Auxin Signal Pathway in Actinidia arguta. Hortic. Plant J. 2025. [Google Scholar] [CrossRef]
- Wang, P.; Luo, Y.; Huang, J.; Gao, S.; Zhu, G.; Dang, Z.; Gai, J.; Yang, M.; Zhu, M.; Zhang, H.; et al. The Genome Evolution and Domestication of Tropical Fruit Mango. Genome Biol. 2020, 21, 60. [Google Scholar] [CrossRef]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.-H.; Hu, S. Evolview v2: An Online Visualization and Management Tool for Customized and Annotated Phylogenetic Trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Yang, C.; Wang, X.; Zhu, W.; Weng, Z.; Li, F.; Zhang, Y.; Wu, H.; Zhou, K.; Strid, Å.; Qian, M. Metabolomic and Transcriptomic Analyses Reveal the Regulation Mechanism of Postharvest Light-Induced Phenolics Accumulation in Mango Peel. LWT 2024, 213, 117050. [Google Scholar] [CrossRef]
- Ni, J.; Liao, Y.; Zhang, M.; Pan, C.; Yang, Q.; Bai, S.; Teng, Y. Blue Light Simultaneously Induces Peel Anthocyanin Biosynthesis and Flesh Carotenoid/Sucrose Biosynthesis in Mango Fruit. J. Agric. Food Chem. 2022, 70, 16021–16035. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; He, Y.; Li, S.; Shi, S.; Li, L.; Liu, Y.; Chen, H. Genome-Wide Characterization and Expression Analysis of AP2/ERF Genes in Eggplant (Solanum melongena L.). Plant Physiol. Biochem. 2021, 167, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Zafar, M.M.; Rehman, A.; Razzaq, A.; Parvaiz, A.; Mustafa, G.; Sharif, F.; Mo, H.; Youlu, Y.; Shakeel, A.; Ren, M. Genome-Wide Characterization and Expression Analysis of Erf Gene Family in Cotton. BMC Plant Biol. 2022, 22, 134. [Google Scholar] [CrossRef]
- Sharoni, A.M.; Nuruzzaman, M.; Satoh, K.; Shimizu, T.; Kondoh, H.; Sasaya, T.; Choi, I.-R.; Omura, T.; Kikuchi, S. Gene Structures, Classification and Expression Models of the AP2/EREBP Transcription Factor Family in Rice. Plant Cell Physiol. 2011, 52, 344–360. [Google Scholar] [CrossRef]
- Wei, Y.; Kong, Y.; Li, H.; Yao, A.; Han, J.; Zhang, W.; Li, X.; Li, W.; Han, D. Genome-Wide Characterization and Expression Profiling of the AP2/ERF Gene Family in Fragaria vesca L. Int. J. Mol. Sci. 2024, 25, 7614. [Google Scholar] [CrossRef]
- He, S.; Hao, X.; He, S.; Hao, X.; Zhang, P.; Chen, X. Correction to: Genome-Wide Identification, Phylogeny and Expression Analysis of AP2/ERF Transcription Factors Family in Sweet Potato. BMC Genom. 2022, 23, 139. [Google Scholar] [CrossRef]
- Cheng, C.; An, L.; Li, F.; Ahmad, W.; Aslam, M.; Ul Haq, M.Z.; Yan, Y.; Ahmad, R.M. Wide-Range Portrayal of AP2/ERF Transcription Factor Family in Maize (Zea mays L.) Development and Stress Responses. Genes 2023, 14, 194. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis among Eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-Binding Specificity of the ERF/AP2 Domain of Arabidopsis DREBs, Transcription Factors Involved in Dehydration- and Cold-Inducible Gene Expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Giri, M.K.; Swain, S.; Gautam, J.K.; Singh, S.; Singh, N.; Bhattacharjee, L.; Nandi, A.K. The Arabidopsis Thaliana At4g13040 Gene, a Unique Member of the AP2/EREBP Family, Is a Positive Regulator for Salicylic Acid Accumulation and Basal Defense against Bacterial Pathogens. J. Plant Physiol. 2014, 171, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Krizek, B.A.; Bantle, A.T.; Heflin, J.M.; Han, H.; Freese, N.H.; Loraine, A.E. AINTEGUMENTA and AINTEGUMENTA-LIKE6 Directly Regulate Floral Homeotic, Growth, and Vascular Development Genes in Young Arabidopsis Flowers. J. Exp. Bot. 2021, 72, 5478–5493. [Google Scholar] [CrossRef] [PubMed]
- Giraudat, J.; Hauge, B.M.; Valon, C.; Smalle, J.; Parcy, F.; Goodman, H.M. Isolation of the Arabidopsis ABI3 Gene by Positional Cloning. Plant Cell 1992, 4, 1251–1261. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.; Yamasaki, K.; Sarai, A.; Ohme-Takagi, M. Determinants in the Sequence Specific Binding of Two Plant Transcription Factors, CBF1 and NtERF2, to the DRE and GCC Motifs. Biochemistry 2002, 41, 4202–4208. [Google Scholar] [CrossRef]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 Encodes an AP2 Domain-Containing Transcriptional Activator That Binds to the C-Repeat/DRE, a Cis-Acting DNA Regulatory Element That Stimulates Transcription in Response to Low Temperature and Water Deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two Transcription Factors, DREB1 and DREB2, with an EREBP/AP2 DNA Binding Domain Separate Two Cellular Signal Transduction Pathways in Drought- and Low-Temperature-Responsive Gene Expression, Respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef]
- Vision, T.J.; Brown, D.G.; Tanksley, S.D. The Origins of Genomic Duplications in Arabidopsis. Science 2000, 290, 2114–2117. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, X.; Qu, Z.; Guo, W.; van Nocker, S.; Zhang, C. Grapevine VviERF105 Promotes Tolerance to Abiotic Stress and Is Degraded by the E3 Ubiquitin Ligase VviPUB19. Environ. Exp. Bot. 2022, 201, 105001. [Google Scholar] [CrossRef]
- Jung, S.E.; Bang, S.W.; Kim, S.H.; Seo, J.S.; Yoon, H.-B.; Kim, Y.S.; Kim, J.-K. Overexpression of OsERF83, a Vascular Tissue-Specific Transcription Factor Gene, Confers Drought Tolerance in Rice. Int. J. Mol. Sci. 2021, 22, 7656. [Google Scholar] [CrossRef]
- Neogy, A.; Garg, T.; Kumar, A.; Dwivedi, A.K.; Singh, H.; Singh, U.; Singh, Z.; Prasad, K.; Jain, M.; Yadav, S.R. Corrigendum to: Genome-Wide Transcript Profiling Reveals an Auxin-Responsive Transcription Factor, OsAP2/ERF-40, Promoting Rice Adventitious Root Development. Plant Cell Physiol. 2021, 62, 1786. [Google Scholar] [CrossRef]
- Nie, J.; Wen, C.; Xi, L.; Lv, S.; Zhao, Q.; Kou, Y.; Ma, N.; Zhao, L.; Zhou, X. The AP2/ERF Transcription Factor CmERF053 of Chrysanthemum Positively Regulates Shoot Branching, Lateral Root, and Drought Tolerance. Plant Cell Rep. 2018, 37, 1049–1060. [Google Scholar] [CrossRef] [PubMed]
- Marsch-Martinez, N.; Greco, R.; Becker, J.D.; Dixit, S.; Bergervoet, J.H.W.; Karaba, A.; de Folter, S.; Pereira, A. BOLITA, an Arabidopsis AP2/ERF-like Transcription Factor That Affects Cell Expansion and Proliferation/Differentiation Pathways. Plant Mol. Biol. 2006, 62, 825–843. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Wang, S.; Ning, K.; Chen, Z.; Wang, Y.; Yang, J.; Qi, M.; Wang, Q. The APETALA2 Transcription Factor LsAP2 Regulates Seed Shape in Lettuce. J. Exp. Bot. 2021, 72, 2463–2476. [Google Scholar] [CrossRef]
- Wu, T.; Liu, H.-T.; Zhao, G.-P.; Song, J.-X.; Wang, X.-L.; Yang, C.-Q.; Zhai, R.; Wang, Z.-G.; Ma, F.-W.; Xu, L.-F. Jasmonate and Ethylene-Regulated Ethylene Response Factor 22 Promotes Lanolin-Induced Anthocyanin Biosynthesis in “Zaosu” Pear (Pyrus bretschneideri Rehd.) Fruit. Biomolecules 2020, 10, 278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, Y.; Zhang, Z.; Guo, D.; Zhao, X.; Gao, W.; Zhang, J.; Song, B. StRAV1 Negatively Regulates Anthocyanin Accumulation in Potato. Sci. Hortic. 2022, 295, 110817. [Google Scholar] [CrossRef]
- Yao, G.; Ming, M.; Allan, A.C.; Gu, C.; Li, L.; Wu, X.; Wang, R.; Chang, Y.; Qi, K.; Zhang, S.; et al. Map-Based Cloning of the Pear Gene MYB114 Identifies an Interaction with Other Transcription Factors to Coordinately Regulate Fruit Anthocyanin Biosynthesis. Plant J. 2017, 92, 437–451. [Google Scholar] [CrossRef]
- Koyama, T.; Sato, F. The Function of ETHYLENE RESPONSE FACTOR Genes in the Light-Induced Anthocyanin Production of Arabidopsis thaliana Leaves. Plant Biotechnol. 2018, 35, 87–91. [Google Scholar] [CrossRef]
- Ma, H.; Yang, T.; Li, Y.; Zhang, J.; Wu, T.; Song, T.; Yao, Y.; Tian, J. The Long Noncoding RNA MdLNC499 Bridges MdWRKY1 and MdERF109 Function to Regulate Early-Stage Light-Induced Anthocyanin Accumulation in Apple Fruit. Plant Cell 2021, 33, 3309–3330. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, W.; Tahir, M.M.; Zhou, K.; Deng, Q.; Qian, M. AP2/ERF Gene Family in Mango: Genome-Wide Identification and Transcription Analysis During Anthocyanin Biosynthesis. Horticulturae 2025, 11, 1500. https://doi.org/10.3390/horticulturae11121500
Zhu W, Tahir MM, Zhou K, Deng Q, Qian M. AP2/ERF Gene Family in Mango: Genome-Wide Identification and Transcription Analysis During Anthocyanin Biosynthesis. Horticulturae. 2025; 11(12):1500. https://doi.org/10.3390/horticulturae11121500
Chicago/Turabian StyleZhu, Wencan, Muhammad Mobeen Tahir, Kaibing Zhou, Qin Deng, and Minjie Qian. 2025. "AP2/ERF Gene Family in Mango: Genome-Wide Identification and Transcription Analysis During Anthocyanin Biosynthesis" Horticulturae 11, no. 12: 1500. https://doi.org/10.3390/horticulturae11121500
APA StyleZhu, W., Tahir, M. M., Zhou, K., Deng, Q., & Qian, M. (2025). AP2/ERF Gene Family in Mango: Genome-Wide Identification and Transcription Analysis During Anthocyanin Biosynthesis. Horticulturae, 11(12), 1500. https://doi.org/10.3390/horticulturae11121500






