Abstract
Sugars function as essential signaling molecules and metabolic substrates in plant growth, development, yield formation, and fruit quality. The aril of litchi (Litchi chinensis Sonn.) accumulates high levels of hexoses, primarily glucose and fructose; however, the molecular mechanisms underlying this process remain poorly characterized. This study aimed to systematically identify the monosaccharide transporter (MST) gene family in litchi and elucidate its role in aril sugar accumulation. Through a comprehensive analysis of the litchi genome, we identified a total of 45 LcMST genes, which were classified into seven distinct subfamilies: STP, ERD6L, PLT, INT, pGlcT, TMT, and VGT. Analysis of gene structure and conserved motifs revealed notable conservation among members within the same subfamily. Collinearity and gene duplication analyses suggested that the LcMST family expanded through both tandem and whole-genome duplication events, a process primarily governed by purifying selection. Expression profiling across diverse tissues demonstrated that LcMST genes exhibit distinct tissue-specific expression patterns. During fruit development in the hexose-dominant cultivar ‘Tianshuili’, the expression of the tonoplast monosaccharide transporter gene LcTMT1 exhibited a significant positive correlation with the accumulation of fructose, glucose, and total sugars. Heterologous functional complementation assays in yeast confirmed the ability of LcTMT1 to transport both glucose and fructose. In conclusion, this study presents the first genome-wide identification and characterization of the MST gene family in litchi, and identifies LcTMT1 as a key contributor of hexose accumulation in the aril. These findings establish a foundation for elucidating the molecular mechanisms of sugar accumulation in litchi fruit and for guiding future genetic improvement of fruit quality.
1. Introduction
In higher plants, sugars such as monosaccharides, sucrose, and polyols are not merely the end products of photosynthesis but also fulfill a multitude of indispensable physiological roles [1]. They serve as the primary substrates for cellular respiration, provide the fundamental carbon skeletons for the biosynthesis of macromolecules, and act as osmotic regulators to maintain cellular turgor [2]. Furthermore, sugars and their metabolic intermediates function as key signaling molecules within complex regulatory networks that modulate developmental processes, including seed germination, flowering, and fruit ripening, and mediate responses to biotic and abiotic stresses, such as drought, low temperature, and pathogen infection [3,4]. In horticultural crops, the relative proportions of sucrose, glucose, and fructose are critical determinants of consumer acceptance, as they directly influence core quality attributes like sweetness, flavor, color, and texture [5].
Sugars are primarily synthesized in source organs, such as mature leaves, with sucrose serving as the major carbohydrate translocated through the phloem to distant sink organs like roots, stems, fruits, and seeds [6,7]. The translocation of sugars from source to sink tissues is a tightly regulated process that encompasses unloading and compartmentalization. This directional transport across various membrane systems, including the plasma membrane, tonoplast, and chloroplast envelope, is facilitated by specific sugar transporter proteins [8,9]. Consequently, these transporters serve as critical regulatory nodes that integrate processes of sugar synthesis, translocation, partitioning, storage, and utilization, thereby playing a decisive role in determining source–sink relationships, plant productivity, and fruit quality, as well as stress tolerance [10,11].
The plant sugar transporter system primarily comprises three major families: sucrose transporters (SUT or SUC), monosaccharide transporters (MSTs), and sugar will eventually be exported transporters (SWEETs) [1]. The MST family encompasses seven functionally specialized subfamilies including sugar transporter proteins (STPs, also named hexose transporters, HTs), tonoplast monosaccharide transporters (TMTs, or tonoplast sugar transporters, TSTs), polyol/monosaccharide transporters (PLTs/PMTs), vacuolar glucose transporters (VGTs), inositol transporters (INTs), plastidic glucose transporters (pGlcTs), and early responsive-to-dehydration six-like proteins (ERD6L, also termed sugar facilitator proteins, SFPs) [12]. These specialized MST proteins transport specific sugar substrates. For instance, the tonoplast sugar transporter MdERDL6.1 facilitates glucose efflux from the vacuole to the cytoplasm in apple, and its overexpression enhances sugar accumulation by upregulating other TST genes [5].
Litchi (Litchi chinensis Sonn.), a member of the Sapindaceae family, is an economically important subtropical fruit tree. Its aril accumulates substantial levels of soluble sugars, constituting 15–20% of the fresh weight at maturity, which are primarily composed of sucrose, glucose, and fructose [13,14]. Based on the hexose-to-sucrose ratio, litchi cultivars are categorized as hexose-dominant (ratio > 2), sucrose-dominant (ratio < 1), or intermediate (ratio 1–2) [15]. To date, research on sugar transporters in litchi has predominantly focused on the SUT and SWEET families. Among the five identified LcSUT genes, LcSUT4 exhibits the highest expression level in the aril, and its expression pattern correlates with sucrose accumulation dynamics, suggesting a role in apoplastic sucrose transport [16]. Similarly, of the 16 identified LcSWEET genes, LcSWEET10 is the most highly expressed in the aril. Its expression is positively correlated with sucrose content and is significantly higher in the high-sugar cultivar ‘Wuheli’ than in ‘Feizixiao’, indicating a potential key role in sucrose unloading and accumulation [17].
In contrast to most fruits, the litchi aril lacks vascular tissue, implying that sucrose unloaded from the phloem in the funiculus is transported via the apoplastic pathway [16]. Consequently, hexoses (glucose and fructose) derived from the hydrolysis of sucrose must be transported into aril cells by plasma membrane-localized MST proteins [18]. Previous research has demonstrated that sucrose synthase activity increases in accordance with the hexose-to-sucrose ratio across different litchi cultivars, exhibiting a significant positive linear correlation [15]. However, the molecular mechanisms governing hexose accumulation in the litchi aril, as well as the characteristics of the corresponding MST gene family, have not yet been systematically investigated. Thus, a comprehensive analysis of the MST gene family and its functional role in aril sugar accumulation is of considerable importance.
This study was designed to characterize the MST gene family in litchi, with the following objectives: (1) to comprehensively analyze the gene structures, conserved motifs, and chromosomal distributions of LcMST genes; (2) to investigate the expression patterns of LcMST genes across various tissues as well as in the aril at different developmental stages; and (3) to validate the transport functions of select candidate genes through heterologous functional complementation assays in yeast. The findings are expected to provide new insights into the roles of MSTs in aril development and sugar accumulation, thereby establishing a molecular foundation for the genetic improvement of fruit quality in litchi.
2. Materials and Methods
2.1. Plant Materials and Sampling
The hexose-accumulating litchi cultivar ‘Tianshuili’ was used in this study. The experimental trees, which were approximately 20 years old and exhibited consistent annual fruiting, were cultivated at the National Litchi and Banana Germplasm Repository in Guangzhou, China. The orchard is located in a region with a marine subtropical monsoon climate and features red clay soil, where consistent management practices and sustained soil fertility guarantee an annual cycle of flowering and fruiting [14].
Fruit samples were collected at 60, 67, 74, 81, and 88 days after flowering (DAF). For each time point, twenty fruits were randomly harvested from each of three individual trees, with the arils from each tree pooled to form one biological replicate (n = 3). After the removal of the pericarp and seed, the arils were immediately frozen in liquid nitrogen and stored at −80 °C for subsequent RNA extraction and sugar content analysis.
2.2. Bioinformatic Analyses
2.2.1. Identification of LcMST Genes
The amino acid sequences of Arabidopsis thaliana monosaccharide transporters (MSTs) were obtained from The Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org/, accessed on 11 January 2025) and relevant literature [19] and used as query sequences. Candidate MST genes in L. chinensis were identified by performing a BLASTP search against the ‘Feizixiao’ lychee genome (BioProject: PRJNA747875) with an E-value threshold of 1.0 × 10−5. To reduce false positives, the resulting sequences were further validated by querying the UniProtKB database (https://www.uniprot.org/, accessed on 19 March 2025). Conserved domains were analyzed using both the CDD and Pfam databases, with a focus on retaining only those LcMST members that contained the characteristic Sugar_tr (PF00083) domain.
The physicochemical properties of the LcMST proteins, including molecular weight (MW) and isoelectric point (pI), were predicted using the ExPASy online tool (https://web.expasy.org/protparam/, accessed on 22 July 2025). Transmembrane helices were predicted using TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/, accessed on 22 July 2025), and subcellular localization was inferred using WoLF PSORT (http://wolfpsort.hgc.jp, accessed on 22 July 2025).
2.2.2. Phylogenetic Analysis
A maximum likelihood (ML) phylogenetic tree was constructed from an alignment of the full-length MST protein sequences using the ‘One Step Build ML Tree’ module in TBtools II [20]. The multiple sequence alignment was performed using default parameters, and branch support was assessed with 1000 bootstrap replicates. The resulting tree was visualized and annotated using FigTree.
2.2.3. Gene Structure and Conserved Motif Analysis
To characterize the structural features of the MST gene family in litchi, exon–intron information was extracted from the lychee genome annotation file (PRJNA747875). The ‘Gene Structure View’ function in TBtools was employed to visualize the alignment between coding sequences (CDS) and genomic sequences, illustrating the number and position of exons and introns, as well as the distribution of untranslated regions (UTRs) for each gene [20]. Conserved protein motifs were identified using the MEME Suite with the following parameters: a motif width ranging from 15–60 amino acids, a maximum number of motifs of 10, and all other options set to default [21]. The resulting XML output from MEME was processed using the ‘Motif Analysis’ module in TBtools to generate a schematic diagram depicting the type, order, and conservation of motifs within each protein sequence.
2.2.4. Chromosomal Localization and Collinearity Analysis
Chromosomal location information for each identified gene was obtained from the litchi genome annotation file (PRJNA747875) and visualized using the ‘Visualize Gene Location’ function in TBtools (v2.330) [20]. To investigate gene duplication events within the LcMST gene family and genomic collinearity between litchi and the reference species A. thaliana and Oryza sativa, the ‘One Step MCScanX’ tool in TBtools was applied. The resulting syntenic relationships were visualized using the ‘Dual Synteny Plotter’ utility [20]. Furthermore, the rates of synonymous (Ks) and nonsynonymous (Ka) substitutions were calculated for duplicated gene pairs. The Ka/Ks ratios were computed using TBtools, and the divergence time (T) for each duplication event was estimated with the formula T = Ks/(2 × 6.1 × 10−9) × 10−6 million years ago (MYA) [22].
2.3. Expression Profile
2.3.1. RNA-Seq Analysis
To investigate the expression profiles of LcMSTs in different tissues of litchi, RNA-seq data of ‘Feizixiao’ cultivar were obtained from the NCBI database (Accession Number: PRJNA747875). After quality control of the raw sequencing data using Trimmomatic [23], the clean reads were aligned to the reference genome with HISAT2 [24] using default parameters. Gene expression levels were quantified as FPKM (Fragments Per Kilobase of transcript per Million mapped reads) values using Cufflinks [25]. The expression patterns of LcMSTs across eight selected tissues (root, leaf, panicle, aril, seed, pericarp, episperm, and embryo) were visualized in a heatmap generated with TBtools [20].
2.3.2. qRT-PCR Analysis
Total RNA was extracted from 0.1 g of a homogenate prepared from the pooled arils of 20 fruits per biological replicate, using the RNAprep Pure Plant Kit (TIANGEN, Beijing, China). Subsequently, 1 μg of purified RNA was reverse-transcribed into cDNA using the HiScript III All-in-one RT SuperMix for qPCR (Vazyme, Nanjing, China). qRT-PCR was performed using Taq Pro Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) on a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The thermal cycling protocol was as follows: initial denaturation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 20 s. A melt curve analysis was generated by heating from 65 °C to 95 °C in 0.5 °C increments with a 5 s hold at each step. The LcACTIN gene was used as an internal reference to normalize target gene expression levels, which were calculated using the 2−△△Ct method [26]. Three biological replicates were included for all gene expression analyses. The primer sequences used for qRT-PCR are listed in Supplementary Table S1.
2.4. Sugar Content Measurement
Soluble sugars were extracted according to a previously described method [14] with minor modifications. Briefly, aril samples were ground into fine powder in liquid nitrogen. Exactly 0.5 g of the powder was weighed into a 10 mL centrifuge tube, mixed with 8 mL of hot 50% (v/v) acetonitrile aqueous solution, and subjected to ultrasonic extraction for 10 min. After cooling to room temperature, the mixture was centrifuged at 13,000 rpm for 5 min. The supernatant was filtered through a 0.22 μm membrane and diluted threefold with deionized water prior to analysis.
Sugar composition was analyzed using an Agilent 1260 II HPLC system equipped with a G7162A Refractive Index Detector (RID) and a ZORBAX Carbohydrate Analysis Column (4.6 mm × 250 mm, 5 μm). The mobile phase consisted of acetonitrile–water (75:25, v/v) at a flow rate of 1.0 mL/min. Both the column and detector were maintained at 30 °C, and the injection volume was 3 μL.
For quantification, a series of standard solutions of sucrose, glucose, and fructose (Sigma-Aldrich, Saint-Louis, MO, USA) was analyzed to establish external standard calibration curves. The limit of detection (LOD) and limit of quantification (LOQ) for each sugar were determined at signal-to-noise ratios of 3 and 10, respectively. The detailed parameters are summarized in Supplementary Table S2.
2.5. Functional Characterization in Yeast
To functionally characterize candidate sugar transporters that are highly expressed in litchi fruit, two representative genes were selected for heterologous functional complementation assays in yeast. The full-length coding sequences of the target genes were cloned into the yeast shuttle vector pDR196 using the ClonExpress Ultra One Step Cloning Kit (Vazyme, Nanjing, China) and transformed into the hexose transporter-deficient yeast mutant strain EBY.VW4000 (Coolaber, Beijing, China). Yeast cells transformed with the empty pDR196 vector served as a negative control. Transformed yeast cells were pre-cultured in liquid synthetic defined (SD)–Ura medium with 2% (w/v) maltose as the sole carbon source until the optical density at 600 nm (OD600) reached 1.0–2.0. Cells were harvested, washed three times with sterile deionized water (ddH2O), and resuspended to an OD600 of 1.0. Serial 10-fold dilutions (10−1, 10−2, and 10−3) were prepared, and 5 μL of each dilution was spotted onto solid SD-Ura plates containing 2% (w/v) maltose or 2% (w/v) of other monosaccharide substrates (glucose, fructose). Yeast growth was observed after incubation at 28 °C for 3 days.
3. Results
3.1. Genome-Wide Identification and Characterization of LcMST Genes
A total of 45 genes encoding monosaccharide transporters (MSTs) were identified in the litchi genome. Phylogenetic analysis classified these proteins into seven distinct subfamilies: STP (14 members), ERD6L (13 members), PLT (6 members), INT (4 members), pGlcT (4 members), TMT (2 members), and VGT (2 members) (Figure 1). The STP and ERD6L subfamilies were the most abundant, consistent with their status as the major branches of the MST family. To investigate their evolutionary relationships, a phylogenetic tree was constructed using MST protein sequences from litchi, A. thaliana, and O. sativa (Supplementary Figure S1 and Table S4). The tree topology revealed that members of each litchi MST subfamily clustered closely with their orthologs from Arabidopsis and rice, indicating conserved evolutionary relationships. The identified genes were subsequently named according to their phylogenetic relationships and orthology with Arabidopsis MST genes.
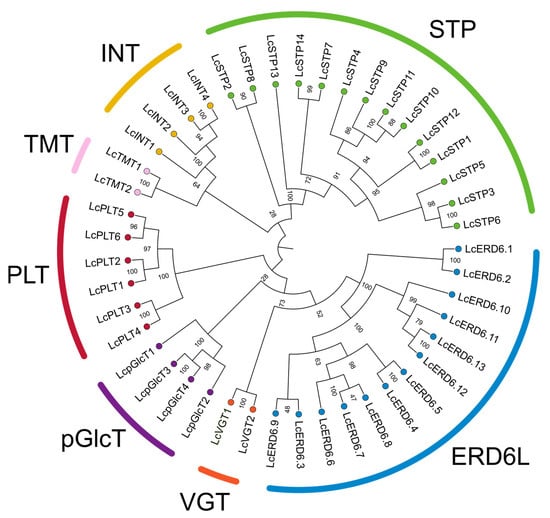
Figure 1.
Phylogenetic tree of litchi MST proteins. The phylogenetic tree was constructed based on multiple sequence alignment of the MST domains using the maximum likelihood method. Bootstrap values were calculated from 1000 replicates, and numbers on the nodes represent the credibility values of each clade. The litchi proteins were named according to their homologous counterparts in A. thaliana. Proteins belonging to seven distinct subfamilies are indicated by different colors. The evolutionary relationships among litchi, Arabidopsis, and rice MSTs are shown in Supplementary Figure S1.
The physicochemical properties of the LcMSTs are summarized in Supplementary Table S3. The proteins ranged from 463 to 741 amino acids in length, with predicted molecular weights between 50.20 and 79.47 kDa and theoretical pI values from 5.02 to 9.82. Transmembrane domains analysis predicted that most LcMSTs possess 10 to 12 transmembrane helices (TMs). Subcellular localization prediction using WoLF PSORT suggested that 34 proteins are localized to the plasma membrane, 10 to the tonoplast, and one to the chloroplast.
To further elucidate the evolutionary origins of the LcMST gene family, we analyzed the exon–intron architecture of each gene. The results revealed substantial variation in exon numbers, which ranged from 2 to 19 exons across the family. Furthermore, the exon–intron structures exhibited marked divergence between different subfamilies (Figure 2A,B). In contrast, genes within the same subfamily showed highly conserved exon–intron structures with similar numbers and lengths of exons (Figure 2A,B). For instance, members of the PLT subfamily generally possessed simplified gene structures with only 2–3 exons, whereas the ERD6L subfamily displayed complex structures containing 18–19 exons (e.g., LcERD6.11 contains 19 exons). This high degree of structural conservation within subfamilies is consistent with the phylogenetic clustering, suggesting that the evolution of gene structure is strongly constrained by sequence homology.
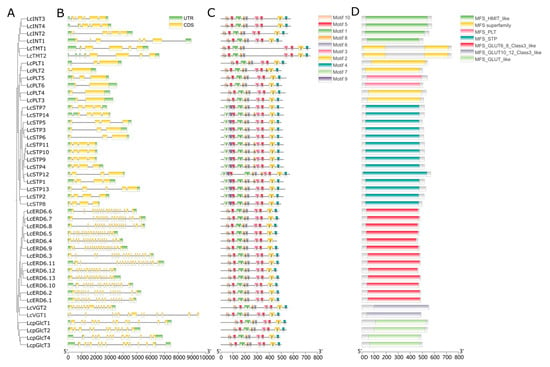
Figure 2.
Analysis of phylogenetic relationship, gene structure, and conserved motifs of LcMST genes. (A) Maximum likelihood tree showing the evolutionary relationships. (B) Exon–intron structures, where green boxes represent untranslated regions (UTRs), yellow boxes represent coding sequences (CDS), and black lines represent introns. (C) Ten conserved motifs identified using the MEME Suite tool. (D) The conserved domains in LcMSTs were identified with NCBI-CDD.
To analyze characteristic regions of the monosaccharide transporters, conserved motifs were identified using the MEME suite (Figure 2C). A total of seven conserved motifs (Motifs 1–6 and 8) were identified in all MST family members. Among these, Motif 5 was duplicated within the functional domains of all sugar transporters, implying its critical role in monosaccharide transport. Motif 10 was present in all subfamilies except LcSTP, while Motifs 7 and 9 were exclusively detected in the LcSTP subfamily, occupying positions analogous to those of Motif 10 in other subfamilies. Analysis using NCBI-CDD predicted that all members contain a conserved Major Facilitator Superfamily (MSF) domain (Figure 2D). Overall, the common conserved motif and domain architectures within subfamilies suggest evolutionarily conserved functions.
3.2. Chromosomal Distribution, Duplication Events, and Collinearity of LcMST Genes
The 45 identified LcMST genes were unevenly distributed across 13 of the 15 litchi chromosomes. Chromosomes 1 and 6 harbored the highest number of genes, with 9 each, followed by chromosome 9 with 6 genes. Chromosomes 4, 6, 8, and 13 each contained 3 genes; chromosomes 5, 11, and 12 contained 2 genes each; and chromosomes 3, 10, and 14 each harbored only one LcMST gene. No LcMST genes were located on chromosomes 2 or 15 (Supplementary Figure S2).
To investigate the expansion mechanisms of the MST family, we analyzed gene duplication events, categorizing them as either tandem duplication (TD) or whole-genome/segmental duplication (WGD/SD). A total of nine tandemly duplicated gene pairs were identified in litchi (Supplementary Figure S2): LcPLT1-LcPLT2, LcERD6.12-LcERD6.13, LcSTP10-LcSTP11, LcERD6.9-LcERD6.5, LcERD6.5-LcERD6.4, LcERD6.8-LcERD6.6, LcERD6.10-LcERD6.3, LcSTP3-LcSTP6, and LcINT3-LcINT4. Additionally, six whole-genome/segmental duplication events were detected (Figure 3), including: LcpGlcT4-LcpGlcT3, LcPLT6-LcPLT5, LcPLT4-LcPLT3, LcERD6.1-LcERD6.2, LcERD6.11-LcERD6.10, and LcTMT2-LcTMT1.

Figure 3.
Collinearity analysis of the MST family in litchi. The two outermost rings represent the gene density of the litchi genome; the third ring indicates the chromosome numbers of litchi. Syntenic relationships among LcMST genes are connected by red lines, representing gene pairs predicted to be formed by WGD or segmental duplication.
Ka/Ks analysis of the duplicated gene pairs revealed that the Ka/Ks ratios of tandemly duplicated pairs ranged from 0.12 to 1.04, averaging 0.20, while the ratios for WGD/SD pairs ranged from 0.13 to 0.22, with a mean of 0.16 (Supplementary Table S5). With the exception of LcSTP3–LcSTP6 pair (Ka/Ks = 1.04), all gene pairs exhibited Ka/Ks values significantly less than 1, indicating that the LcMST family has predominantly evolved under strong purifying selection. The estimated divergence times of these duplication events ranged from 1.23 to 150.54 MYA, with an average of 66.60 MYA (Supplementary Table S5).
To further explore the evolutionary relationships of the MST family, we constructed syntenic maps between litchi and two reference species—A. thaliana and O. sativa (Figure 4). The analysis revealed that 35 LcMST genes exhibited syntenic relationships with Arabidopsis, whereas only 13 syntenic gene pairs were identified with rice.

Figure 4.
Synteny analysis of MST genes between L. chinensis, A. thaliana, and O. sativa. The gray lines in the background represent syntenic blocks between the litchi genome and the genomes of the other two plant species. The blue lines highlight syntenic gene pairs involving MST genes.
3.3. Tissue-Specific Expression Profiles and Developmental Dynamics of LcMST Genes
To assess the differential expression of LcMST genes, we analyzed publicly available RNA-seq data from Litchi chinensis cv. Feizixiao (Supplementary Table S6). A heatmap was generated to illustrate the expression patterns of all 45 LcMST genes (Figure 5). The results revealed distinct divergence in expression among these sugar transporter genes across different tissues. A subset of genes, including LcSTP5, LcINT1, LcpGlcT1, LcpGlcT4, LcTMT1, LcTMT2, and LcVGT1, were ubiquitously expressed. In contrast, several genes exhibited pronounced tissue specificity. For instance, LcPLT5 was highly expressed in the roots and pericarp, LcVGT2 was predominantly expressed in leaves, and LcSTP9 showed specific expression in the embryo (Figure 5; Supplementary Table S6).
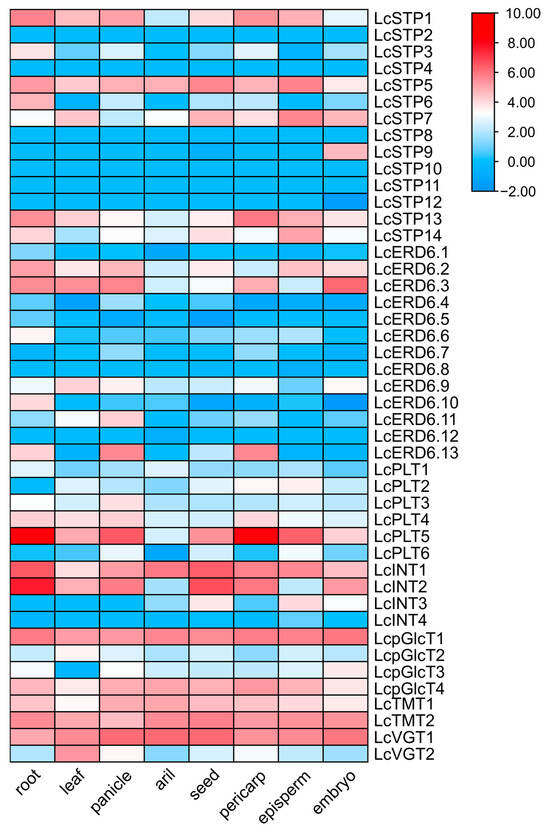
Figure 5.
Expression patterns of LcMST genes in different tissues. The heatmap was generated based on log2-transformed fold-change values. Gene expression levels are represented by a color scale, with red indicating higher expression and blue indicating lower expression.
To investigate the potential roles of LcMST genes in aril sugar accumulation, the expression profiles of seven LcMST genes with relatively high expression in the aril were analyzed via qRT-PCR during the fruit maturation in the ‘Tianshuili’ cultivar (Figure 6). The results revealed that only LcINT1 and LcTMT1 showed progressively increasing expression during fruit maturation, whereas the expression of LcpGlcT1 progressively decreased. In contrast, LcSTP5 expression peaked at 60 DAF and subsequently declined. The expression levels of LcpGlcT4, LcVGT1, and LcTMT2 did not show significant variations throughout the developmental period examined.

Figure 6.
Expression patterns of seven LcMST genes during fruit development in the litchi cultivar ‘Tianshuili’. Values are presented as mean ± standard deviation (SD) (n = 3). Different lowercase letters above the bars indicate significant differences among developmental stages (p < 0.05, LSD test).
3.4. Sugar Accumulation During Fruit Development and Its Correlation with Candidate LcMST Genes
The dynamic changes in sugar content were analyzed during fruit development in ‘Tianshuili’ litchi cultivar (Figure 7). The results demonstrated a continuous accumulation of both fructose and glucose. Fructose content rose significantly from 6.69 g/kg FW at 60 DAF to 146.97 g/kg FW at 88 DAF, representing an approximately 21.9-fold increase (Figure 7A). Similarly, glucose content increased from 10.14 g/kg FW at 60 DAF to 189.85 g/kg FW at 88 DAF, an approximately 18.7-fold increase (Figure 7B). In contrast, sucrose content exhibited an initial increase followed by a decline: it increased from 16.75 g/kg FW at 60 DAF, peaked at 115.55 g/kg FW at 74 DAF, and subsequently decreased to 82.74 g/kg FW by 88 DAF (Figure 7C). Beginning at 81 DAF, the ratio of hexoses (fructose and glucose) to sucrose exceeded 2, indicating a transition in the sugar accumulation pattern from sucrose-dominant to hexose-dominant (Figure 7D).

Figure 7.
Dynamic changes in sugar concentrations and composition during fruit development in the litchi cultivar ‘Tianshuili. (A) Fructose content. (B) Glucose content. (C) Sucrose content. (D) Ratio of hexoses to sucrose. Data are presented as mean ± SD (n = 3). Different lowercase letters above the bars indicate significant differences among developmental stages (p < 0.05, LSD test).
To explore the potential roles of specific transporters, a Pearson correlation analysis was conducted between the expression of these seven candidate LcMST genes and the sugar accumulation profiles (Supplementary Table S7). The expression of LcINT1 showed a significant positive correlation with fructose content. LcTMT1 expression was significantly positively correlated with fructose, glucose, and total sugar contents. In contrast, LcpGlcT1 expression was negatively correlated with fructose, glucose, and total sugar contents. Collectively, these findings suggest that LcINT1 and LcTMT1 may play crucial roles in monosaccharide accumulation during litchi fruit development.
3.5. Functional Characterization of LcTMT1 and LcINT1 in a Yeast System
To determine the transport ability capabilities of two key candidates, LcINT1 and LcTMT1, their coding sequences were independently expressed in the sugar transporter-deficient yeast strain EBY.VW4000. Heterologous functional complementation assays were performed on synthetic defined (SD) media with different carbon sources. As expected, all yeast strains, including the empty vector (pDR196) control, grew well on SD medium containing 2% (w/v) maltose, which confirms the viability of all strains under non-selective conditions (Figure 8). Yeast expressing LcINT1 did not support growth on media with either glucose or fructose, similarly to the negative control, suggesting that LcINT1 cannot effectively transport these two monosaccharides under the tested conditions. In contrast, yeast cells expressing LcTMT1 restored growth on media containing 2% (w/v) glucose or fructose, indicating that LcTMT1 functions as a transporter for both monosaccharides (Figure 8).

Figure 8.
Functional complementation analysis of representative MST genes in the hexose transporter-deficient Saccharomyces cerevisiae strain EBY.VW4000.
4. Discussion
4.1. Identification and Evolutionary Features of the MST Family in Litchi
MSTs play crucial roles in carbohydrate partitioning and yield formation in higher plants [10]. As a characteristic fruit tree, litchi is notable for accumulating high levels of fructose, glucose, and sucrose in its aril [14]. However, the MST gene family in litchi had not been systematically characterized. In this study, we identified 45 LcMST genes in the litchi genome, a number smaller than that in A. thaliana (53) [12], O. sativa (65) [27], Malus domestica (64–78) [28], and Pyrus spp. (69) [29], but comparable to that in Dimocarpus longan (46) [30]. Phylogenetic analysis classified the LcMST genes into seven subfamilies—STP, ERD6L, PLT, INT, pGlcT, TMT, and VGT—which aligns with the established classification of MST families in other higher plants [13,27], thereby supporting the reliability of our phylogenetic inference.
Regarding subfamily composition, STP (14 members) and ERD6L (13 members) were the two largest subfamilies, collectively accounting for approximately 60% of all members. Notably, the STP subfamily is the largest in rice, whereas ERD6L is the largest in Arabidopsis [12], suggesting species-specific expansion patterns of the MST family across plants. Collinearity analysis revealed more syntenic gene pairs between litchi and the dicot Arabidopsis (35 pairs) than with the monocot rice (13 pairs), which is consistent with their closer evolutionary relationship.
Further analysis of gene structure and conserved motifs indicated that members within the same subfamily shared similar characteristics, while exon numbers varied considerably among subfamilies. Members of the PLT, TMT, INT, and STP subfamilies were relatively conserved, containing 2–6 exons, whereas genes in the ERD6L, pGlcT, and VGT subfamilies contained more exons. This trend aligns with previous reports in Arabidopsis and rice [13,27]. All LcMST proteins contained conserved Motifs 1–6 and 8, suggesting that all subfamilies not only share evolutionary proximity but also maintain structural and functional conservation. The subfamily-specific Motifs 7 and 9, unique to the STP subfamily, may be associated with its specialized role in apoplastic hexose uptake, providing clues for future investigations into the functional divergence of this subfamily.
4.2. Gene Duplication Contributed to the Expansion of the LcMST Gene Family
Gene duplication is a major mechanism driving the expansion and functional diversification of plant gene families, primarily through TD, WGD, and segmental duplication [31]. In this study, we identified 9 TD and 6 WGD/segmental duplicated gene pairs within the LcMST family, suggesting that TD played a more prominent role in the expansion of this family in litchi than WGD. This finding aligns with previous reports indicating that approximately 30% of sugar transporter genes in Arabidopsis and rice originated from tandem duplication [13,32], with this proportion reaching about 60% in sugarcane [33]. Several duplicated gene pairs—such as LcERD6.12-LcERD6.13, LcSTP10-LcSTP11, LcSTP3-LcSTP6, LcINT3-LcINT4, LcpGlcT4-LcpGlcT3, LcERD6.1-LcERD6.2, and LcTMT2-LcTMT1—exhibited highly similar exon–intron structures and conserved motifs (Figure 2), implying potential functional redundancy. However, structural variations arising during evolution may have led to the divergence of expression patterns and functions among these duplicated genes. For example, LcERD6.12 showed low expression across seven tissues, whereas LcERD6.13 was highly expressed in the panicle and pericarp; LcpGlcT3 was only weakly expressed in the embryo, while LcpGlcT4 was expressed in all seven examined tissues at significantly higher levels.
Furthermore, we performed Ka/Ks analysis to estimate the divergence times and selective pressures acting on the duplicated genes. A Ka/Ks ratio significantly greater than 1 indicates positive selection, a ratio equal to 1 suggests neutral evolution, and a ratio less than 1 signifies purifying selection [34]. Most duplicated gene pairs in this study had Ka/Ks values less than 1 (with average values of 0.20 for TD and 0.16 for WGD duplicates), indicating that the LcMST gene family has evolved under strong purifying selection. The estimated average divergence time for these duplication events was approximately 66.6 MYA, which is largely consistent with the divergence time of the Sapindaceae family (approximately 65–70 MYA) [20]. This temporal coincidence suggests that key duplication events in the LcMST family occurred around the period of Sapindaceae speciation, providing an important temporal framework for understanding the evolution of sugar transport mechanisms in litchi.
4.3. Coordinated Regulation of Sugar Accumulation by LcMST Genes
Litchi fruit development typically follows a sigmoidal growth curve and comprises two distinct phases [13]. The aril initiates development approximately 21–35 DAA, exhibiting slow initial growth that accelerates markedly during fruit maturation, accompanied by a sharp rise in sugar content [18]. In the hexose-dominant cultivar ‘Tianshuili’, the contents of glucose and fructose increased continuously throughout maturation, while sucrose content increased initially and then declined. By 81 DAF, the hexose-to-sucrose ratio exceeded 2, confirming the transition to a hexose-dominant composition—a trend consistent with other hexose-dominant cultivars such as ‘Feizixiao’ [35].
To elucidate the molecular basis of this sugar accumulation pattern, we identified seven LcMST genes with high expression in the aril and analyzed the correlation between their expression and sugar content. The results revealed that the expression of LcINT1 and LcTMT1 was significantly positively correlated with fructose, and with fructose, glucose, and total sugar, respectively, whereas LcpGlcT1 expression was negatively correlated with all these sugar components. This complex pattern, involving both positive and negative correlations among different MST family members, has also been observed in other fruit trees. For instance, in apple, a comprehensive study revealed that MSTs exhibited divergent correlations with sugar content: MdERDL6.1/6.2/6.5 and MdTST1/2 showed significant positive correlations with fructose and/or total sugar, whereas MdERDL6.7-6.11 and MdTST4/6 were negatively correlated [28]. Similarly, in citrus, the protein abundance of ERD6L-5/6/7, TMT2, and SUT4 was highly consistent with sugar content variations between species, and their mRNA levels increased alongside sugar accumulation during fruit development [36]. These cross-species comparisons reinforce that sugar accumulation is not governed by a single transporter but by a network where different members, even within the same subfamily, may play opposing or specialized roles.
4.4. Functional Insights and Practical Implications of LcTMT1 in Litchi Sugar Accumulation
Our functional complementation assays unequivocally demonstrated that LcTMT1 can transport both glucose and fructose, establishing it as a key candidate gene responsible for hexose accumulation in the litchi aril. TMTs are localized to the vacuolar membrane and mediate sugar transport between the cytoplasm and the vacuole, making them core components in determining a cell’s capacity for sugar storage [37]. Their importance has been confirmed in numerous crops. For example, silencing three TST genes in sugarcane led to a dramatic 80% reduction in soluble stem sugars [38], and, in tomato, SlTST1 was identified as the primary transporter influencing fruit sugar content [39]. Collectively, these findings underscore the fundamental role of TMT/TST genes in determining sugar composition and content, providing strong support for the central role of LcTMT1 in litchi.
The functional confirmation of LcTMT1 suggests new strategies for litchi quality improvement. First, molecular markers based on favorable allelic variations in the LcTMT1 gene can be developed for the marker-assisted selection (MAS). This would allow for the early identification of elite germplasm with high sugar accumulation potential, significantly accelerating the breeding process for high-quality litchi cultivars. Second, LcTMT1 represents an ideal target for genetic engineering. Modulating its expression could enhance vacuolar sugar storage, potentially leading to new cultivars with superior sweetness and flavor.
5. Conclusions
This study presents a comprehensive genome-wide identification and functional analysis of the MST gene family in litchi. A total of 45 LcMST genes were identified and classified into seven subfamilies. Evolutionary analysis revealed that the expansion of the LcMST family was driven by both tandem and segmental duplications, with purifying selection acting as the dominant evolutionary force. Tissue-specific expression profiling and temporal analysis during fruit development demonstrated a significant positive correlation between the expression of the tonoplast monosaccharide transporter gene LcTMT1 and hexose accumulation. Heterologous functional complementation assays in yeast confirmed that LcTMT1 transports both glucose and fructose. These findings collectively establish LcTMT1 as a key regulator of hexose accumulation in the litchi aril. This study provides a valuable genetic resource and a solid foundation for elucidating the molecular mechanisms of fruit quality formation and for facilitating molecular breeding in litchi.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae11101252/s1, Figure S1: Phylogenetic analysis of monosaccharide transporters (MSTs) in litchi, Arabidopsis thaliana, and Oryza sativa; Figure S2: Chromosomal distribution of LcMST genes in litchi; Table S1: PCR primer sequences used in this study; Table S2. Standard curves, correlation coefficients, limits of detection, and limits of quantification for sugars; Table S3: Characteristics of the identified monosaccharide transporters in the litchi genome; Table S4: Protein sequences used to generate the phylogenetic tree; Table S5: Identification of substitution rates in gene pairs of the monosaccharide transporter superfamily in litchi; Table S6: FPKM data used for expression heat map; Table S7: Correlation between sugar content and MST expression in mature fruit of ‘Tianshuili’.
Author Contributions
Conceptualization, Y.W. and Q.Y.; methodology, Y.W.; software, Y.W. and H.L.; validation, Y.J. and F.S.; investigation, Y.W. and H.Z.; resources, Q.Y.; data curation, H.Z.; writing—original draft preparation, Y.W.; writing—review and editing, Y.W. and Q.Y.; supervision, Q.Y.; project administration, Y.W. and Q.Y.; funding acquisition, Q.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Guangdong Provincial Rural Revitalization Strategy Special Project (grant number: 2024CXTD19), Guangzhou Science and Technology Program Project (grant number: 2023B01J2002), the Introduction of Scientific and Technological Talents of Guangdong Academy of Agricultural Sciences (grant number: R2022YJ-YB3028) and the Open Project Program of Key Laboratory of South Subtropical Fruit Biology and Genetic Resource Utilization, Ministry of Agriculture and Rural Affairs (grant number: 249302).
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Acknowledgments
We thank Rui Xia from South China Agricultural University for kindly providing the ‘Feizixiao’ genome assembly and the curated annotation files.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Liu, W.; Jiang, H.; Zeng, F. The sugar transporter proteins in plants: An elaborate and widespread regulation network-A review. Int. J. Biol. Macromol. 2025, 294, 139252. [Google Scholar] [CrossRef]
- Li, J.; Wu, L.; Foster, R.; Ruan, Y. Molecular regulation of sucrose catabolism and sugar transport for development, defence and phloem function. J. Integr. Plant Biol. 2017, 59, 322–335. [Google Scholar] [CrossRef]
- Mishra, B.S.; Sharma, M.; Laxmi, A. Role of sugar and auxin crosstalk in plant growth and development. Physiol. Plant. 2022, 174, e13546. [Google Scholar] [CrossRef]
- Yoon, J.; Cho, L.H.; Tun, W.; Jeon, J.S.; An, G. Sucrose signaling in higher plants. Plant Sci. 2021, 302, 110703. [Google Scholar] [CrossRef]
- Zhu, L.; Li, B.; Wu, L.; Li, H.; Wang, Z.; Wei, X.; Ma, B.; Zhang, Y.; Ma, F.; Ruan, Y.L.; et al. MdERDL6-mediated glucose efflux to the cytosol promotes sugar accumulation in the vacuole through up-regulating TSTs in apple and tomato. Proc. Natl. Acad. Sci. USA 2021, 118, e2022788118. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, V.K.; Rivas-Ubach, A.; Winkler, T.; Mitchell, H.; Moran, J.; Ahkami, A.H. Modulation of polar auxin transport identifies the molecular determinants of source–sink carbon relationships and sink strength in poplar. Tree Physiol. 2024, 44, 82–101. [Google Scholar] [CrossRef]
- Chen, L.Q.; Cheung, L.S.; Feng, L.; Tanner, W.; Frommer, W.B. Transport of sugars. Annu. Rev. Biochem. 2015, 84, 865–894. [Google Scholar] [CrossRef]
- Braun, D.M. Phloem loading and unloading of sucrose: What a long, strange trip from source to sink. Annu. Rev. Plant Biol. 2022, 73, 553–584. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Lan, J.; Zhao, T.; Li, M.; Ruan, Y.L. How vacuolar sugar transporters evolve and control cellular sugar homeostasis, organ development and crop yield. Nat. Plants 2025, 11, 1102–1115. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Neuhaus, H.E.; Cheng, J.; Bie, Z. Contributions of sugar transporters to crop yield and fruit quality. J. Exp. Bot. 2022, 73, 2275–2289. [Google Scholar] [CrossRef]
- Ren, Y.; Liao, S.; Xu, Y. An update on sugar allocation and accumulation in fruits. Plant Physiol. 2023, 193, 888–899. [Google Scholar] [CrossRef]
- Buettner, M. The monosaccharide transporter(-like) gene family in Arabidopsis. FEBS Lett. 2007, 581, 2318–2324. [Google Scholar] [CrossRef]
- Fan, S.; Wang, D.; Xie, H.; Wang, H.; Qin, Y.; Hu, G.; Zhao, J. Sugar transport, metabolism and signaling in fruit development of Litchi chinensis Sonn: A review. Int. J. Mol. Sci. 2021, 22, 11231. [Google Scholar] [CrossRef]
- Yan, Q.; Feng, J.; Chen, J.; Wen, Y.; Jiang, Y.; Mai, Y.; Huang, K.; Liu, H.; Liu, H.; Shi, F.; et al. Comprehensive genomic and phenotypic analyses reveal the genetic basis of fruit quality in litchi. Genome Biol. 2025, 26, 222. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, T.; Wang, H.; Huang, X.; Qin, Y.; Hu, G. Patterns of enzyme activities and gene expressions in sucrose metabolism in relation to sugar accumulation and composition in the aril of Litchi chinensis Sonn. J. Plant Physiol. 2013, 170, 731–740. [Google Scholar] [CrossRef]
- Wang, T.D.; Zhang, H.F.; Wu, Z.C.; Li, J.G.; Huang, X.M.; Wang, H.C. Sugar uptake in the aril of litchi fruit depends on the apoplasmic post-phloem transport and the activity of proton pumps and the putative transporter LcSUT4. Plant Cell Physiol. 2015, 56, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Wang, D.; Qin, Y.; Ma, A.; Fu, J.; Qin, Y.; Hu, G.; Zhao, J. Genome-wide identification and expression analysis of SWEET gene family in Litchi chinensis reveal the involvement of LcSWEET2a/3b in early seed development. BMC Plant Biol. 2019, 19, 499. [Google Scholar] [CrossRef]
- Wang, D.; Zhao, J.; Qin, Y.; Qin, Y.; Hu, G. Molecular cloning, characterization and expression profile of the sucrose synthase gene family in Litchi chinensis. Hortic. Plant J. 2021, 7, 520–528. [Google Scholar] [CrossRef]
- Huang, Z.; Gao, C.; Xu, Y.; Liu, J.; Kang, J.; Ren, Z.; Cui, Q.; Li, D.; Ma, S.; Xia, Y.; et al. Identification and expression analysis of putative sugar transporter gene family during bulb formation in lilies. Int. J. Mol. Sci. 2024, 25, 3483. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Chan, C.-K.K. Analysis of RNA-Seq data using TopHat and Cufflinks. Plant Bioinform. Methods Protoc. 2016, 1374, 339–361. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Johnson, D.A.; Thomas, M.A. The monosaccharide transporter gene family in Arabidopsis and rice: A history of duplications, adaptive evolution, and functional divergence. Mol. Biol. Evol. 2007, 24, 2412–2423. [Google Scholar] [CrossRef]
- Zhu, L.; Tian, X.; Peng, Y.; Su, J.; Li, B.; Yang, N.; Ma, F.; Li, M. Comprehensive identification of sugar transporters in the Malus spp. genomes reveals their potential functions in sugar accumulation in apple fruits. Sci. Hortic. 2022, 303, 111232. [Google Scholar] [CrossRef]
- Li, J.M.; Zheng, D.M.; Li, L.T.; Qiao, X.; Wei, S.W.; Bai, B.; Zhang, S.; Wu, J. Genome-wide function, evolutionary characterization and expression analysis of sugar transporter family genes in pear (Pyrus bretschneideri Rehd.). Plant Cell Physiol. 2015, 56, 1721–1737. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Peng, Y.; Rao, Y.; Li, S.H.; Zeng, L.H. Genome-wide identification and expression analysis of sugar transporter (ST) gene family in longan (Dimocarpus longan L.). Plants 2020, 9, 342. [Google Scholar] [CrossRef]
- Iñiguez, L.P.; Hernández, G. The evolutionary relationship between alternative splicing and gene duplication. Front. Genet. 2017, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; An, B.; Zhong, H.; Yang, J.; Kong, W.; Li, Y. A novel insight into functional divergence of the MST gene family in rice based on comprehensive expression patterns. Genes 2019, 10, 239. [Google Scholar] [CrossRef]
- Zhang, Q.; Hua, X.; Liu, H.; Yuan, Y.; Shi, Y.; Wang, Z.; Zhang, M.; Ming, R.; Zhang, J. Evolutionary expansion and functional divergence of sugar transporters in Saccharum (S. spontaneum and S. officinarum). Plant J. 2020, 105, 884–906. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, S.; He, F.; Zhu, J.; Hu, S.; Yu, J. How do variable substitution rates influence Ka and Ks calculations? Genom. Proteom. Bioinform. 2009, 7, 116–127. [Google Scholar] [CrossRef]
- Peng, J.; Du, J.; Ma, W.; Chen, T.; Shui, X.; Liao, H.; Lin, X.; Zhou, K. Transcriptomics-based analysis of the causes of sugar receding in Feizixiao litchi (Litchi chinensis Sonn.) pulp. Front. Plant Sci. 2022, 13, 1083753. [Google Scholar] [CrossRef]
- Mao, Z.; Wang, Y.; Li, M.; Zhang, S.; Zhao, Z.; Xu, Q.; Liu, J.-H.; Li, C. Vacuolar proteomic analysis reveals tonoplast transporters for accumulation of citric acid and sugar in citrus fruit. Hortic. Res. 2024, 11, uhad249. [Google Scholar] [CrossRef] [PubMed]
- Wormit, A.; Trentmann, O.; Feifer, I.; Lohr, C.; Tjaden, J.; Meyer, S. Schmidt, U., Martinoia, E.; Neuhaus, H.E. Molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport. Plant Cell 2006, 18, 3476–3490. [Google Scholar] [CrossRef]
- Tang, M.; Wang, J.; Kannan, B.; Koukoulidis, N.M.; Lin, Y.H.; Altpeter, F.; Chen, L.Q. Tonoplast sugar transporters as key drivers of sugar accumulation, a case study in sugarcane. Hortic. Res. 2025, 12, uhae312. [Google Scholar] [CrossRef]
- Cai, H.; Liang, M.; Qin, X.; Dong, R.; Wang, X.; Wang, H.; Sun, S.; Cui, X.; Yang, W.; Li, R. Tonoplast sugar transporters coordinately regulate tomato fruit development and quality. Plant Commun. 2025, 6, 101314. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).