Identification of Reliable Reference Genes for qRT-PCR Normalization in Tomato Genotypes with Contrasting Salinity Tolerance
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Total RNA Extraction and cDNA Synthesis
2.3. Primer Design and Calibration
2.4. Quantitative Real-Time PCR
2.5. Determination of Electrolyte Leakage and Ion Absorption
2.6. Water Content
2.7. Statistical Analysis
3. Results
3.1. Primer Efficiency and Ct Variation
3.2. Stability of Reference Genes Using the ∆Ct Algorithm
3.3. Stability of Reference Genes Using BestKeeper Analysis
3.4. Stability of Reference Genes Using the geNorm Algorithm
3.5. Stability of Reference Genes Using NormFinder Analysis
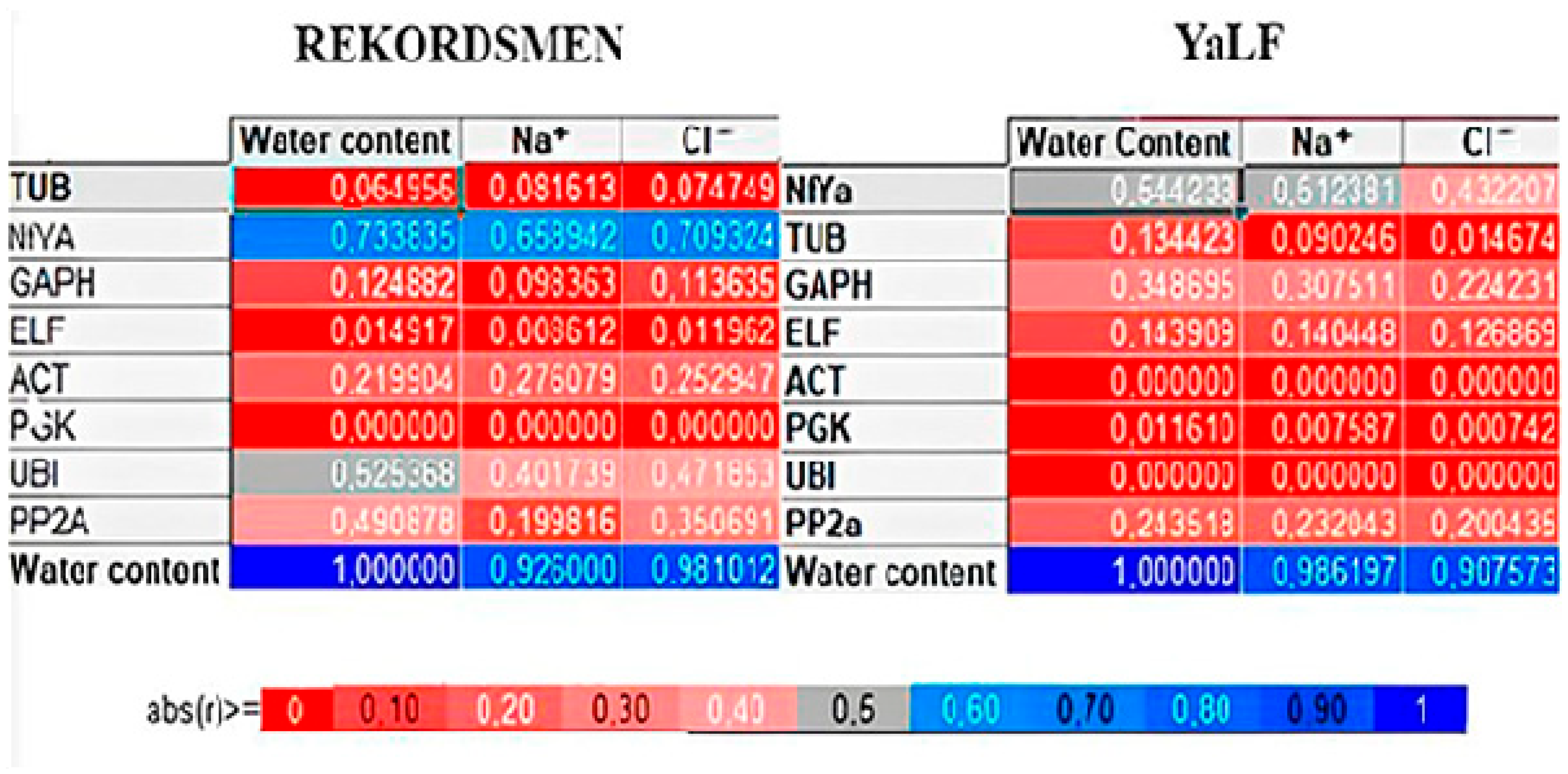
3.6. RWC and Ion Accumulation Correlation with Candidate Gene Expression Level
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DW | Drought weight |
| EtBr | Ethidium bromide |
| MMLV | Moloney murine leukemia virus |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| OD260/280 | Optical density |
| RWC | Relative water content |
| FAO | Food and Agriculture Organization |
| FW | Fresh weight |
| HKG | Housekeeping gene |
Appendix A
| Name | Accession Number | Forward Primer (Sequence 5′-3′) | Reverse Primer (Primer Sequence 5′-3′) | Primer Efficiency (R2) | Amplicon Length (bp) | Tm |
|---|---|---|---|---|---|---|
| ELF | X53043.1 | GGAACTTGAGAAGGAGCCTAAG | CAACACCAACAGCAACAGTCT | 0.98 | 158 | 58 |
| ACT | BT013707.1 | TCCTTACCTGAACGCCTGTCA | ATACGCATCCTTCTGTCCCATTCCGA | 0.97 | 107 | 58 |
| UBI | NM_001346406.1 | TCGTAAGGAGTGCCCTAATGCTGA | CAATCGCCTCCAGCCTTGTTGTAA | 0.91 | 120 | 50 |
| NTFY | XM_026030313.2 | GGATCTGGCATGGGAACACT | TCATCGGCATTCACCAA | 0.96 | 126 | 50 |
| TUB | NM_001247878.2 | AACCTCCATTCAGGAGATGTTT | TCTGCTGTAGCATCCTGGTATT | 0.86 | 180 | 50 |
| GAPH | NM_001247874.2 | ACCACAAATTGCCTTGCTCCCTTG | ATCAACGGTCTTCTGAGTGGCTGT | 0.9 | 111 | 50 |
| PP2a | NM_001247587.2 | CGATGTGTGATCTCCTATGGTC | AAGCTGATGGGCTCTAGAAATC | 0.9 | 149 | 50 |
| PGK | XM_004243920.4 | CTTCCTCCTTAAAACTCCTCTCC | CTAAGGTCTCCAACGCTCTTCT | 0.85 | 162 | 50 |
| TUB | NFT-Y | GAPH | ELF | ACT | PGK | UBI | PP2a | |
|---|---|---|---|---|---|---|---|---|
| StDev + Cp (1) | 3.17 | 0.83 | 0.56 | 0.99 | 0.81 | 1.25 | 0.85 | 0.79 |
| StDev + fold (1) | 12.84 | 3.21 | 4.41 | 4.58 | 6.79 | 4.64 | 3.19 | 2.98 |
| StDev + Cp (1) | 1.27 | 1.53 | 0.21 | 2.81 | 0.49 | 1.17 | 0.96 | 1.59 |
| StDev + fold (2) | 2.42 | 2.88 | 1.16 | 7.02 | 1.41 | 2.25 | 1.95 | 3.00 |
References
- Xu, T.A.; Kohler, T.M.; Lenton, T.M.; Svenning, J.; Scheffer, M. Future of the human climate niche. Proc. Natl. Acad. Sci. USA 2020, 117, 11350–11355. [Google Scholar] [CrossRef] [PubMed]
- Reisinger, A.; Fuglestvedt, J.S.; Pirani, A.; Geden, O.; Jones, C.D.; Maharaj, S.; Poloczanska, E.S.; Morelli, A.; Johansen, T.G.; Adler, C.; et al. Overshoot: A Conceptual Review of Exceeding and Returning to Global Warming of 1.5 °C. Annu. Rev. Environ. Resour. 2025, 50, 185–217. [Google Scholar] [CrossRef]
- Shiogama, H.; Stone, D.; Emori, S.; Takahashi, K.; Mori, S.; Maeda, A.; Ishizaki, Y.; Allen, M.R. Predicting future uncertainty constraints on global warming projections. Sci. Rep. 2016, 6, 18903. [Google Scholar] [CrossRef]
- Basak, N.; Shit, P.K.; Adhikary, P.P.; Bhunia, G.S.; Sengupta, D. Salt Affected Soils: Global Perspectives. In Soil Health and Environmental Sustainability; Environmental Science and Engineering; Springer: Cham, Switzerland, 2022; pp. 107–129. [Google Scholar]
- Spencer, T.; Magnan, A.; Donner, S.; Garschagen, M.; Ford, J. Habitability of low-lying socio-ecological systems under a changing climate. Clim. Change 2024, 177, 14. [Google Scholar] [CrossRef]
- Løvdal, T.; Lillo, C. Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal. Biochem. 2009, 387, 238–242. [Google Scholar] [CrossRef]
- Yourieva, N.O.; Voronkov, A.S.; Tereshonok, D.V.; Osipova, E.S.; Platonova, E.V.; Belyaev, D.V. An assay for express screening of potato transformants by GFP fluorescence. Mosc. Univ. Biol. Sci. Bull. 2018, 73, 69–75. [Google Scholar] [CrossRef]
- Wieczorek, P.; Wrzesińska, B.; Obrępalska-Stęplowska, A. Assessment of reference gene stability influenced by extremely divergent disease symptoms in Solanum lycopersicum L. J. Virol. Methods. 2013, 194, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, Q.; Zhang, B. Evaluation and selection of reliable reference genes for gene expression under abiotic stress in cotton (Gossypium hirsutum L.). Gene 2013, 530, 44–50. [Google Scholar] [CrossRef]
- Kashyap, S.P.; Kumari, N.; Mishra, P.; Moharana, D.P.; Aamir, M. Tapping the potential of Solanum lycopersicum L. pertaining to salinity tolerance: Perspectives and challenges. Genet. Resour. Crop Evol. 2021, 68, 2207–2233. [Google Scholar] [CrossRef]
- Deanda-Tovar, A.A.; Rodríguez-Pérez, J.E.; Sahagún-Castellanos, J.; Colinas-y-León, M.T.B.; Pérez-Rodríguez, P.; Paredes-Cervantes, A.E. Tomato Lines Tolerant to Sodium Chloride at Early Growth Stages. Horticulturae. 2025, 11, 532. [Google Scholar] [CrossRef]
- Pailles, Y.; Awlia, M.; Julkowska, M.; Passone, L.; Zemmouri, K.; Negrão, S.; Schmöckel, S.M.; Tester, M. Diverse traits contribute to salinity tolerance of wild tomato seedlings from the Galapagos Islands. Plant Physiol. 2020, 182, 534–546. [Google Scholar] [CrossRef]
- Almeida, P.; Feron, R.; de Boer, G.J.; de Boer, A.H. Role of Na+, K+, Cl−, proline and sucrose concentrations in determining salinity tolerance and their correlation with the expression of multiple genes in tomato. AoB Plants 2014, 6, plu039. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Sun, C.; Deng, L.; Zhou, M.; Li, J.; Du, Y.; Ye, Z.; Huang, S.; Li, T.; Yu, J.; et al. Molecular breeding of tomato: Advances and challenges. J. Integr. Plant Biol. 2025, 67, 669–721. [Google Scholar] [CrossRef]
- Tanveer, K.; Gilani, S.; Hussain, Z.; Ishaq, R.; Adeel, M.; Ilyas, N. Effect of salt stress on tomato plant and the role of calcium. J. Plant Nutr. 2020, 43, 1, 28–35. [Google Scholar]
- Wei, A.M.; Wais, A.H.; Ditta, A.; Islam, M.R.; Chowdhury, M.K.; Pramanik, M.H.; Ismaan, H.N.; Soufan, W.; El Sabah, A.; Islam, M.S. Seed Germination and Early Seedling Growth of Sorghum (Sorghum bicolor L. Moench) Genotypes Under Salinity Stress. Pol. Environ. Stud. 2024, 33, 3019–3032. [Google Scholar] [CrossRef]
- Khaliluev, M.R.; Bogoutdinova, L.R.; Raldugina, G.N.; Baranova, E.N. A Simple and Effective Bioassay Method Suitable to Comparative In Vitro Study of Tomato Salt Tolerance at Early Development Stages. Methods Protoc. 2022, 5, 11. [Google Scholar] [CrossRef]
- Gachon, C.; Mingam, A.; Charrier, B. Real-time PCR: What relevance to plant studies? J. Exp. Bot. 2004, 55, 1445–1454. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Gao, Y.; Tian, L.; Ding, J.; Shao, G.; Zeng, Z.; Wang, J.; Su, D. Saline water irrigation induced differences in sodium ion distribution and tomato yield following drought hardening pre-treatment. Sci. Hortic. 2025, 350, 114285. [Google Scholar] [CrossRef]
- Guo, J.; Ling, H.; Wu, Q.; Xu, L.; Que, Y. The choice of reference genes for assessing gene expression in sugarcane under salinity and drought stresses. Sci. Rep. 2014, 4, 7042. [Google Scholar] [CrossRef]
- Souza, B.C.O.Q.; Andrade, E.S.; Ribeiro, C.C.; Santos, R.P.S.S.; Costa, L.C.; Tsehaye, Y.; Silva, M.L.S.; Araújo, W.L.; Marchiori, P.E.R.; Nascimento, V.L. The ethylene-insensitive tomato mutant Never ripe exhibits enhanced growth and phosphorus use efficiency under phosphorus stress. Plant Physiol. Biochem. 2025, 228, 110213. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Mestdagh, P.; Van Vlierberghe, P.; De Weer, A.; Muth, D.; Westermann, F.; Speleman, F.; Vandesompeleet, J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009, 10, R64. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Ben-Amar, A.; Allel, D.; Haouel, A.; Daldoul, S.; Zaghden, H.; Mliki, A. Overexpression of Grapevine Raffinose Synthase-Like Seed Imbibition Protein VvRS/SIP Gene Confers Salt Tolerance in Transgenic Tomato. J. Plant Growth Regul. 2025, 1–20. [Google Scholar] [CrossRef]
- Song, Y.; Wang, Y.; Guo, D.; Jing, L. Selection of reference genes for quantitative real-time PCR normalization in the plant pathogen Puccinia helianthi Schw. BMC Plant Biol. 2019, 19, 20. [Google Scholar] [CrossRef]
- Zhao, L.; Lu, J.; Zhang, J.; Wu, P.Y.; Yang, S.; Wu, K. Identification and characterization of histone deacetylases in tomato (Solanum lycopersicum). Front. Plant Sci. 2015, 5, 760. [Google Scholar] [CrossRef]
- Özdemir, Ö.Ü.; Yurt, K.; Pektaş, A.N.; Berk, Ş. Evaluation and normalization of a set of reliable reference genes for quantitative sgk-1 gene expression analysis in Caenorhabditis elegans-focused cancer research. Nucleosides Nucleotides Nucleic Acids 2025, 44, 91–110. [Google Scholar] [CrossRef]
- Yuan, Y.; Liu, C.; Bao, J.; Li, F. Selection and Validation of Appropriate Reference Genes for qRT-PCR Analysis of Iris germanica L. Under Various Abiotic Stresses. Food Sci. Nutr. 2025, 13, e4765. [Google Scholar] [CrossRef] [PubMed]
- Barrs, H.D. Determination of water deficits in plant tissue. In Water Deficits and Plant Growth, 1st ed.; Academic Press: New York, NY, USA, 1968; pp. 235–368. [Google Scholar]
- Papoui, E.; Koukounaras, A. Evaluation of Ascophyllum nodosum and Sargassum spp. Seaweed Extracts’ Effect on Germination of Tomato Under Salinity Stress. Horticulturae. 2025, 11, 290. [Google Scholar] [CrossRef]
- Tani, E.; Sarri, E.; Goufa, M.; Asimakopoulou, G.; Psychogiou, M.; Bingham, E.; Skaracis, G.N.; Abraham, E.M. Seedling growth and transcriptional responses to salt shock and stress in Medicago sativa L., Medicago arborea L., and their hybrid (Alborea). Agronomy 2018, 8, 231. [Google Scholar] [CrossRef]
- Alaguero-Cordovilla, A.; Gran-Gómez, F.J.; Tormos-Moltó, S.; Pérez-Pérez, J.M. Morphological characterization of root system architecture in diverse tomato genotypes during early growth. Int. J. Mol. Sci. 2018, 19, 3888. [Google Scholar] [CrossRef]
- Tal, M.; Shannon, M.C. Salt tolerance in the wild relatives of the cultivated tomato: Responses of Lycopersicon esculentum, L. cheesmanii, L. peruvianum, Solanum pennellii and F1 hybrids to high salinity. Aust. J. Plant Biol. 1983, 10, 109–117. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Kumar, S.; Muthukumar, M.; Bajpai, A.; Kushwaha, A.K.; Ahmad, I.; Bajpai, Y.; Singh, A.; Damodaran, T.; Trivedi, M. Selection and validation of stable reference genes in guava (Psidium guajava L.) for reliable and consistent gene expression analysis. Electron. J. Biotechnol. 2025, 75, 49–56. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Liu, X.; Tang, S.; Gao, Y.; Zhang, X.; Dong, G.; Zhou, J.; Zhou, Y.; Yang, Z.; Huang, J.; Yao, Y. Rice Reference Genes: Redefining reference genes in rice by mining RNA-seq datasets. Plant Cell Physiol. 2025, 66, 120–132. [Google Scholar] [CrossRef]
- Shi, C.; Liu, B.; Song, L.; Tang, Y.; Chen, J.; Shen, X.; Peng, Y.; Yang, C.; Lu, G.; Deng, X.; et al. Physio-biochemical and dynamic transcriptome comparison of heat tolerance in wild and cultivated tomato accessions. Plant Growth Regul. 2025, 105, 1105–1123. [Google Scholar] [CrossRef]
- Bogoutdinova, L.R.; Lazareva, E.M.; Chaban, I.A.; Kononenko, N.V.; Dilovarova, T.; Khaliluev, M.R.; Kurenina, L.V.; Gulevich, A.A.; Smirnova, E.A.; Baranova, E.N. Salt Stress-Induced Structural Changes Are Mitigated in Transgenic Tomato Plants Over-Expressing Superoxide Dismutase. Biology 2020, 9, 297. [Google Scholar] [CrossRef]
- Chaban, I.A.; Kononenko, N.V.; Gulevich, A.A.; Bogoutdinova, L.R.; Khaliluev, M.R.; Baranova, E.N. Morphological Features of the Anther Development in Tomato Plants with Non-Specific Male Sterility. Biology 2020, 9, 32. [Google Scholar] [CrossRef]
- Singh, H.; Kumar, P.; Kumar, A.; Kyriacou, M.C.; Colla, G.; Rouphael, Y. Grafting Tomato as a Tool to Improve Salt-Tolerance. Agronomy 2020, 10, 263. [Google Scholar] [CrossRef]
- Dong, S.; Ling, J.; Song, L.; Zhao, L.; Wang, Y.; Zhao, T. Transcriptomic Profiling of Tomato Leaves Identifies Novel Transcription Factors Responding to Dehydration Stress. Int. J. Mol. Sci. 2023, 24, 9725. [Google Scholar] [CrossRef]
- Wei, K.; Li, X.; Cao, X.; Li, S.; Zhang, L.; Lu, F.; Wang, X. Candidate Gene Identification and Transcriptome Analysis of Tomato male sterile-30 and Functional Marker Development for ms-30 and Its Alleles, ms-33, 7B-1, and stamenless-2. Int. J. Mol. Sci. 2024, 25, 3331. [Google Scholar] [CrossRef]
- Guo, M.; Wang, X.S.; Guo, H.D.; Bai, S.Y.; Khan, A.; Wang, X.M.; Li, J.S. Tomato salt tolerance mechanisms and their potential applications for fighting salinity: A review. Front. Plant Sci. 2022, 13, 949541. [Google Scholar] [CrossRef]
- Bazihizina, N.; Colmer, T.D.; Cuin, T.A.; Mancuso, S.; and Shabala, S. Friend or foe? Chloride patterning in halophytes. Trends Plant Sci. 2019, 24, 142–151. [Google Scholar] [CrossRef]
- Joseph, J.T.; Poolakkalody, N.J.; Shah, J.M. Plant reference genes for development and stress response studies. J. Biosci. 2018, 43, 173–187. [Google Scholar] [CrossRef] [PubMed]
- López-Méndez, A.G.; Rodríguez-Pérez, J.E.; Mascorro-Gallardo, J.O.; Sahagún-Castellanos, J.; Lobato-Ortiz, R. Sodium chloride tolerance during germination and seedling stages of tomato (Solanum lycopersicum L.) lines native to Mexico. Horticulturae 2024, 10, 466. [Google Scholar] [CrossRef]
- Habibi, N.; Terada, N.; Sanada, A.; Koshio, K. Alleviating salt stress in tomatoes through seed priming with polyethylene glycol and sodium chloride combination. Stresses. 2024, 4, 210–224. [Google Scholar] [CrossRef]
- Sootahar, R.K.; Sootahar, M.K.; Rais, N.; Jamro, G.M.; Rais, M.U.N.; Iqbal, R.; Kumarasamy, V. In Vitro Early Vegetative Growth of Tomato (Solanum lycopersicum L.) Cultivars Under Salt Stress. Pol. J. Environ. Stud. 2024, 33, 5879–5885. [Google Scholar] [CrossRef]
- Sun, F.; Wang, Y.; Liu, G.; Fang, D.; Sun, M.; Bao, Z.; Ma, F. Salt stress induces SFT expression to promote early flowering and inhibits floral organ development by disturbing cell cycle in tomato. Veg. Res. 2024, 4, e019. [Google Scholar] [CrossRef]
- Sang, Q.; Shan, X.; An, Y.; Shu, S.; Sun, J.; Guo, S. Proteomic Analysis Reveals the Positive Effect of Exogenous Spermidine in Tomato Seedlings’ Response to High-Temperature Stress. Front. Plant Sci. 2017, 8, 120. [Google Scholar] [CrossRef] [PubMed]
- Al Hosni, A.; Joyce, D.C.; Hunter, M.; Perkins, M.; Al Yahyai, R. Altered calcium and potassium distribution maps in tomato tissues cultivated under salinity: Studies using X-ray fluorescence (XFM) microscopy. Irrig. Sci. 2025, 43, 613–636. [Google Scholar] [CrossRef]
- Garcia-Daga, S.; Roy, S.J.; Gilliham, M. Redefining the role of sodium exclusion within salt tolerance. Trends Plant Sci. 2025, 30, 137–146. [Google Scholar] [CrossRef]
- Du, J.; Xu, H.; Zhang, D.X.; Feng, S. Chelation and nanoparticle delivery of monomeric dopamine to increase plant salt stress resistance. Nat. Commun. 2025, 16, 4157. [Google Scholar] [CrossRef]
- Zhang, D.; Xu, F.; Wang, F.; Le, L.; Pu, L. Synthetic biology and artificial intelligence in crop improvement. Plant Commun. 2025, 6, 101220. [Google Scholar] [CrossRef]
- Wu, J.; Wang, J.; Pan, C.; Guan, X.; Wang, Y.; Liu, S.; He, Y.; Chen, J.; Chen, L.; Lu, G. Genome-wide identification of MAPKK and MAPKKK gene families in tomato and transcriptional profiling analysis during development and stress response. PLoS ONE 2014, 9, e103032. [Google Scholar] [CrossRef]
- Zhang, Y.; Han, X.; Chen, S.; Zheng, L.; He, X.; Liu, M.; Qiao, G.; Wang, Y.; Zhuo, R. Selection of suitable reference genes for quantitative real-time PCR gene expression analysis in Salix matsudana under different abiotic stresses. Sci. Rep. 2017, 7, 40290. [Google Scholar] [CrossRef]
- Bogoutdinova, L.R.; Khaliluev, M.R.; Chaban, I.A.; Gulevich, A.A.; Shelepova, O.V.; Baranova, E.N. Salt Tolerance Assessment of Different Tomato Varieties at the Seedling Stage. Horticulturae 2024, 10, 598. [Google Scholar] [CrossRef]
- Romaniuk-Drapała, A.; Kosicka-Noworzyń, K.; Sheng, Y.H.; Yohn, C.; Brunetti, L.; Kagan, L. Evaluation of reference genes for qPCR in human liver and kidney tissue from individuals with obesity. Sci. Rep. 2025, 15, 5347. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Zhang, L.; Zheng, H.; Zhang, Z.; Luo, Y.; Liu, Y.; Wang, L. ZmmiR169q/ZmNF-YA8 is a module that homeostatically regulates primary root growth and salt tolerance in maize. Front. Plant Sci. 2023, 14, 1163228. [Google Scholar] [CrossRef]
- Dang, T.V.T.; Cho, H.S.; Lee, S.; Hwang, I. Salt stress-accelerated proteasomal degradation of LBD11 suppresses ROS-mediated meristem development and root growth in Arabidopsis. Plant Commun. 2025, 6, 101241. [Google Scholar] [CrossRef]
- Negi, Y.; Kumar, K. OsWNK9 mitigates salt stress by promoting root growth and stomatal closure in rice. Physiol. Plant. 2025, 177, e70129. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Zhang, J.; Sun, P.; Li, M.; Hy, J. Selection of reliable reference genes for gene expression analysis under abiotic stresses in desert biomass willow, Salix psammophila. Front. Plant Sci. 2016, 7, 1505. [Google Scholar] [CrossRef]
- Hashemipetroudi, S.H.; Rezaei, A.; Kuhlmann, M. Validation of Reference Genes for Accurate RT-qPCR Normalization in Aeluropus littoralis Under Drought, Cold, and ABA Treatments. Agronomy 2025, 15, 1596. [Google Scholar] [CrossRef]
- Yang, X.; Xiong, J.; Du, X.; Sun, M.; Ding, L.; Mei, W.; Sun, Z.; Feng, N.; Zheng, D.; Shen, X. Effects of Exogenous Spermidine on Seed Germination and Physiological Metabolism of Rice Under NaCl Stress. Plants 2024, 13, 3599. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, C.; Lin, Z.; Li, H.; Di, X.; Yue, X.; Gao, Z. Systematic identification and validation of the reference genes from 447 transcriptome datasets of moso bamboo (Phyllostachys edulis). Hortic. Plant J. 2025, 11, 1353–1363. [Google Scholar] [CrossRef]
- Ghosh, U.K.; Islam, M.N.; Siddiqui, M.N.; Cao, X.; Khan, M.A.R. Proline, a multifaceted signalling molecule in plant responses to abiotic stress: Understanding the physiological mechanisms. Plant Biol. 2022, 24, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Manan, A.; Ayyub, C.M.; Ahmad, R.; Bukhari, M.A.; Mustafa, Z. Salinity Induced Deleterious Effects on Biochemical and Physiological Processes of Tomato. Pak. J. Life Soc. Sci. 2016, 14, 83–90. [Google Scholar]
- Polash, M.A.S.; Sakil, M.A.; Tahjib-Ul-Arif, M.; Hossain, M. An Effect of salinity on osmolytes and relative water content of selected rice genotypes. Trop. Plant Res. 2018, 5, 227–232. [Google Scholar] [CrossRef]
- Kul, R.; Arjumend, T.; Ekinci, M.; Yildirim, E.; Turan, M.; Argin, S. Biochar as an organic soil conditioner for mitigating salinity stress in tomato. Soil Sci. Plant Nutr. 2021, 67, 693–706. [Google Scholar] [CrossRef]
- Fu, S.; Li, Y.; Liu, F.; Lu, C.; Liu, T.; Tian, S.; Chen, L. Structural and functional insights into NAC transcription factors in tomato stress responses and development. Biol. plantarum. 2025, 69, 49–57. [Google Scholar] [CrossRef]
- Filichkin, S.A.; Ansariola, M.; Fraser, V.N.; Megraw, M. Identification of transcription factors from NF-Y, NAC, and SPL families responding to osmotic stress in multiple tomato varieties. Plant Sci. 2018, 274, 441–450. [Google Scholar] [CrossRef]
- Chen, Y.P.; Hu, C.C.; Tsai, S.; Wen, Z.H.; Lin, C. Identification of housekeeping gene for future studies exploring effects of cryopreservation on gene expression in shrimp. Sci. Rep. 2025, 15, 11046. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, X.; Yin, Z.; Zhao, T.; Du, D.; Li, J.; Zhu, M.; Sun, Y.; Pan, Y. Genome- and Transcriptome-Wide Characterization and Expression Analyses of bHLH Transcription Factor Family Reveal Their Relevance to Salt Stress Response in Tomato. Plants 2025, 14, 200. [Google Scholar] [CrossRef]
- Ait Hammou, R.; El Caid, M.B.; Harrouni, C.; Daoud, S. Germination enhancement, antioxidant enzyme activity, and metabolite changes in late Argania spinosa kernels under salinity. J. Arid. Environ. 2023, 219, 105095. [Google Scholar] [CrossRef]
- Mascia, T.; Santovito, E.; Gallitelli, D.; Cillo, F. Evaluation of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in infected tomato plants. Mol. Plant Pathol. 2020, 11, 805–816. [Google Scholar] [CrossRef]
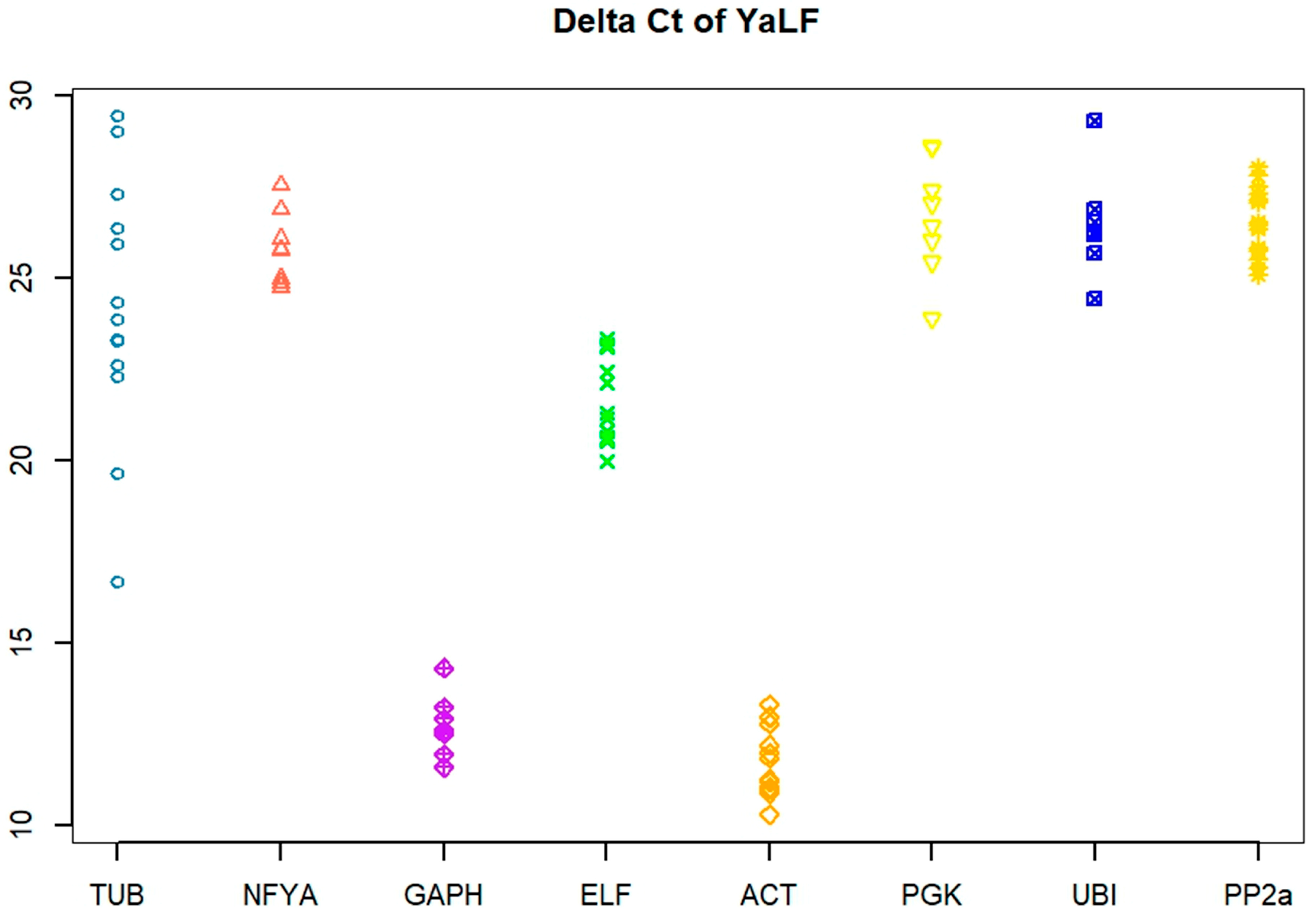
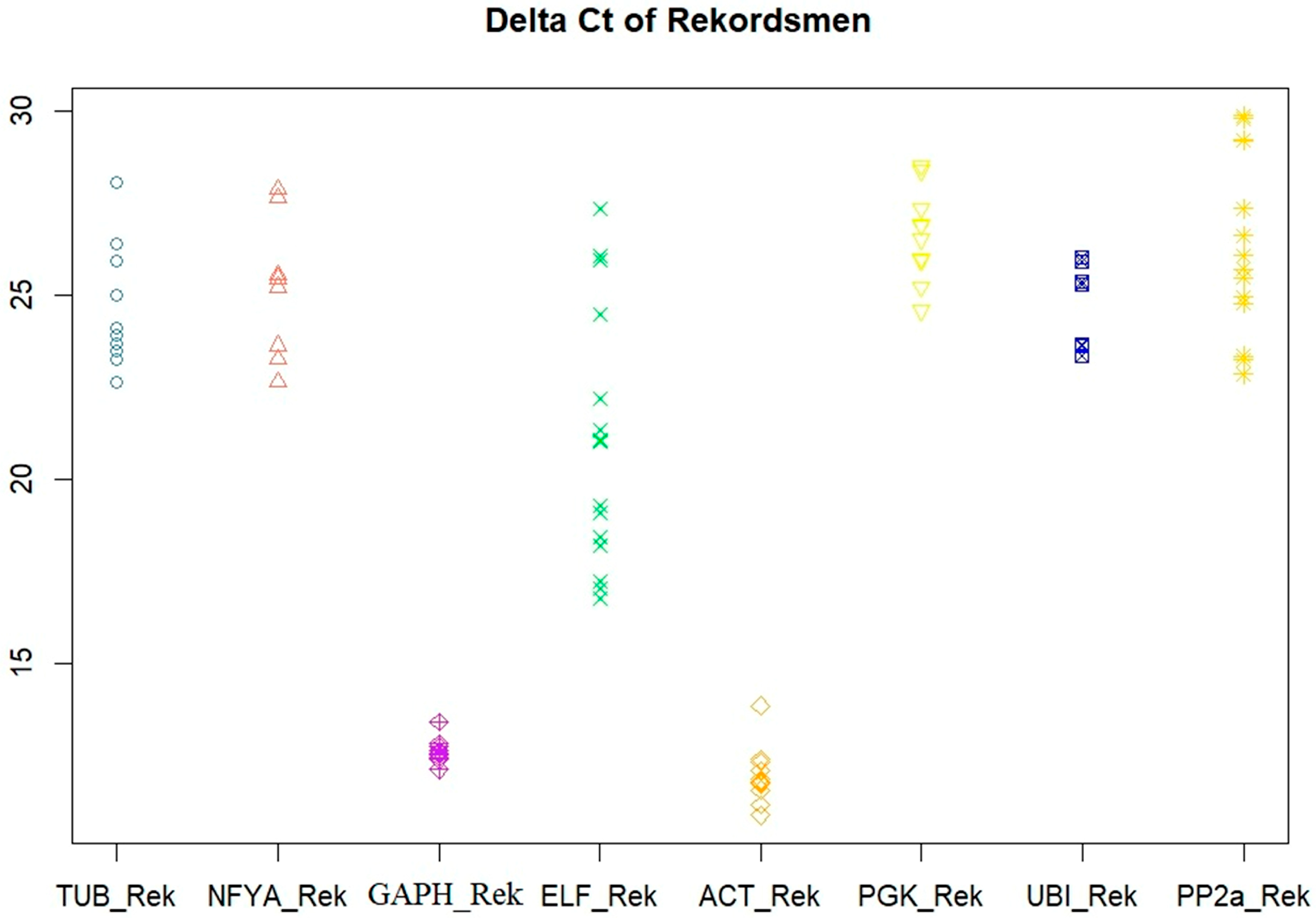

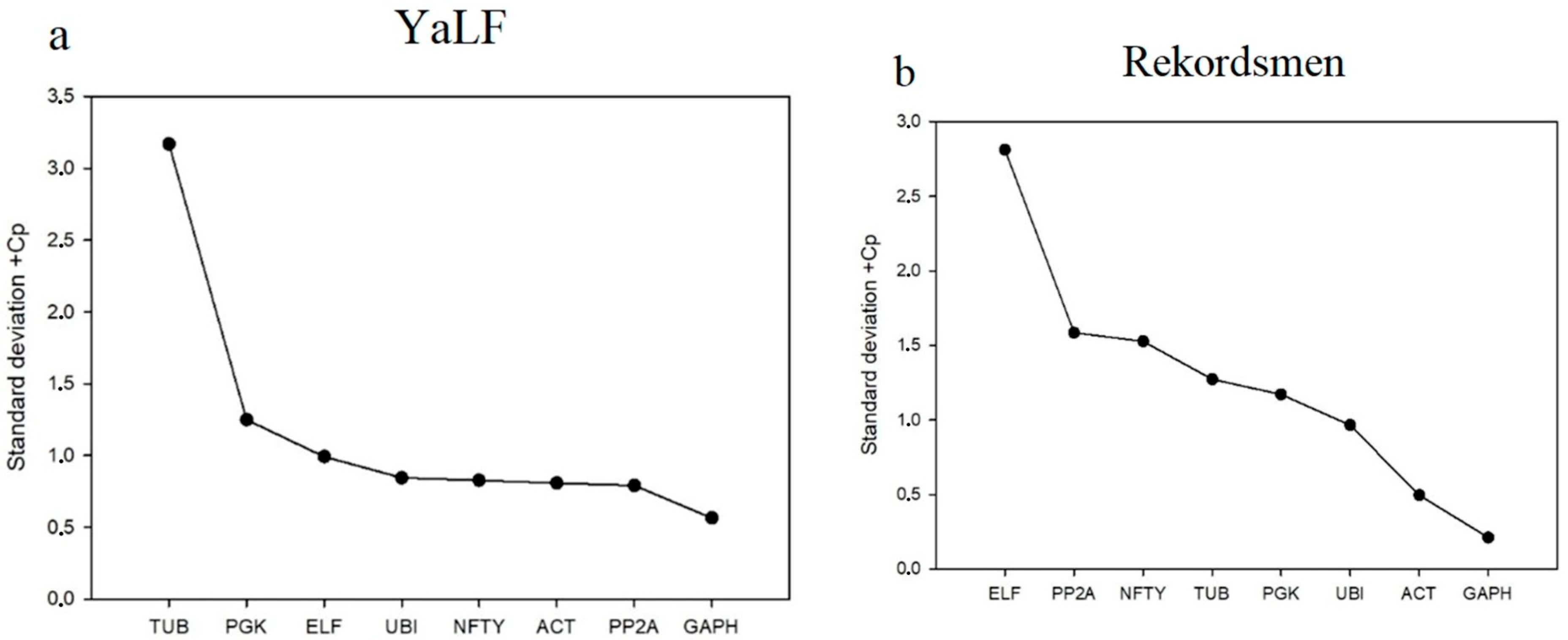



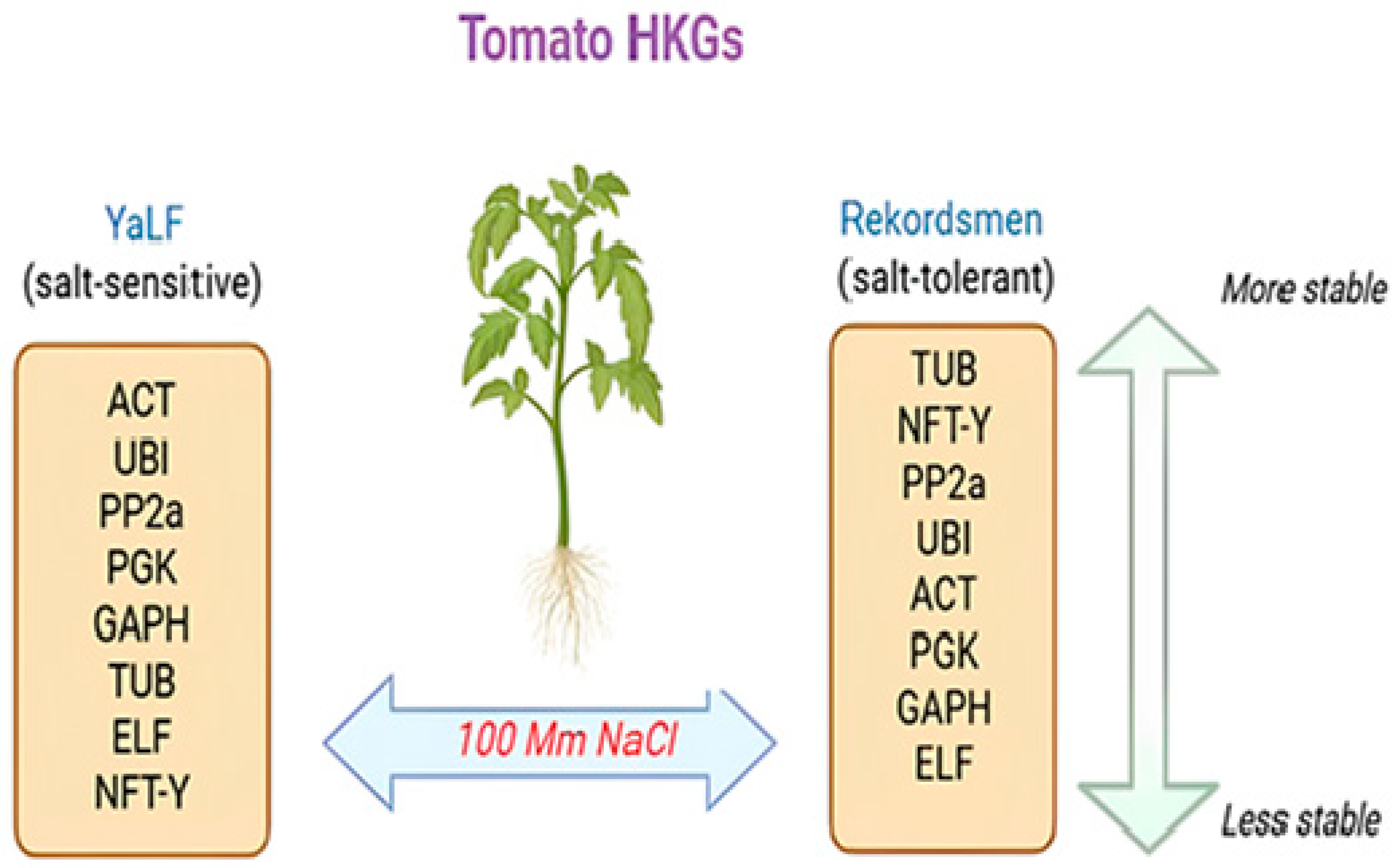
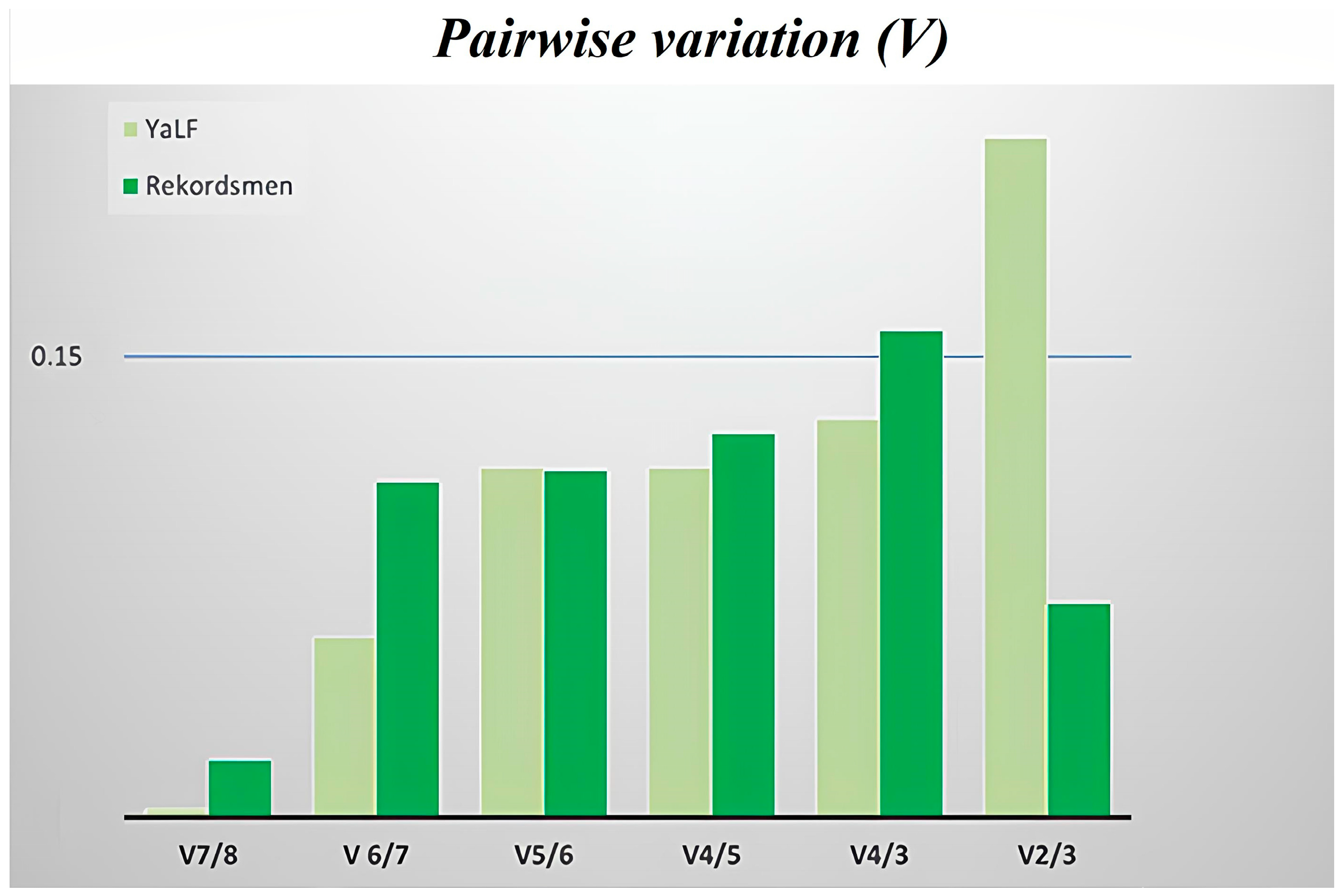
| Gene | Changed | SD, BestKeeper | GeNorm | NormFinder | Final Rank |
|---|---|---|---|---|---|
| TUB | 0.801 | 3.17 | 9.72 | 9 | 4.45 |
| PP2a | 0.67 | 0.79 | 6.13 | 7 | 3.31 |
| PGK | 0.64 | 1.25 | 7.87 | 6.63 | 3.83 |
| ELF | 0.41 | 0.99 | 10.64 | 7.26 | 5.47 |
| GAPH | 0.39 | 0.56 | 11.28 | 6.7 | 4.35 |
| NFT-Y | 0.36 | 0.83 | 10.49 | 8.95 | 6.5 |
| UBI | 0.36 | 0.85 | 6.13 | 6.51 | 2.3 |
| ACT | 0.25 | 0.81 | 10.25 | 4.35 | 1.68 |
| Gene | SD, ∆Ct | SD, BestKeeper | GeNorm | NormFinder | Final Rank |
|---|---|---|---|---|---|
| NFT-Y | 0.86 | 1.53 | 9.85 | 6.96 | 2.63 |
| TUB | 0.38 | 1.27 | 9.38 | 6.16 | 1.41 |
| ELF | 0.27 | 2.81 | 7.97 | 8.39 | 6.48 |
| PP2a | 0.19 | 1.59 | 3.38 | 7.54 | 2.78 |
| ACT | 0.19 | 0.49 | 6.58 | 7.39 | 3.93 |
| PGK | 0.13 | 1.17 | 4.56 | 8.52 | 5.6 |
| GAPH | 0.065 | 0.21 | 8.67 | 11.11 | 6.26 |
| UBI | 0.039 | 0.96 | 3.38 | 8.07 | 3.63 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rostovtseva, H.I.; Bogoutdinova, L.R.; Raldugina, G.N.; Baranova, E.N. Identification of Reliable Reference Genes for qRT-PCR Normalization in Tomato Genotypes with Contrasting Salinity Tolerance. Horticulturae 2025, 11, 1249. https://doi.org/10.3390/horticulturae11101249
Rostovtseva HI, Bogoutdinova LR, Raldugina GN, Baranova EN. Identification of Reliable Reference Genes for qRT-PCR Normalization in Tomato Genotypes with Contrasting Salinity Tolerance. Horticulturae. 2025; 11(10):1249. https://doi.org/10.3390/horticulturae11101249
Chicago/Turabian StyleRostovtseva, Helen I., Liliya R. Bogoutdinova, Galina N. Raldugina, and Ekaterina N. Baranova. 2025. "Identification of Reliable Reference Genes for qRT-PCR Normalization in Tomato Genotypes with Contrasting Salinity Tolerance" Horticulturae 11, no. 10: 1249. https://doi.org/10.3390/horticulturae11101249
APA StyleRostovtseva, H. I., Bogoutdinova, L. R., Raldugina, G. N., & Baranova, E. N. (2025). Identification of Reliable Reference Genes for qRT-PCR Normalization in Tomato Genotypes with Contrasting Salinity Tolerance. Horticulturae, 11(10), 1249. https://doi.org/10.3390/horticulturae11101249






