Complete Chloroplast Genome of Hygrophila polysperma (Acanthaceae): Insights into Its Genetic Features and Phylogenetic Relationships
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, DNA Extraction, and Plastid Genome Sequencing
2.2. Cp Genome Assembly and Annotation
2.3. Repeat Sequences and Codon Usage Analysis
2.4. Inverted-Repeats Contraction and Expansion
2.5. Comparative Analysis of Genome Structure
2.6. Phylogenetic Analysis
3. Results
3.1. Species Identification
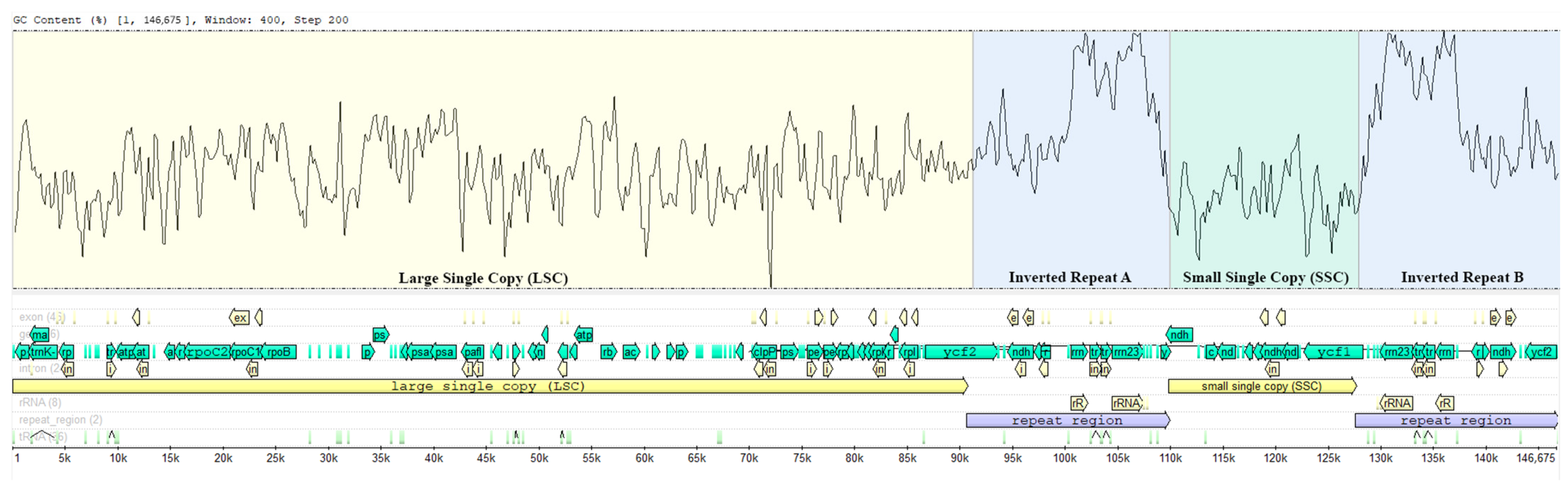
3.2. General Characterization of Genome
3.3. General Comparison of Cp Genome with Another Closely Related Species
3.4. Repeat Sequences
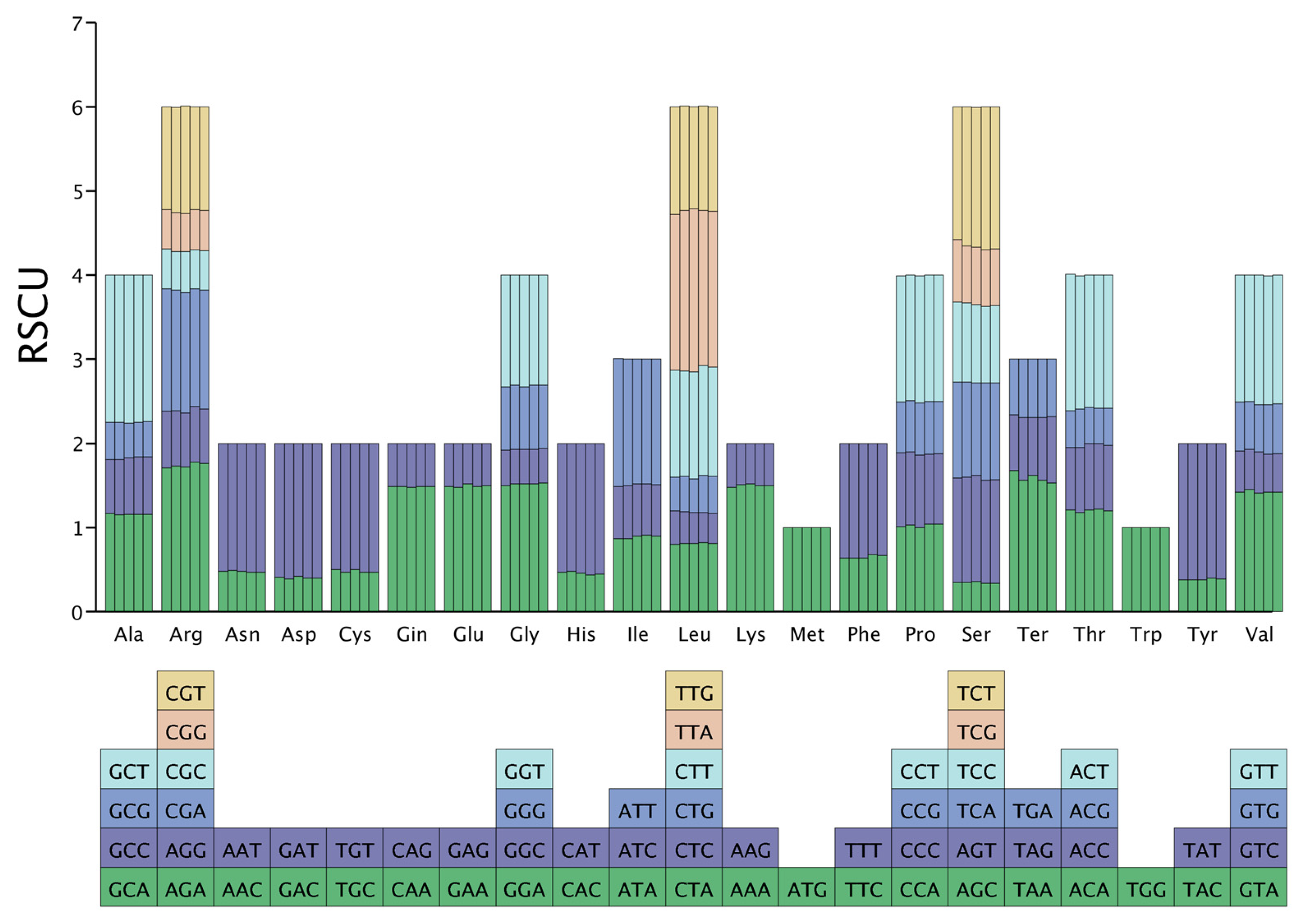
3.5. Codon Usage Analysis
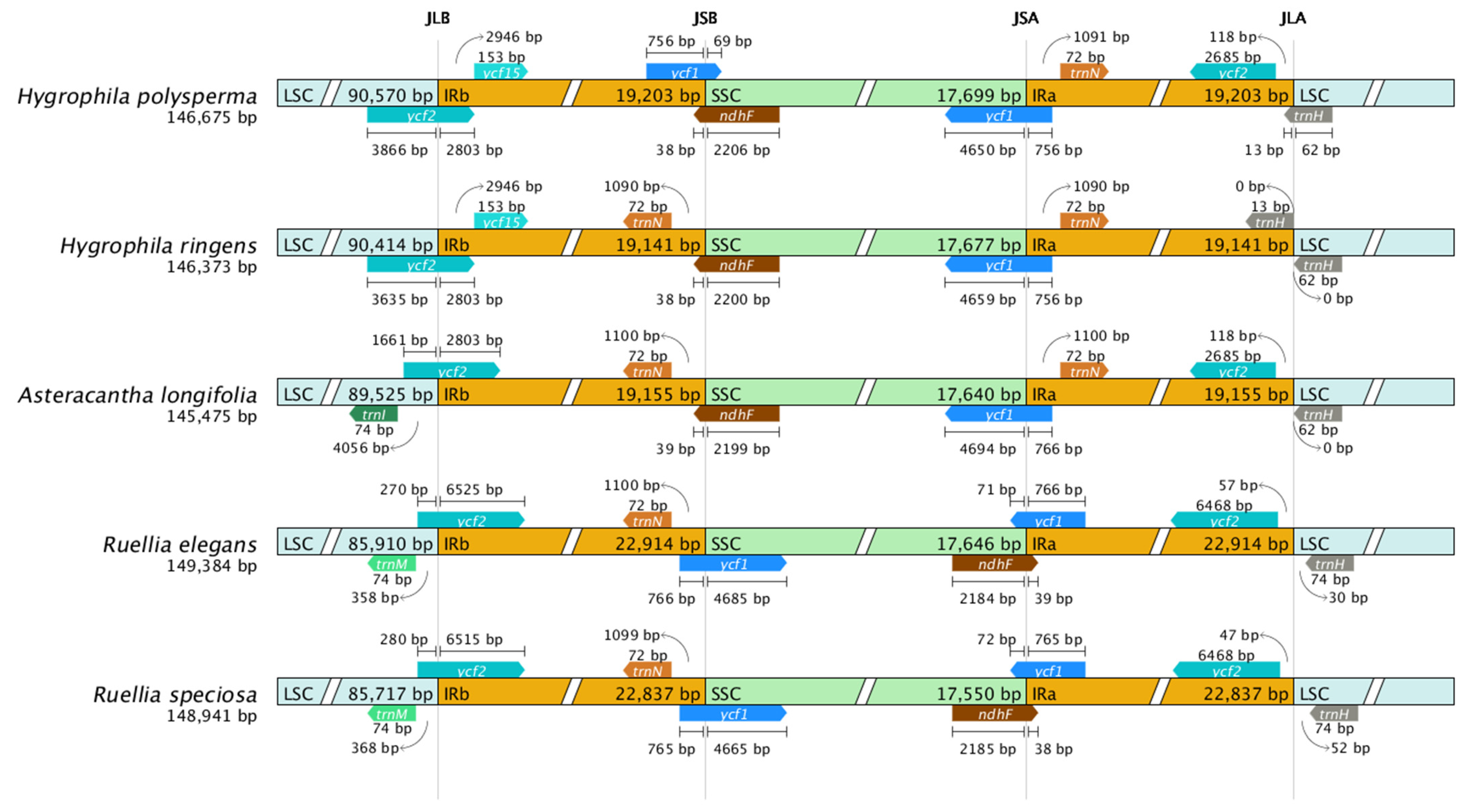
3.6. IR Contraction and Expansion in the cp Genome
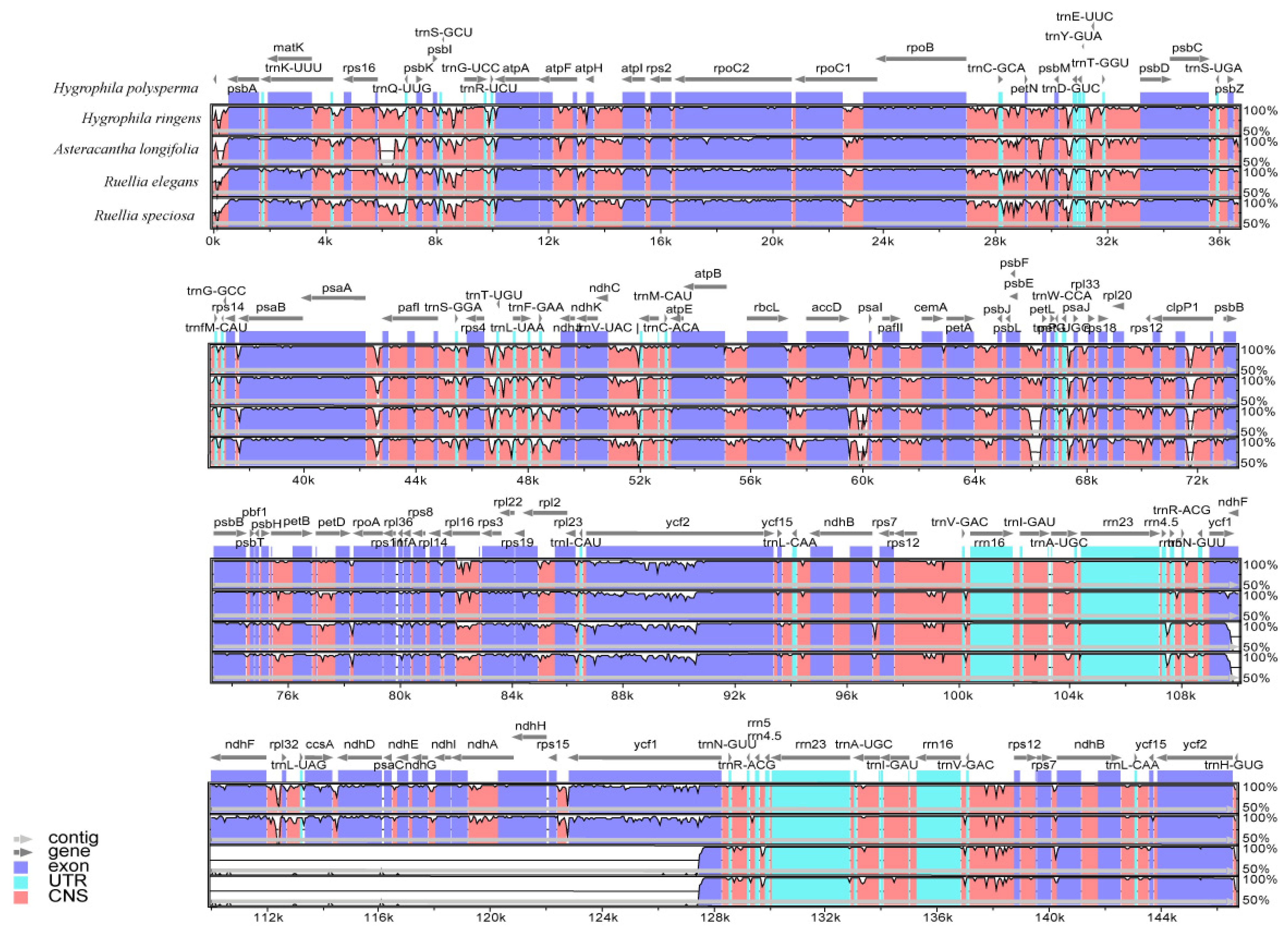
3.7. Comparative Analysis of cp Genome Structure
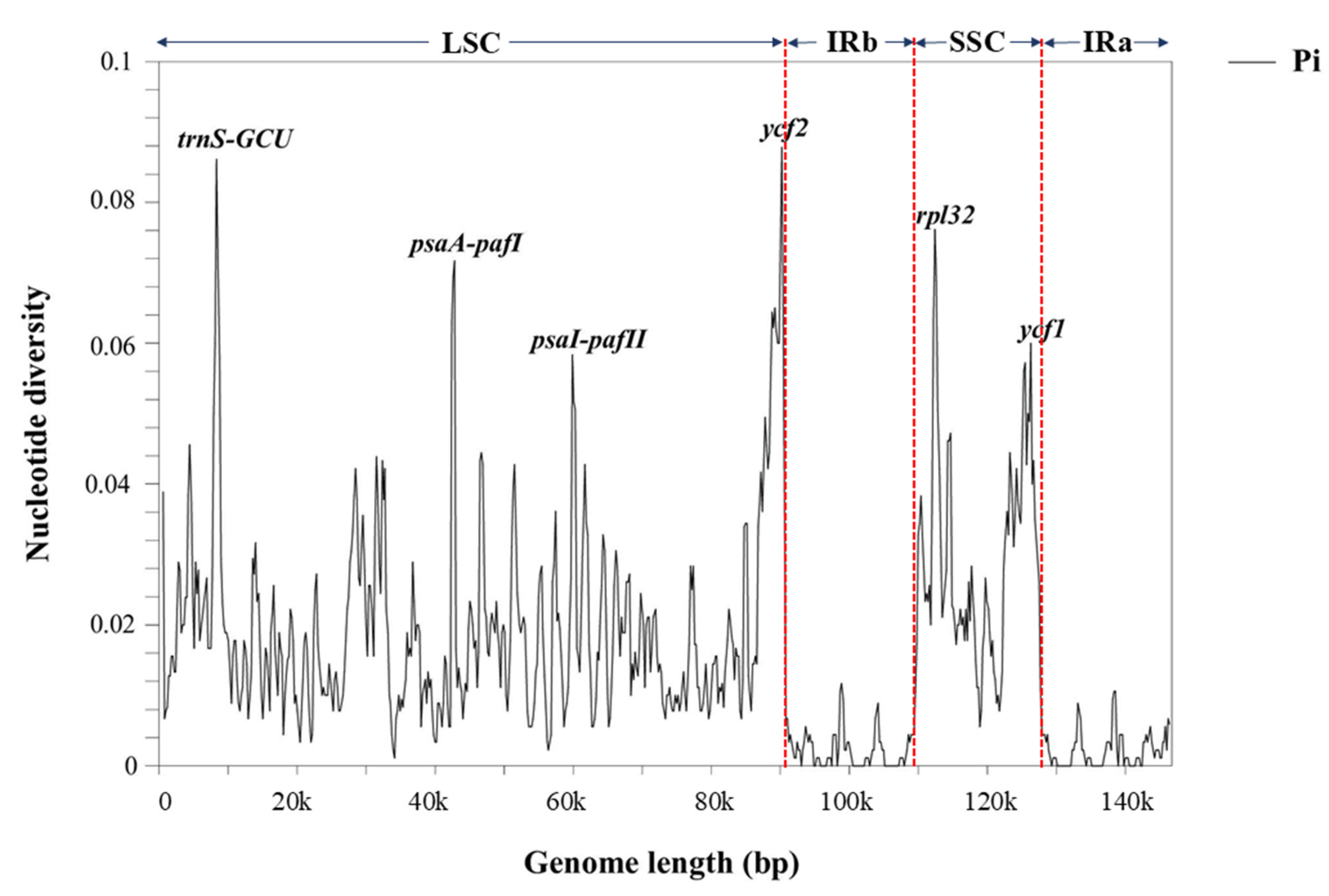
3.8. Nucleotide Diversity (Pi)
3.9. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kew Royal Botanic Gardens. Hygrophila polysperma (Roxb.) T. Anderson. Available online: https://powo.science.kew.org/taxon/urn:lsid:ipni.org:names:49756-1#distributions (accessed on 18 April 2023).
- NCBI. Hygrophila polysperma (Miramar-Weed). Available online: https://pubchem.ncbi.nlm.nih.gov/taxonomy/Hygrophila-polysperma (accessed on 30 March 2025).
- Qiu, X.; Xie, Y. New Records of Acanthaceae to Jiangxi Province. Jiangxi Sci. 2020, 38, 643–644. [Google Scholar]
- Mukherjee, A. Prospects for Classical Biological Control of the Aquatic Invasive Weed Hygrophila polysperma (Acanthaceae). Ph.D. Thesis, University of Florida, Gainesville, FL, USA, 2011. [Google Scholar]
- Doyle, R.D.; Francis, M.D.; Smart, R.M. Interference competition between Ludwigia repens and Hygrophila polysperma: Two morphologically similar aquatic plant species. Aquat. Bot. 2003, 77, 223–234. [Google Scholar] [CrossRef]
- Mukherjee, A.; Williams, D.; Gitzendanner, M.A.; Overholt, W.A.; Cuda, J.P. Microsatellite and chloroplast DNA diversity of the invasive aquatic weed Hygrophila polysperma in native and invasive ranges. Aquat. Bot. 2016, 129, 55–61. [Google Scholar] [CrossRef]
- Kew Royal Botanic Gardens. Hygrophila ringens . Available online: https://powo.science.kew.org/taxon/urn:lsid:ipni.org:names:60454721-2 (accessed on 30 March 2025).
- Li, G.; Zhao, X.; Yang, J.; Hu, S.; Ponnu, J.; Kimura, S.; Hwang, I.; Torii, K.U.; Hou, H. Water wisteria genome reveals environmental adaptation and heterophylly regulation in amphibious plants. Plant Cell Environ. 2024, 47, 4720–4740. [Google Scholar] [CrossRef] [PubMed]
- Tripp, E.; Daniel, T.; Fatimah, S.; McDade, L. Phylogenetic relationships within Ruellieae (Acanthaceae) and a revised classification. Int. J. Plant Sci. 2013, 174, 97–137. [Google Scholar] [CrossRef]
- Jiao, H.; Chen, Q.; Xiong, C.; Wang, H.; Ran, K.; Dong, R.; Dong, X.; Guan, Q.; Wei, S. Chloroplast genome profiling and phylogenetic insights of the “Qixiadaxiangshui” pear (Pyrus bretschneideri Rehd.1). Horticulturae 2024, 10, 744. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) Version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-Web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yu, H.; Wang, Z.; Xu, W. CPStools: A package for analyzing chloroplast genome sequences. iMetaOmics 2024, 1, e25. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Li, H.; Guo, Q.; Xu, L.; Gao, H.; Liu, L.; Zhou, X. CPJSdraw: Analysis and visualization of junction sites of chloroplast genomes. PeerJ 2023, 11, e15326. [Google Scholar] [CrossRef]
- Mayor, C.; Brudno, M.; Schwartz, J.R.; Poliakov, A.; Rubin, E.M.; Frazer, K.A.; Pachter, L.S.; Dubchak, I. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 2000, 16, 1046–1047. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Xue, S.; Shi, T.; Luo, W.; Ni, X.; Iqbal, S.; Ni, Z.; Huang, X.; Yao, D.; Shen, Z.; Gao, Z. Comparative analysis of the complete chloroplast genome among Prunus mume, P. armeniaca, and P. salicina. Hortic. Res. 2019, 6, 89. [Google Scholar] [CrossRef]
- Raman, G.; Park, K.T.; Kim, J.-H.; Park, S. Characteristics of the completed chloroplast genome sequence of Xanthium spinosum: Comparative analyses, identification of mutational hotspots and phylogenetic implications. BMC Genom. 2020, 21, 855. [Google Scholar] [CrossRef]
- Boudreau, E.; Takahashi, Y.; Lemieux, C.; Turmel, M.; Rochaix, J. The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 1997, 16, 6095–6104. [Google Scholar] [CrossRef]
- Naver, H.; Boudreau, E.; Rochaix, J.D. Functional studies of ycf3: Its role in assembly of photosystem I and interactions with some of its subunits. Plant Cell 2001, 13, 2731–2745. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, Z.; Zhang, Y.; Zhou, W.; Zhang, A.; Lu, C. Pentatricopeptide repeat protein PHOTOSYSTEM I BIOGENESIS FACTOR2 is required for splicing of ycf3. J. Integr. Plant Biol. 2020, 62, 1741–1761. [Google Scholar] [CrossRef] [PubMed]
- Krech, K.; Ruf, S.; Masduki, F.F.; Thiele, W.; Bednarczyk, D.; Albus, C.A.; Tiller, N.; Hasse, C.; Schöttler, M.A.; Bock, R. The plastid genome-encoded ycf4 protein functions as a nonessential assembly factor for photosystem I in higher plants. Plant Physiol. 2012, 159, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Al-Andal, A. Comparative analysis of chloroplast genomes in Commiphora gileadensis from Saudi Arabia and Oman reveal evolutionary genetic divergence. Cogent Food Agric. 2025, 11, 2477796. [Google Scholar] [CrossRef]
- Yan, M.; Dong, S.; Gong, Q.; Xu, Q.; Ge, Y. Comparative chloroplast genome analysis of four Polygonatum species insights into DNA barcoding, evolution, and phylogeny. Sci. Rep. 2023, 13, 16495. [Google Scholar] [CrossRef]
- Wu, M.; Lan, S.; Cai, B.; Chen, S.; Chen, H.; Zhou, S. The complete chloroplast genome of Guadua angustifolia and comparative analyses of neotropical-paleotropical bamboos. PLoS ONE 2015, 10, e0143792. [Google Scholar] [CrossRef]
- Gao, L.-Z.; Liu, Y.-L.; Zhang, D.; Li, W.; Gao, J.; Liu, Y.; Li, K.; Shi, C.; Zhao, Y.; Zhao, Y.-J.; et al. Evolution of Oryza chloroplast genomes promoted adaptation to diverse ecological habitats. Commun. Biol. 2019, 2, 278. [Google Scholar] [CrossRef]
- Majeran, W.; Wollman, F.A.; Vallon, O. Evidence for a role of clpP in the degradation of the chloroplast cytochrome b6f complex. Plant Cell 2000, 12, 137–149. [Google Scholar] [CrossRef]
- Kuroda, H.; Maliga, P. The plastid clpP1 protease gene is essential for plant development. Nature 2003, 425, 86–89. [Google Scholar] [CrossRef]
- Ramundo, S.; Rochaix, J.-D. Chloroplast unfolded protein response, a new plastid stress signaling pathway? Plant Signal. Behav. 2014, 9, e972874. [Google Scholar] [CrossRef]
- Shikanai, T.; Shimizu, K.; Ueda, K.; Nishimura, Y.; Kuroiwa, T.; Hashimoto, T. The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in Tobacco. Plant Cell Physiol. 2001, 42, 264–273. [Google Scholar] [CrossRef]
- Nishimura, K.; van Wijk, K.J. Organization, function and substrates of the essential Clp protease system in plastids. Biochim. Biophys. Acta BBA Bioenerg. 2015, 1847, 915–930. [Google Scholar] [CrossRef] [PubMed]
- Torabi, S.; Umate, P.; Manavski, N.; Plöchinger, M.; Kleinknecht, L.; Bogireddi, H.; Herrmann, R.G.; Wanner, G.; Schröder, W.P.; Meurer, J. psbN is required for assembly of the photosystem II reaction center in Nicotiana tabacum. Plant Cell 2014, 26, 1183–1199. [Google Scholar] [CrossRef] [PubMed]
- Plöchinger, M.; Schwenkert, S.; von Sydow, L.; Schröder, W.P.; Meurer, J. Functional update of the auxiliary proteins psbW, psbY, HCF136, psbN, terC and ALB3 in maintenance and assembly of PSII. Front. Plant Sci. 2016, 7, 423. [Google Scholar] [CrossRef] [PubMed]
- Lang, Z.; Wills, D.M.; Lemmon, Z.H.; Shannon, L.M.; Bukowski, R.; Wu, Y.; Messing, J.; Doebley, J.F. Defining the role of Prolamin-Box Binding Factor1 gene during maize domestication. J. Hered. 2014, 105, 576–582. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Yao, X.; Ye, Q.; Kang, M.; Huang, H. Microsatellite analysis reveals interpopulation differentiation and gene flow in the endangered tree Changiostyrax dolichocarpa (Styracaceae) with fragmented distribution in central China. New Phytol. 2007, 176, 472–480. [Google Scholar] [CrossRef]
- Yao, H.; Xu, Y.; Lan, Y.; Xiang, D.; Jiao, P.; Xu, H.; Qiao, D.; Cao, Y. Codon usage bias analysis in the chloroplast genomes of diatoms within the family Thalassiosiraceae and Skeletonemataceae. Plant Mol. Biol. Rep. 2025. [Google Scholar] [CrossRef]
- Huang, Y.; Li, J.; Yang, Z.; An, W.; Xie, C.; Liu, S.; Zheng, X. Comprehensive analysis of complete chloroplast genome and phylogenetic aspects of ten Ficus species. BMC Plant Biol. 2022, 22, 253. [Google Scholar] [CrossRef]
- Dong, W.; Xu, C.; Li, C.; Sun, J.; Zuo, Y.; Shi, S.; Cheng, T.; Guo, J.; Zhou, S. ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 2015, 5, 8348. [Google Scholar] [CrossRef]
- Manzitto-Tripp, E.A.; Darbyshire, I.; Daniel, T.F.; Kiel, C.A.; McDade, L.A. Revised classification of Acanthaceae and worldwide dichotomous keys. TAXON 2022, 71, 103–153. [Google Scholar] [CrossRef]
- Wood, J.R.I. Nees, Arnott and some forgotten Acanthaceae types from Asia. Edinb. J. Bot. 1994, 51, 103–115. [Google Scholar] [CrossRef]
- McDade, L.; Moody, M. Phylogenetic relationships among Acanthaceae: Evidence from noncoding trnL-trnF chloroplast DNA sequences. Am. J. Bot. 1999, 86, 70–80. [Google Scholar] [CrossRef]
- McDade, L.; Daniel, T.; Masta, S.; Riley, K. Phylogenetic relationships within the tribe Justicieae (Acanthaceae): Evidence from molecular sequences, morphology, and cytology. Ann. Mo. Bot. Gard. 2000, 87, 435. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, H.; Xu, W.; Wan, X.; Zhu, K. Comparative analysis of chloroplast genome of Lonicera japonica cv. Damaohua. Open Life Sci. 2024, 19, 20220984. [Google Scholar] [CrossRef]
- Shukla, N.; Kuntal, H.; Shanker, A.; Sharma, S.N. Mining and analysis of simple sequence repeats in the chloroplast genomes of genus Vigna. Biotechnol. Res. Innov. 2018, 2, 9–18. [Google Scholar] [CrossRef]

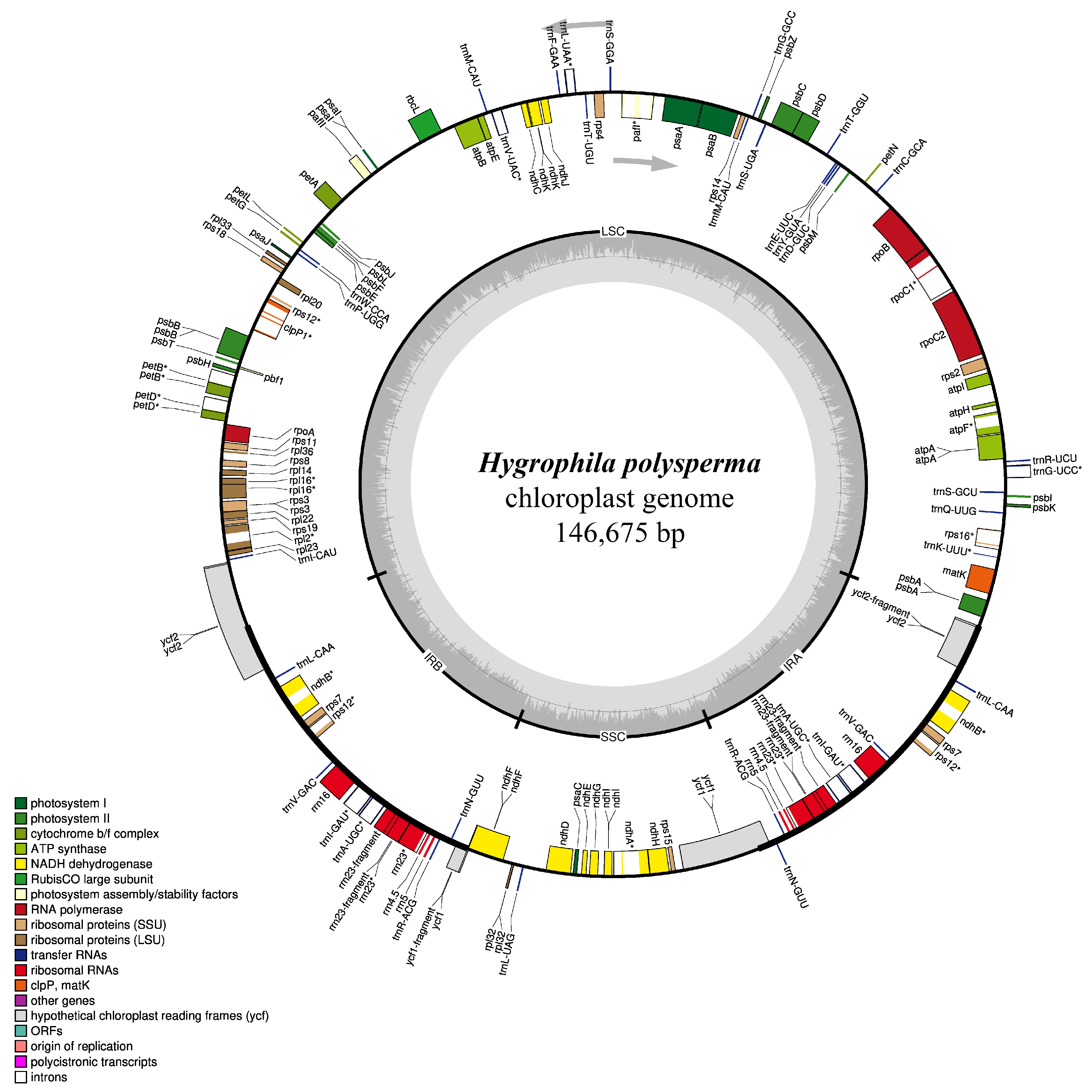

| Genome Features | H. polysperma |
|---|---|
| Genome size (bp) | 146,675 |
| LSC size (bp) | 90,570 |
| SSC size (bp) | 17,699 |
| IR size (bp) | 19,203 |
| GC content (%) | 38.3 |
| No. of genes | 130 |
| No. of PCGs | 86 |
| No. of tRNA | 36 |
| No. of rRNA | 8 |
| Category | Gene Group | Gene Name | Quantity |
|---|---|---|---|
| Photosynthesis | Subunits of photosystem I | psaB, psaA, psaI, psaJ, psaC | 5 |
| Subunits of photosystem II | psbA, psbK, psbI, psbM, psbD, psbC, psbZ, psbJ, psbL, psbF, psbE, psbB, psbT, psbH | 14 | |
| Subunits of cytochrome b/f complex | petN, petA, petL, petG, petB, petD | 6 | |
| NADH dehydrogenase subunit | ndhJ, ndhK, ndhC, ndhB (×2), ndhD, ndhF, ndhE, ndhG, ndhI, ndhA, ndhH | 12 | |
| Subunits of ATP synthase | atpA, atpF, atpH, atpI, atpE, atpB | 6 | |
| Large subunits of rubisco | rbcL | 1 | |
| Self-replication | Proteins of large ribosomal subunit | rpl33, rpl20, rpl36, rpl14, rpl16, rpl22, rpl2, rpl23, rpl32 | 9 |
| Proteins of small ribosomal subunit | rps12 (×2), rps16, rps2, rps14, rps4, rps18, rps11, rps8, rps3, rps19, rps7 (×2), rps15 | 14 | |
| Subunits of RNA polymerase | rpoC2, rpoC1, rpoB, rpoA | 4 | |
| Ribosomal RNAs | rrn16 (×2), rrn23 (×2), rrn4.5 (×2), rrn5 (×2) | 8 | |
| Transfer RNAs | trnH-GUG, trnK-UUU, trnQ-UUG, trnS-GCU, trnG-UCC, trnR-UCU, trnC-GCA, trnD-GUC, trnY-GUA, trnE-UUC, trnT-GGU, trnS-UGA, trnG-GCC, trnfM-CAU, trnS-GGA, trnT-UGU, trnL-UAA, trnF-GAA, trnV-UAC | trnC-ACA, trnM-CAU, trnW-CCA, trnP-UGG, trnI-CAU, trnL-CAA (×2), trnV-GAC (×2), trnI-GAU (×2), trnA-UGC (×2), trnR-ACG (×2), trnN-GUU (×2), trnL-UAG | 37 | |
| Other genes | Maturase | matK | 1 |
| Protease | clpP1 | 1 | |
| Acetyl-CoA carboxylase | accD | 1 | |
| c-type cytochrome synthesis gene | ccsA | 1 | |
| Translation initiation factor | infA | 1 | |
| Envelope membrane carbon uptake protein | cemA | ||
| Other | pafI, pafII, pbf1 | 3 | |
| Genes of unknown function | Conserved hypothetical chloroplast ORF | ycf1 (×2), ycf2 (×2), ycf15 (×2) | 6 |
| Region | Length (bp) | A (%) | T (%) | C (%) | G (%) | GC (%) |
|---|---|---|---|---|---|---|
| LSC | 90,570 | 31.3 | 32.2 | 18.7 | 17.7 | 36.4 |
| SSC | 17,699 | 33.8 | 33.5 | 17.1 | 15.5 | 32.7 |
| IRa | 19,203 | 27.8 | 27.0 | 21.4 | 23.8 | 45.2 |
| IRb | 19,203 | 27.0 | 27.8 | 23.8 | 21.4 | 45.2 |
| Total genome | 146,675 | 30.6 | 31.1 | 19.5 | 18.8 | 38.3 |
| CDS | 74,493 | 30.2 | 31.4 | 17.9 | 20.5 | 38.4 |
| No. | Type | Quantity | Start | End | Loc | Loc Type |
|---|---|---|---|---|---|---|
| 1 | TA | 6 | 4631 | 4642 | trnK-UUU_1-rps16_2 | IGS |
| 2 | C | 15 | 4651 | 4665 | trnK-UUU_1-rps16_2 | IGS |
| 3 | A | 11 | 6152 | 6162 | rps16_1-trnQ-UUG | IGS |
| 4 | TA | 6 | 7039 | 7050 | trnQ-UUG-psbK | IGS |
| 5 | A | 12 | 12,283 | 12,294 | atpF_2-atpF_1 | Intron |
| 6 | T | 10 | 12,375 | 12,384 | atpF_2-atpF_1 | Intron |
| 7 | T | 11 | 13,082 | 13,092 | atpF_1-atpH | IGS |
| 8 | A | 10 | 16,360 | 16,369 | rps2-rpoC2 | IGS |
| 9 | TCAA | 4 | 29,279 | 29,294 | petN-psbM | IGS |
| 10 | T | 10 | 30,457 | 30,466 | psbM-trnD-GUC | IGS |
| 11 | T | 12 | 35,749 | 35,760 | psbC-trnS-UGA | IGS |
| 12 | A | 11 | 36,529 | 36,539 | psbZ-trnG-GCC | IGS |
| 13 | A | 10 | 36,998 | 37,007 | trnG-GCC-trnfM-CAU | IGS |
| 14 | TA | 6 | 42,637 | 42,648 | psaA-pafI_3 | IGS |
| 15 | A | 11 | 44,614 | 44,624 | pafI_2-pafI_1 | Intron |
| 16 | T | 11 | 70,950 | 70,960 | clpP1_3-clpP1_2 | Intron |
| 17 | A | 15 | 71,099 | 71,113 | clpP1_3-clpP1_2 | Intron |
| 18 | A | 11 | 71,587 | 71,597 | clpP1_2-clpP1_1 | Intron |
| 19 | A | 11 | 77,197 | 77,207 | petD_1-petD_2 | Intron |
| 20 | T | 10 | 78,563 | 78,572 | rpoA | Gene |
| 21 | A | 17 | 81,500 | 81,516 | rpl14-rpl16_2 | IGS |
| 22 | T | 11 | 86,724 | 86,734 | ycf2 | Gene |
| 23 | A | 10 | 86,898 | 86,907 | ycf2 | Gene |
| 24 | T | 11 | 87,248 | 87,258 | ycf2 | Gene |
| 25 | G | 11 | 103,922 | 103,932 | trnA-UGC_1-trnA-UGC_2 | Intron |
| 26 | T | 10 | 112,269 | 112,278 | ndhF-rpl32 | IGS |
| 27 | T | 11 | 124,973 | 124,983 | ycf1 | Gene |
| 28 | T | 11 | 125,270 | 125,280 | ycf1 | Gene |
| 29 | T | 10 | 125,571 | 125,580 | ycf1 | Gene |
| 30 | C | 11 | 133,314 | 133,324 | trnA-UGC_2-trnA-UGC_1 | Intron |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chin, L.-X.; Huang, Q.; Fan, Q.; Tan, H.; Li, Y.; Peng, C.; Deng, Y.; Li, Y. Complete Chloroplast Genome of Hygrophila polysperma (Acanthaceae): Insights into Its Genetic Features and Phylogenetic Relationships. Horticulturae 2025, 11, 1240. https://doi.org/10.3390/horticulturae11101240
Chin L-X, Huang Q, Fan Q, Tan H, Li Y, Peng C, Deng Y, Li Y. Complete Chloroplast Genome of Hygrophila polysperma (Acanthaceae): Insights into Its Genetic Features and Phylogenetic Relationships. Horticulturae. 2025; 11(10):1240. https://doi.org/10.3390/horticulturae11101240
Chicago/Turabian StyleChin, Li-Xuan, Qiurui Huang, Qinglang Fan, Haibo Tan, Yuping Li, Caixia Peng, Yunfei Deng, and Yongqing Li. 2025. "Complete Chloroplast Genome of Hygrophila polysperma (Acanthaceae): Insights into Its Genetic Features and Phylogenetic Relationships" Horticulturae 11, no. 10: 1240. https://doi.org/10.3390/horticulturae11101240
APA StyleChin, L.-X., Huang, Q., Fan, Q., Tan, H., Li, Y., Peng, C., Deng, Y., & Li, Y. (2025). Complete Chloroplast Genome of Hygrophila polysperma (Acanthaceae): Insights into Its Genetic Features and Phylogenetic Relationships. Horticulturae, 11(10), 1240. https://doi.org/10.3390/horticulturae11101240






