Comparative Chloroplast Genomes to Gain Insights into the Phylogenetic Relationships and Evolution of Opisthopappus Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval and Consent to Participate
2.2. Plant Material, DNA Isolation and Sequencing
2.3. O. taihangensis Chloroplast Genome Assembly and Annotation
2.4. Phylogenetic Relationship and Divergence Time of Asteraceae
2.5. Genome Characteristics and Comparative Analyses Between Anthemideae
2.6. Analysis of Codon Usage Bias in Anthemideae
2.7. Opisthopappus Positive Selection Analyses
2.8. Opisthopappus Species RNA Editing Sites
3. Results
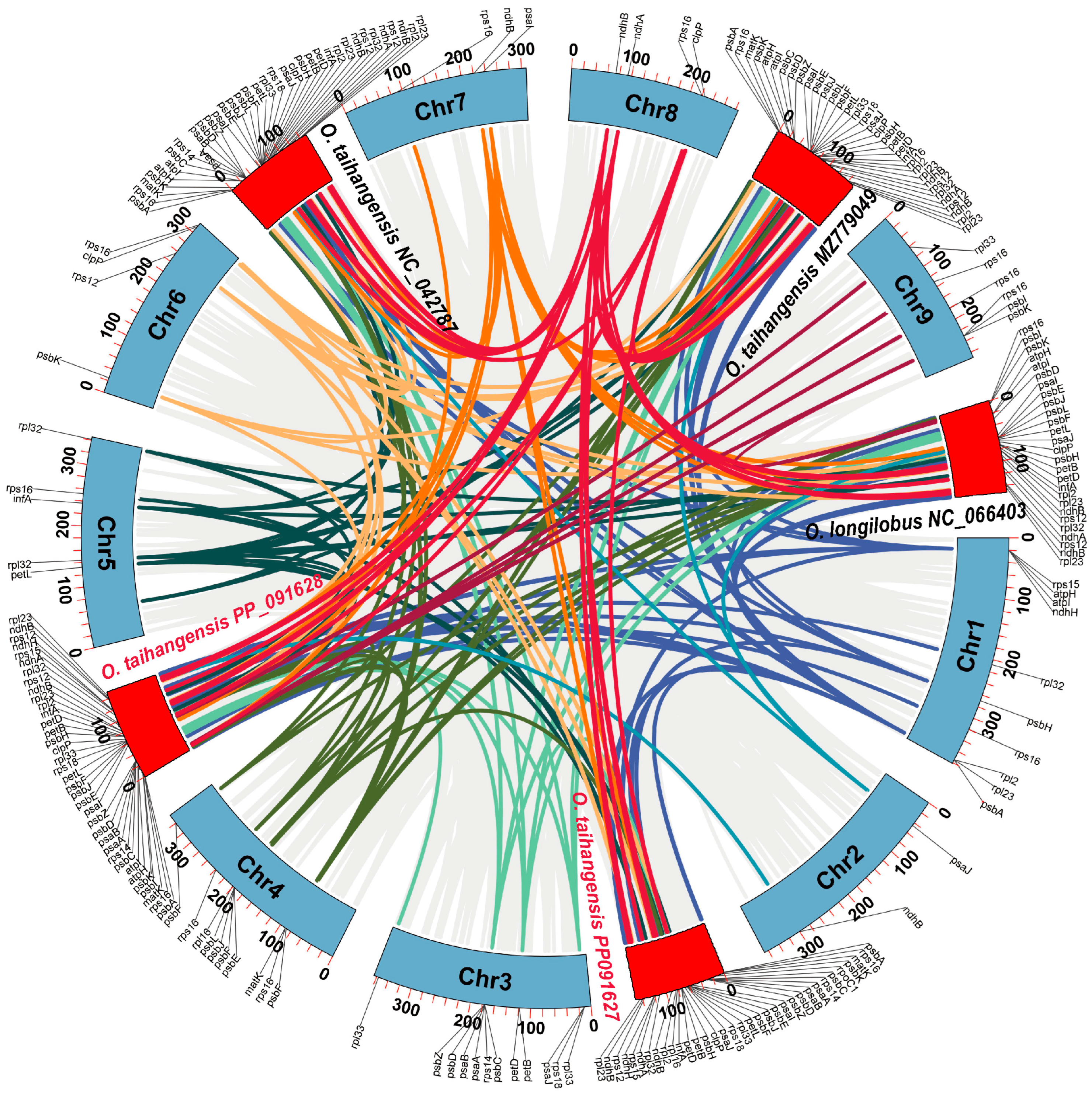
3.1. O. taihangensis Chloroplast Genomes
3.2. Asteraceae Phylogenetic Relationships and Divergence Times
3.3. Comparative Anthemideae Chloroplast Genomes
3.4. Anthemideae Codon Usage Pattern
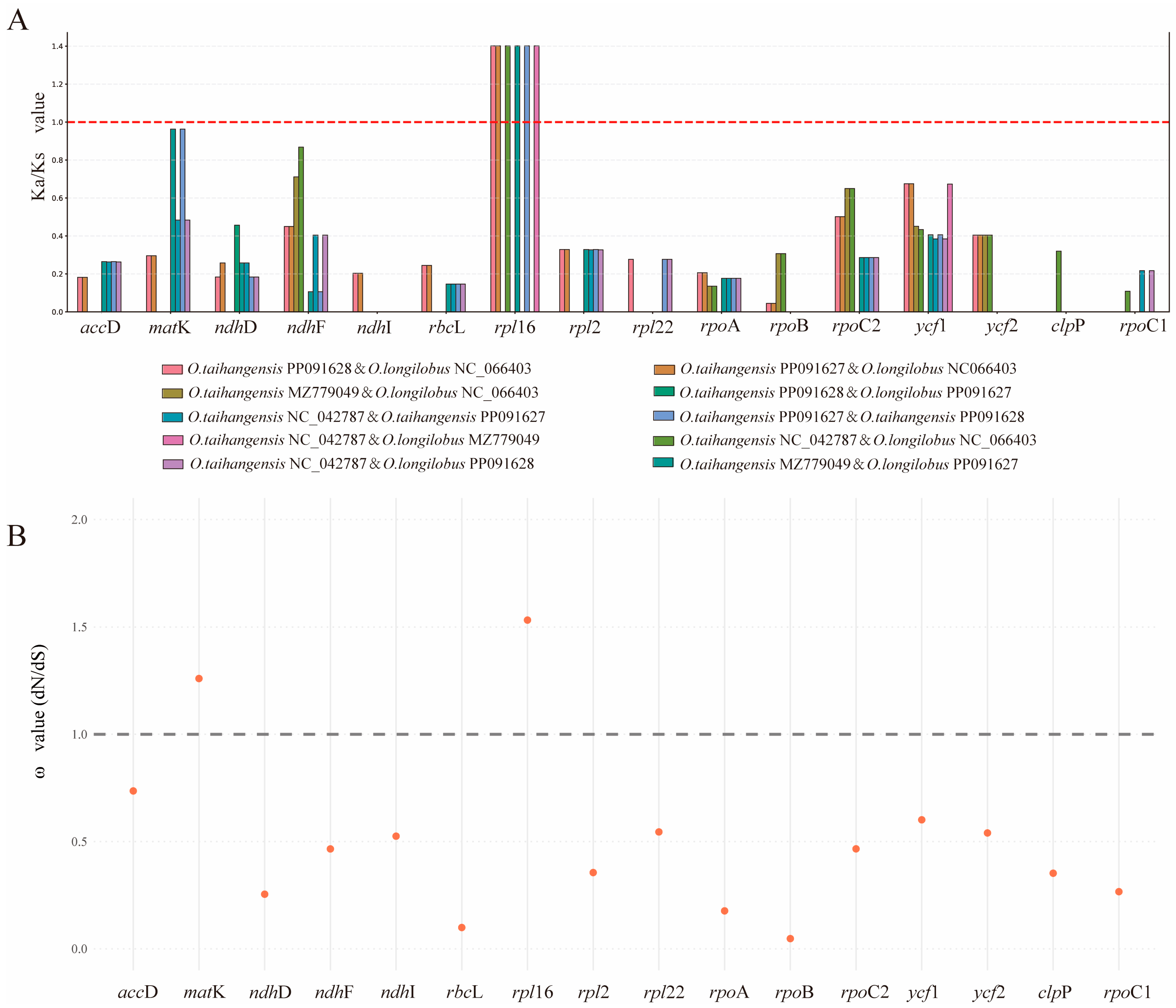
3.5. Opisthopappus Positive Selection
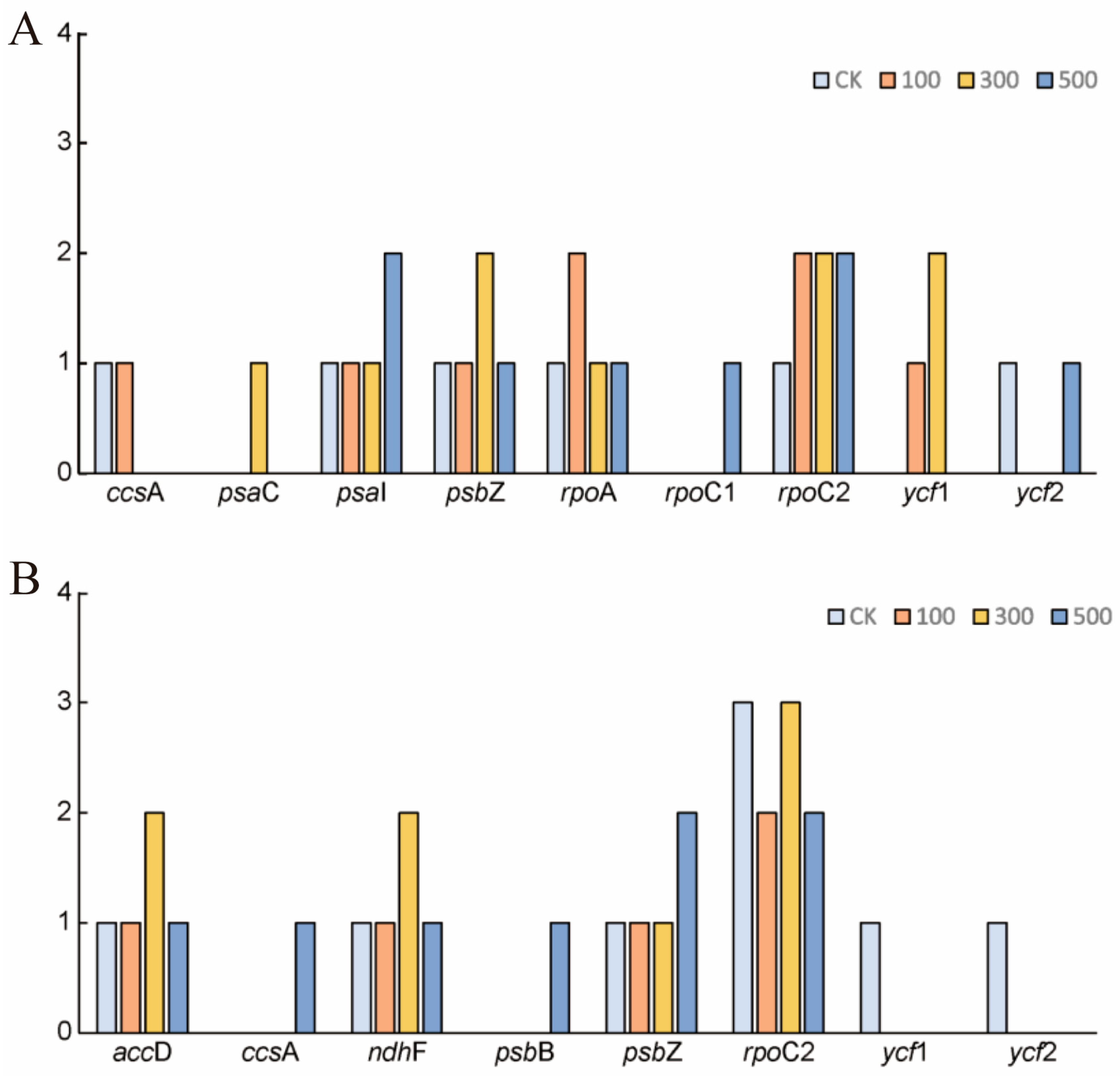
3.6. Opisthopappus RNA Editing Sites
4. Discussion
4.1. Structural Characteristics of O. taihangensis Chloroplast Genome
4.2. Phylogenetic Relationships and Divergence Among Asteraceae Species
4.3. Adaptive Evolution of Opisthopappus Chloroplast Genomes
4.4. RNA Editing Events Between Opisthopappus Species
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| LSC | Large single-copy region |
| SSC | Small single-copy region |
| IR | Inverted repeat region |
| rRNA | Ribosomal RNA |
| tRNA | Transfer RNA |
| FOC | Flora of China |
| ML | Maximum likelihood |
| SSR | Simple sequence repeat |
| Pi | Nucleotide diversity |
| RSCU | Relative synonymous codon usage |
| ENc | Effective number of codons |
| CAI | Codon adaptation index |
| Ser | Serine |
| Arg | Arginine |
| Leu | Leucine |
| Trp | Tryptophan |
| Ka | Non-synonymous |
| Ks | Synonymous |
| Phe | Phenylalanine |
| Ile | Isoleucine |
| ILS | Incomplete lineage sorting |
References
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef]
- Liu, J.; Qi, Z.-C.; Zhao, Y.-P.; Fu, C.-X.; Xiang, Q.-Y. Complete cpDNA genome sequence of Smilax china and phylogenetic placement of Liliales—Influences of gene partitions and taxon sampling. Mol. Phylogenet. Evol. 2012, 64, 545–562. [Google Scholar] [CrossRef]
- Gao, X.; Zhang, X.; Meng, H.; Li, J.; Zhang, D.; Liu, C. Comparative chloroplast genomes of Paris Sect. Marmorata: Insights into repeat regions and evolutionary implications. BMC Genom. 2018, 19, 878. [Google Scholar] [CrossRef]
- Jansen, R.K.; Raubeson, L.A.; Boore, J.L.; dePamphilis, C.W.; Chumley, T.W.; Haberle, R.C.; Wyman, S.K.; Alverson, A.J.; Peery, R.; Herman, S.J.; et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 2005, 395, 348–384. [Google Scholar] [CrossRef]
- Zhang, Y.; Tian, L.; Lu, C. Chloroplast gene expression: Recent advances and perspectives. Plant Commun. 2023, 4, 100611. [Google Scholar] [CrossRef] [PubMed]
- Li, D.M.; Li, J.; Wang, D.R.; Xu, Y.C.; Zhu, G.F. Molecular evolution of chloroplast genomes in subfamily Zingiberoideae (Zingiberaceae). BMC Plant Biol. 2021, 21, 558. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wen, F.; Hong, X.; Li, Z.; Mi, Y.; Zhao, B. Comparative chloroplast genome analyses of Paraboea (Gesneriaceae): Insights into adaptive evolution and phylogenetic analysis. Front. Plant Sci. 2022, 13, 1019831. [Google Scholar] [CrossRef]
- Frailey, D.C.; Chaluvadi, S.R.; Vaughn, J.N.; Coatney, C.G.; Bennetzen, J.L. Gene loss and genome rearrangement in the plastids of five Hemiparasites in the family Orobanchaceae. BMC Plant Biol. 2018, 18, 30. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Qian, S.; Zhang, J.; Kessler, M. Effects of climate and environmental heterogeneity on the phylogenetic structure of regional angiosperm floras worldwide. Nat. Commun. 2024, 15, 1079. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, Y.; Landis, J.B.; Zhang, J.; Yang, L.; Lin, N.; Zhang, H.; Guo, R.; Li, L.; Zhang, Y.; et al. Genomic insights into adaptation to heterogeneous environments for the ancient relictual Circaeaster agrestis (Circaeasteraceae, Ranunculales). New Phytol. 2020, 228, 285–301. [Google Scholar] [CrossRef]
- Wen, J.; Xie, D.F.; Price, M.; Ren, T.; Deng, Y.Q.; Gui, L.J.; Guo, X.L.; He, X.J. Backbone phylogeny and evolution of Apioideae (Apiaceae): New insights from phylogenomic analyses of plastome data. Mol. Phylogenet. Evol. 2021, 161, 107183. [Google Scholar] [CrossRef]
- Jiang, D.; Cai, X.; Gong, M.; Xia, M.; Xing, H.; Dong, S.; Tian, S.; Li, J.; Lin, J.; Liu, Y.; et al. Complete chloroplast genomes provide insights into evolution and phylogeny of Zingiber (Zingiberaceae). BMC Genom. 2023, 24, 30. [Google Scholar] [CrossRef] [PubMed]
- Li, P.W.; Lu, Y.B.; Qin, X.M.; Zhang, Q. Plastome phylogenomics unravels the evolutionary relationships and biogeographic history of Chloranthaceae. BMC Plant Biol. 2025, 25, 543. [Google Scholar] [CrossRef]
- Arella, D.; Dilucca, M.; Giansanti, A. Codon usage bias and environmental adaptation in microbial organisms. Mol. Genet. Genom. MGG 2021, 296, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, T.; Firoz, A.; Ramadan, A.M. RNA Editing in Chloroplast: Advancements and Opportunities. Curr. Issues Mol. Biol. 2022, 44, 5593–5604. [Google Scholar] [CrossRef]
- Zhang, A.; Jiang, X.; Zhang, F.; Wang, T.; Zhang, X. Dynamic response of RNA editing to temperature in grape by RNA deep sequencing. Funct. Integr. Genom. 2020, 20, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Yueqiao, Z.; Yinsheng, M.; Nong, Y.; Wei, S.; Shuwen, D. Cenozoic extensional stress evolution in North China. J. Geodyn. 2003, 36, 591–613. [Google Scholar] [CrossRef]
- Ji, A.; Wang, Y.; Wu, G.; Wu, W.; Yang, H.; Wang, Q. Genetic Diversity and Population Structure of North China Mountain Walnut Revealed by ISSR. Am. J. Plant Sci. 2014, 05, 3194–3202. [Google Scholar] [CrossRef]
- Ye, H.; Wang, Z.; Hou, H.; Wu, J.; Gao, Y.; Han, W.; Ru, W.; Sun, G.; Wang, Y. Localized environmental heterogeneity drives the population differentiation of two endangered and endemic Opisthopappus Shih species. BMC Ecol. Evol. 2021, 21, 56. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, G.; Jiang, Y.; Huang, L. Complete Chloroplast Genome Sequence of Endangered Species in the Genus Opisthopappus C. Shih: Characterization, Species Identification, and Phylogenetic Relationships. Genes 2022, 13, 2410. [Google Scholar] [CrossRef]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Zheng, S.; Poczai, P.; Hyvönen, J.; Tang, J.; Amiryousefi, A. Chloroplot: An Online Program for the Versatile Plotting of Organelle Genomes. Front. Genet. 2020, 11, 576124. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Yoshida, T.; Furihata, H.Y.; Kawabe, A. Patterns of genomic integration of nuclear chloroplast DNA fragments in plant species. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2014, 21, 127–140. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Gao, M.; Huo, X.; Lu, L.; Liu, M.; Zhang, G. Analysis of codon usage patterns in Bupleurum falcatum chloroplast genome. Chin. Herb. Med. 2023, 15, 284–290. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Nielsen, R.; Goldman, N.; Pedersen, A.M. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 2000, 155, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Chen, C.; Arab, D.A.; Du, Z.; He, Y.; Ho, S.Y.W. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019, 9, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, R.; Yang, Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 1998, 148, 929–936. [Google Scholar] [CrossRef]
- Lan, Y.; Sun, J.; Tian, R.; Bartlett, D.H.; Li, R.; Wong, Y.H.; Zhang, W.; Qiu, J.W.; Xu, T.; He, L.S.; et al. Molecular adaptation in the world’s deepest-living animal: Insights from transcriptome sequencing of the hadal amphipod Hirondellea gigas. Mol. Ecol. 2017, 26, 3732–3743. [Google Scholar] [CrossRef]
- Han, M.; Niu, M.; Gao, T.; Shen, Y.; Zhou, X.; Zhang, Y.; Liu, L.; Chai, M.; Sun, G.; Wang, Y. Responsive Alternative Splicing Events of Opisthopappus Species against Salt Stress. Int. J. Mol. Sci. 2024, 25, 1227. [Google Scholar] [CrossRef]
- Wu, S.; Liu, W.; Aljohi, H.A.; Alromaih, S.A.; Alanazi, I.O.; Lin, Q.; Yu, J.; Hu, S. REDO: RNA Editing Detection in Plant Organelles Based on Variant Calling Results. J. Comput. Biol. A J. Comput. Mol. Cell Biol. 2018, 25, 509–516. [Google Scholar] [CrossRef]
- Hu, E.Z.; Lan, X.R.; Liu, Z.L.; Gao, J.; Niu, D.K. A positive correlation between GC content and growth temperature in prokaryotes. BMC Genom. 2022, 23, 110. [Google Scholar] [CrossRef]
- Vinogradov, A.E. DNA helix: The importance of being GC-rich. Nucleic Acids Res. 2003, 31, 1838–1844. [Google Scholar] [CrossRef] [PubMed]
- Foerstner, K.U.; von Mering, C.; Hooper, S.D.; Bork, P. Environments shape the nucleotide composition of genomes. EMBO Rep. 2005, 6, 1208–1213. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, H.; Zang, E.; Qie, Q.; He, S.; Hao, W.; Lan, Y.; Liu, Z.; Sun, G.; Wang, Y. Geographic distribution pattern and ecological niche differentiation of endangered Opisthopappus in Taihang Mountains. Braz. J. Bot. 2023, 46, 217–226. [Google Scholar] [CrossRef]
- Yang, Y.; Ma, X.; Huo, Y.X. Application of codon optimization strategy in heterologous protein expression. Sheng Wu Gong Cheng Xue Bao 2019, 35, 2227–2237. [Google Scholar] [CrossRef]
- Piovesan, A.; Pelleri, M.C.; Antonaros, F.; Strippoli, P.; Caracausi, M.; Vitale, L. On the length, weight and GC content of the human genome. BMC Res. Notes 2019, 12, 106. [Google Scholar] [CrossRef] [PubMed]
- Ping, J.; Hao, J.; Li, J.; Yang, Y.; Su, Y.; Wang, T. Loss of the IR region in conifer plastomes: Changes in the selection pressure and substitution rate of protein-coding genes. Ecol. Evol. 2022, 12, e8499. [Google Scholar] [CrossRef]
- Wu, C.S.; Chaw, S.M. Highly rearranged and size-variable chloroplast genomes in conifers II clade (cupressophytes): Evolution towards shorter intergenic spacers. Plant Biotechnol. J. 2014, 12, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Dobrogojski, J.; Adamiec, M.; Luciński, R. The chloroplast genome: A review. Acta Physiol. Plant. 2020, 42, 98. [Google Scholar] [CrossRef]
- Dong, W.; Xu, C.; Li, C.; Sun, J.; Zuo, Y.; Shi, S.; Cheng, T.; Guo, J.; Zhou, S. ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 2015, 5, 8348. [Google Scholar] [CrossRef] [PubMed]
- Nakai, M. YCF1: A Green TIC: Response to the de Vries et al. Commentary. Plant Cell 2015, 27, 1834–1838. [Google Scholar] [CrossRef]
- Fu, C.N.; Li, H.T.; Milne, R.; Zhang, T.; Ma, P.F.; Yang, J.; Li, D.Z.; Gao, L.M. Comparative analyses of plastid genomes from fourteen Cornales species: Inferences for phylogenetic relationships and genome evolution. BMC Genom. 2017, 18, 956. [Google Scholar] [CrossRef]
- Wen, J.; Wu, B.C.; Li, H.M.; Zhou, W.; Song, C.F. Plastome structure and phylogenetic relationships of genus Hydrocotyle (apiales): Provide insights into the plastome evolution of Hydrocotyle. BMC Plant Biol. 2024, 24, 778. [Google Scholar] [CrossRef]
- Deusch, O.; Landan, G.; Roettger, M.; Gruenheit, N.; Kowallik, K.V.; Allen, J.F.; Martin, W.; Dagan, T. Genes of cyanobacterial origin in plant nuclear genomes point to a heterocyst-forming plastid ancestor. Mol. Biol. Evol. 2008, 25, 748–761. [Google Scholar] [CrossRef]
- Bock, R.; Timmis, J.N. Reconstructing evolution: Gene transfer from plastids to the nucleus. Bioessays 2008, 30, 556–566. [Google Scholar] [CrossRef]
- Filip, E.; Skuza, L. Horizontal Gene Transfer Involving Chloroplasts. Int. J. Mol. Sci. 2021, 22, 4484. [Google Scholar] [CrossRef]
- Robinson, H.E. New supertribes, Helianthodae and Senecionodae, for the subfamily Asteroideae (Asteraceae). Phytologia 2004, 86, 116–120. [Google Scholar] [CrossRef]
- Watson, L.E.; Siniscalchi, C.M.; Mandel, J. Phylogenomics of the hyperdiverse daisy tribes: Anthemideae, Astereae, Calenduleae, Gnaphalieae, and Senecioneae. J. Syst. Evol. 2020, 58, 841–852. [Google Scholar] [CrossRef]
- Panero, J.L.; Freire, S.E.; Ariza Espinar, L.; Crozier, B.S.; Barboza, G.E.; Cantero, J.J. Resolution of deep nodes yields an improved backbone phylogeny and a new basal lineage to study early evolution of Asteraceae. Mol. Phylogenet. Evol. 2014, 80, 43–53. [Google Scholar] [CrossRef]
- Panero, J.L.; Funk, V.A. The value of sampling anomalous taxa in phylogenetic studies: Major clades of the Asteraceae revealed. Mol. Phylogenet. Evol. 2008, 47, 757–782. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, C.; Huang, C.-H.; Ma, H. Phylogenetic Reconstruction of Tribal Relationships in Asteroideae (Asteraceae) with Low-copy Nuclear Genes. Chin. Bull. Bot. 2015, 50, 549–564. [Google Scholar] [CrossRef]
- Karis, P.O. Morphological phylogenetics of theAsteraceae-Asteroideae, with notes on character evolution. Plant Syst. Evol. 1993, 186, 69–93. [Google Scholar] [CrossRef]
- Zhang, C.; Huang, C.H.; Liu, M.; Hu, Y.; Panero, J.L.; Luebert, F.; Gao, T.; Ma, H. Phylotranscriptomic insights into Asteraceae diversity, polyploidy, and morphological innovation. J. Integr. Plant Biol. 2021, 63, 1273–1293. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Chen, S.; Chen, F.; Cheng, X.; Zhang, F. The embryo rescue derived intergeneric hybrid between chrysanthemum and Ajania przewalskii shows enhanced cold tolerance. Plant Cell Rep. 2011, 30, 2177–2186. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, S.; Tang, F.; Jiang, J.; Li, C.; Miao, H.; Chen, F.; Fang, W.; Guo, W. Morphological characteristics and chromosome behaviour in F1, F2 and BC1 progenies between Chrysanthemum x morifolium and Ajania pacifica. Genetika 2012, 48, 951–961. [Google Scholar] [CrossRef]
- Criado Ruiz, D.; Villa Machío, I.; Herrero Nieto, A.; Nieto Feliner, G. Hybridization and cryptic speciation in the Iberian endemic plant genus Phalacrocarpum (Asteraceae-Anthemideae). Mol. Phylogenet. Evol. 2021, 156, 107024. [Google Scholar] [CrossRef]
- Liu, P.L.; Wan, Q.; Guo, Y.P.; Yang, J.; Rao, G.Y. Phylogeny of the genus Chrysanthemum L.: Evidence from single-copy nuclear gene and chloroplast DNA sequences. PLoS ONE 2012, 7, e48970. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.B. Phylogeny of Tribe Anthemideae (Asteraceae) from East Asia and Intergeneric Cross Between Dendranthema × Grandiflorum (Ramat.) Kitam. and Ajania pacifica (Nakai) K. Bremer & Humphries. Ph.D. Dissertation, Nanjing Agricultural University, Nanjing, China, 2007. [Google Scholar]
- Hu, X. Preliminary Studies on Inter-Generic Hybridization Within Chrysanthemum in Broad Sense (III). Master’s Thesis, Beijing Forestry University, Beijing, China, 2008. [Google Scholar]
- Chen, X.; Wang, H.; Jiang, J.; Jiang, Y.; Zhang, W.; Chen, F. Biogeographic and metabolic studies support a glacial radiation hypothesis during Chrysanthemum evolution. Hortic. Res. 2022, 9, uhac153. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, H. Initial Formation and Mesozoic Tectonic Exhumation of an Intracontinental Tectonic Belt of the Northern Part of the Taihang Mountain Belt, Eastern Asia. J. Geol. 2008, 116, 155–172. [Google Scholar] [CrossRef]
- Chi, X.; Zhang, F.; Dong, Q.; Chen, S. Insights into Comparative Genomics, Codon Usage Bias, and Phylogenetic Relationship of Species from Biebersteiniaceae and Nitrariaceae Based on Complete Chloroplast Genomes. Plants 2020, 9, 1605. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Ji, C.; Chen, Z.; Cai, H.; Wu, X.; Shi, C.; Wang, S. Comparative Analysis the Complete Chloroplast Genomes of Nine Musa Species: Genomic Features, Comparative Analysis, and Phylogenetic Implications. Front. Plant Sci. 2022, 13, 832884. [Google Scholar] [CrossRef]
- Xue, S.; Shi, T.; Luo, W.; Ni, X.; Iqbal, S.; Ni, Z.; Huang, X.; Yao, D.; Shen, Z.; Gao, Z. Comparative analysis of the complete chloroplast genome among Prunus mume, P. armeniaca, and P. salicina. Hortic. Res. 2019, 6, 89. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, J.; Hu, B.; Dong, C.; Sun, J.; Li, Z.; Ye, K.; Deng, F.; Wang, L.; Aslam, M.; et al. Assembly, annotation, and comparative analysis of Ipomoea chloroplast genomes provide insights into the parasitic characteristics of Cuscuta species. Front. Plant Sci. 2023, 13, 1074697. [Google Scholar] [CrossRef] [PubMed]
- Hooper, S.D.; Berg, O.G. Gradients in nucleotide and codon usage along Escherichia coli genes. Nucleic Acids Res. 2000, 28, 3517–3523. [Google Scholar] [CrossRef]
- Wen, F.; Wu, X.; Li, T.; Jia, M.; Liu, X.; Liao, L. The complete chloroplast genome of Stauntonia chinensis and compared analysis revealed adaptive evolution of subfamily Lardizabaloideae species in China. BMC Genom. 2021, 22, 161. [Google Scholar] [CrossRef]
- Somaratne, Y.; Guan, D.-L.; Wang, W.-Q.; Zhao, L.; Xu, S.-Q. The Complete Chloroplast Genomes of Two Lespedeza Species: Insights into Codon Usage Bias, RNA Editing Sites, and Phylogenetic Relationships in Desmodieae (Fabaceae: Papilionoideae). Plants 2020, 9, 51. [Google Scholar] [CrossRef]
- Mehmood, F.; Abdullah; Shahzadi, I.; Ahmed, I.; Waheed, M.T.; Mirza, B. Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics 2020, 112, 1522–1530. [Google Scholar] [CrossRef]
- Nie, X.; Deng, P.; Feng, K.; Liu, P.; Du, X.; You, F.M.; Weining, S. Comparative analysis of codon usage patterns in chloroplast genomes of the Asteraceae family. Plant Mol. Biol. Report. 2014, 32, 828–840. [Google Scholar] [CrossRef]
- Li, X.; Liu, L.; Ren, Q.; Zhang, T.; Hu, N.; Sun, J.; Zhou, W. Analysis of synonymous codon usage bias in the chloroplast genome of five Caragana. BMC Plant Biol. 2025, 25, 322. [Google Scholar] [CrossRef]
- Wang, N.; Chen, S.; Xie, L.; Wang, L.; Feng, Y.; Lv, T.; Fang, Y.; Ding, H. The complete chloroplast genomes of three Hamamelidaceae species: Comparative and phylogenetic analyses. Ecol. Evol. 2022, 12, e8637. [Google Scholar] [CrossRef]
- Yang, Q.; Xin, C.; Xiao, Q.S.; Lin, Y.T.; Li, L.; Zhao, J.L. Codon usage bias in chloroplast genes implicate adaptive evolution of four ginger species. Front. Plant Sci. 2023, 14, 1304264. [Google Scholar] [CrossRef]
- Jia, X.; Wei, J.; Chen, Y.; Zeng, C.; Deng, C.; Zeng, P.; Tang, Y.; Zhou, Q.; Huang, Y.; Zhu, Q. Codon usage patterns and genomic variation analysis of chloroplast genomes provides new insights into the evolution of Aroideae. Sci. Rep. 2025, 15, 4333. [Google Scholar] [CrossRef]
- Huang, R.; Xie, X.; Chen, A.; Li, F.; Tian, E.; Chao, Z. The chloroplast genomes of four Bupleurum (Apiaceae) species endemic to Southwestern China, a diversity center of the genus, as well as their evolutionary implications and phylogenetic inferences. BMC Genom. 2021, 22, 714. [Google Scholar] [CrossRef]
- Jiang, P.; Shi, F.X.; Li, M.R.; Liu, B.; Wen, J.; Xiao, H.X.; Li, L.F. Positive Selection Driving Cytoplasmic Genome Evolution of the Medicinally Important Ginseng Plant Genus Panax. Front. Plant Sci. 2018, 9, 359. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, S.; Wang, J.; Shen, H.; Ai, B.; Gao, W.; Zhang, C.; Fei, Q.; Yuan, D.; Wu, Z.; et al. Chloroplast genomes in Populus (Salicaceae): Comparisons from an intensively sampled genus reveal dynamic patterns of evolution. Sci. Rep. 2021, 11, 9471. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Yamazaki, Y.; Ogihara, Y.; Tsunewaki, K. Whole chloroplast genome comparison of rice, maize, and wheat: Implications for chloroplast gene diversification and phylogeny of cereals. Mol. Biol. Evol. 2002, 19, 2084–2091. [Google Scholar] [CrossRef][Green Version]
- Li, J.; Yang, M.; Li, Y.; Jiang, M.; Liu, C.; He, M.; Wu, B. Chloroplast genomes of two Pueraria DC. species: Sequencing, comparative analysis and molecular marker development. FEBS Open Bio 2022, 12, 349–361. [Google Scholar] [CrossRef]
- Baxter, R.M.; White, V.T.; Zahid, N.D. The modification of the peptidyltransferase activity of 50-S ribosomal subunits, LiCl-split proteins and L16 ribosomal protein by pyridoxal phosphate. Eur. J. Biochem. 1980, 110, 161–166. [Google Scholar] [CrossRef]
- Gietler, M.; Nykiel, M.; Orzechowski, S.; Fettke, J.; Zagdańska, B. Proteomic analysis of S-nitrosylated and S-glutathionylated proteins in wheat seedlings with different dehydration tolerances. Plant Physiol. Biochem. 2016, 108, 507–518. [Google Scholar] [CrossRef]
- Song, W.; Chen, Z.; Shi, W.; Han, W.; Feng, Q.; Shi, C.; Engel, M.S.; Wang, S. Comparative Analysis of Complete Chloroplast Genomes of Nine Species of Litsea (Lauraceae): Hypervariable Regions, Positive Selection, and Phylogenetic Relationships. Genes 2022, 13, 1550. [Google Scholar] [CrossRef]
- Hameed, A.; Ahmed, M.Z.; Hussain, T.; Aziz, I.; Ahmad, N.; Gul, B.; Nielsen, B.L. Effects of Salinity Stress on Chloroplast Structure and Function. Cells 2021, 10, 2023. [Google Scholar] [CrossRef]
- Ding, S.; Zhang, Y.; Hu, Z.; Huang, X.; Zhang, B.; Lu, Q.; Wen, X.; Wang, Y.; Lu, C. mTERF5 Acts as a Transcriptional Pausing Factor to Positively Regulate Transcription of Chloroplast psbEFLJ. Mol. Plant 2019, 12, 1259–1277. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Luo, C. Molecular and Functional Diversity of RNA Editing in Plant Mitochondria. Mol. Biotechnol. 2018, 60, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [PubMed]
- Pace, C.N.; Scholtz, J.M. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998, 75, 422–427. [Google Scholar] [CrossRef]
- Tirosh, I.; Reikhav, S.; Levy, A.A.; Barkai, N. A yeast hybrid provides insight into the evolution of gene expression regulation. Science 2009, 324, 659–662. [Google Scholar] [CrossRef]
- Martín, M.; Sabater, B. Plastid ndh genes in plant evolution. Plant Physiol. Biochem. 2010, 48, 636–645. [Google Scholar] [CrossRef]
- Yamamoto, H.; Peng, L.; Fukao, Y.; Shikanai, T. An Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell 2011, 23, 1480–1493. [Google Scholar] [CrossRef]
- Nakaminami, K.; Matsui, A.; Shinozaki, K.; Seki, M. RNA regulation in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 149–153. [Google Scholar] [CrossRef]
- Kavuri, N.R.; Ramasamy, M.; Qi, Y.; Mandadi, K. Applications of CRISPR/Cas13-Based RNA Editing in Plants. Cells 2022, 11, 2665. [Google Scholar] [CrossRef]
- Zhao, Y.; Gao, R.; Zhao, Z.; Hu, S.; Han, R.; Jeyaraj, A.; Arkorful, E.; Li, X.; Chen, X. Genome-wide identification of RNA editing sites in chloroplast transcripts and multiple organellar RNA editing factors in tea plant (Camellia sinensis L.): Insights into the albinism mechanism of tea leaves. Gene 2023, 848, 146898. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, W.; Wu, Y.; Maliga, P.; Messing, J. RNA Editing in Chloroplasts of Spirodela polyrhiza, an Aquatic Monocotelydonous Species. PLoS ONE 2015, 10, e0140285. [Google Scholar] [CrossRef]
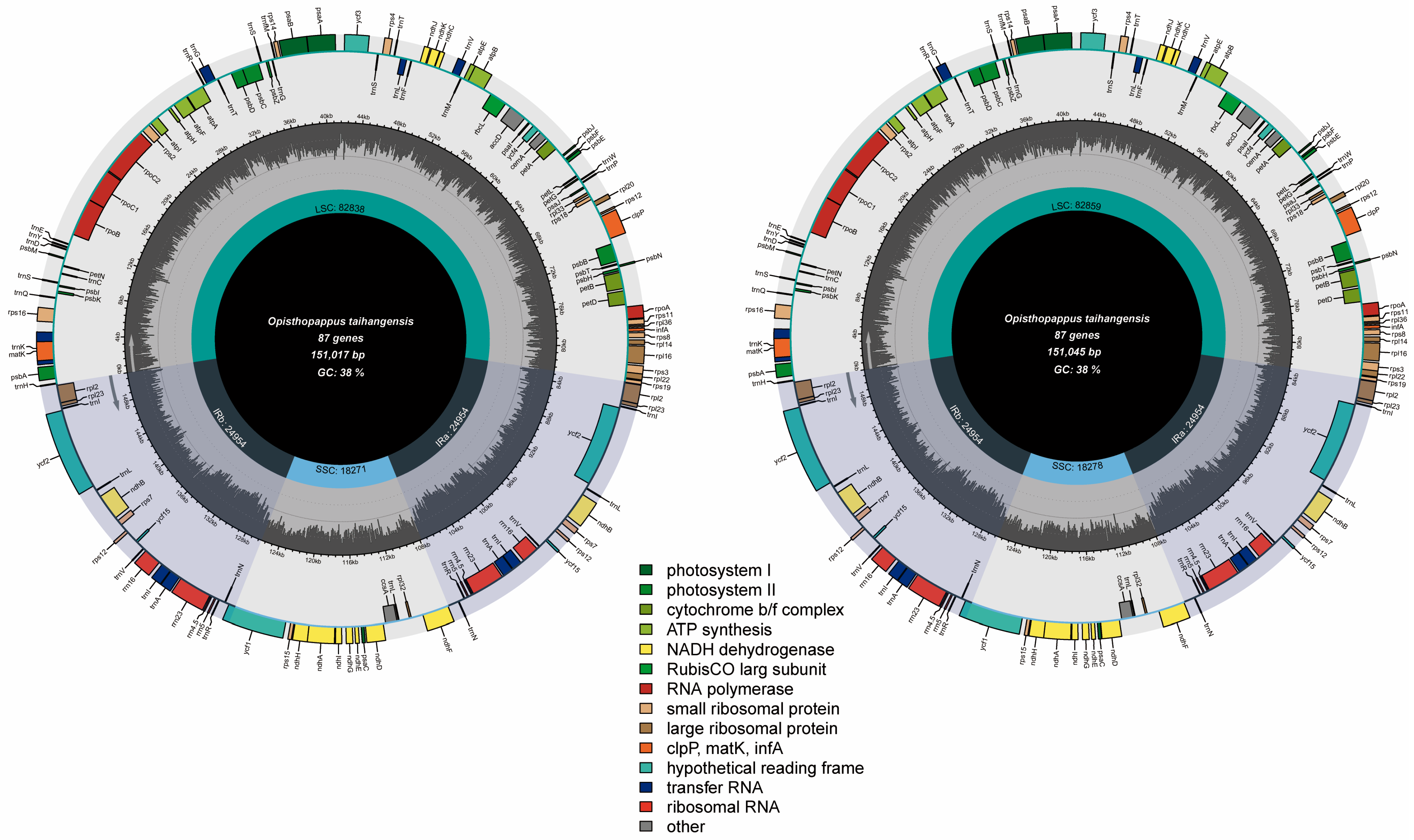
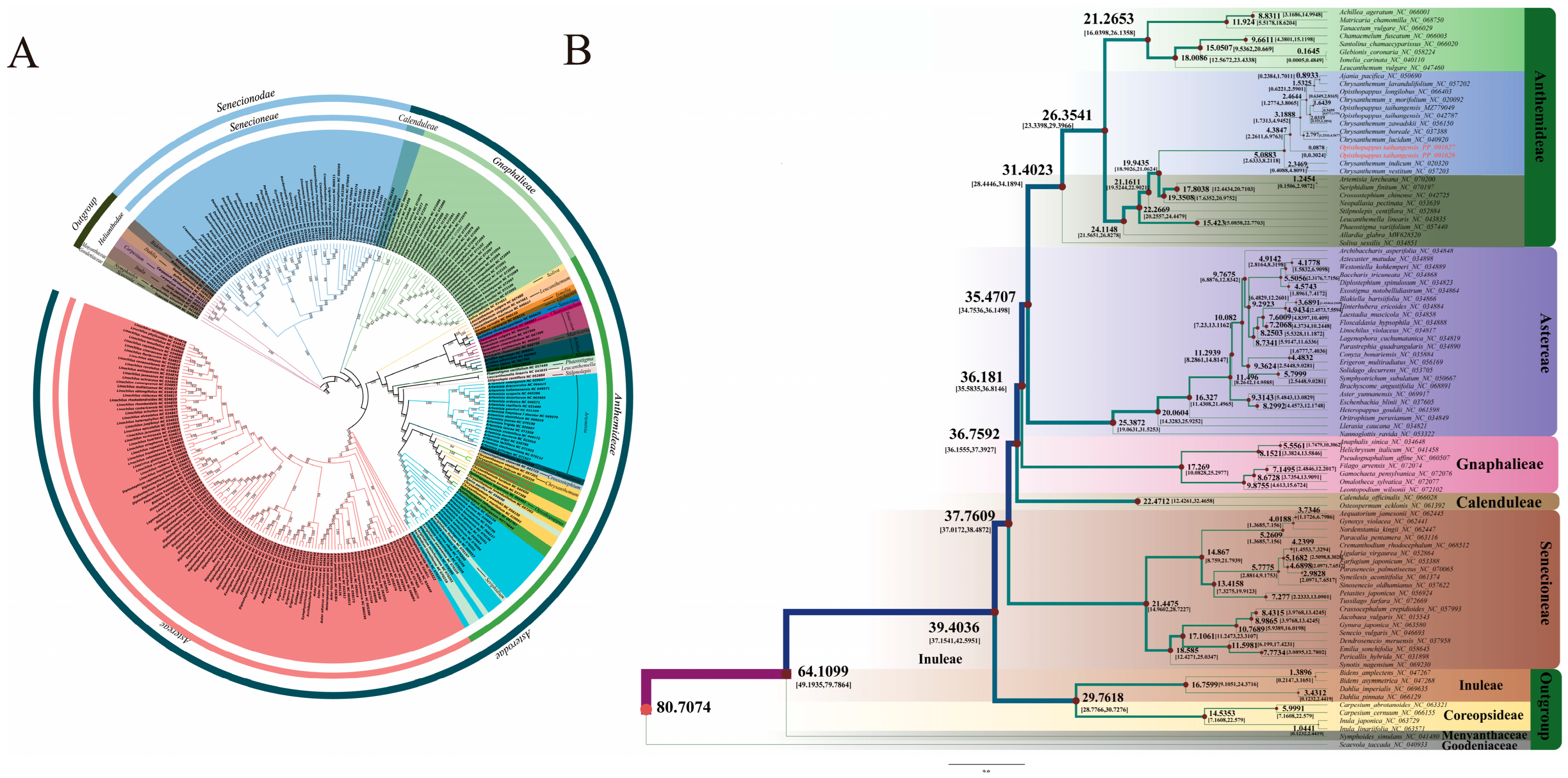
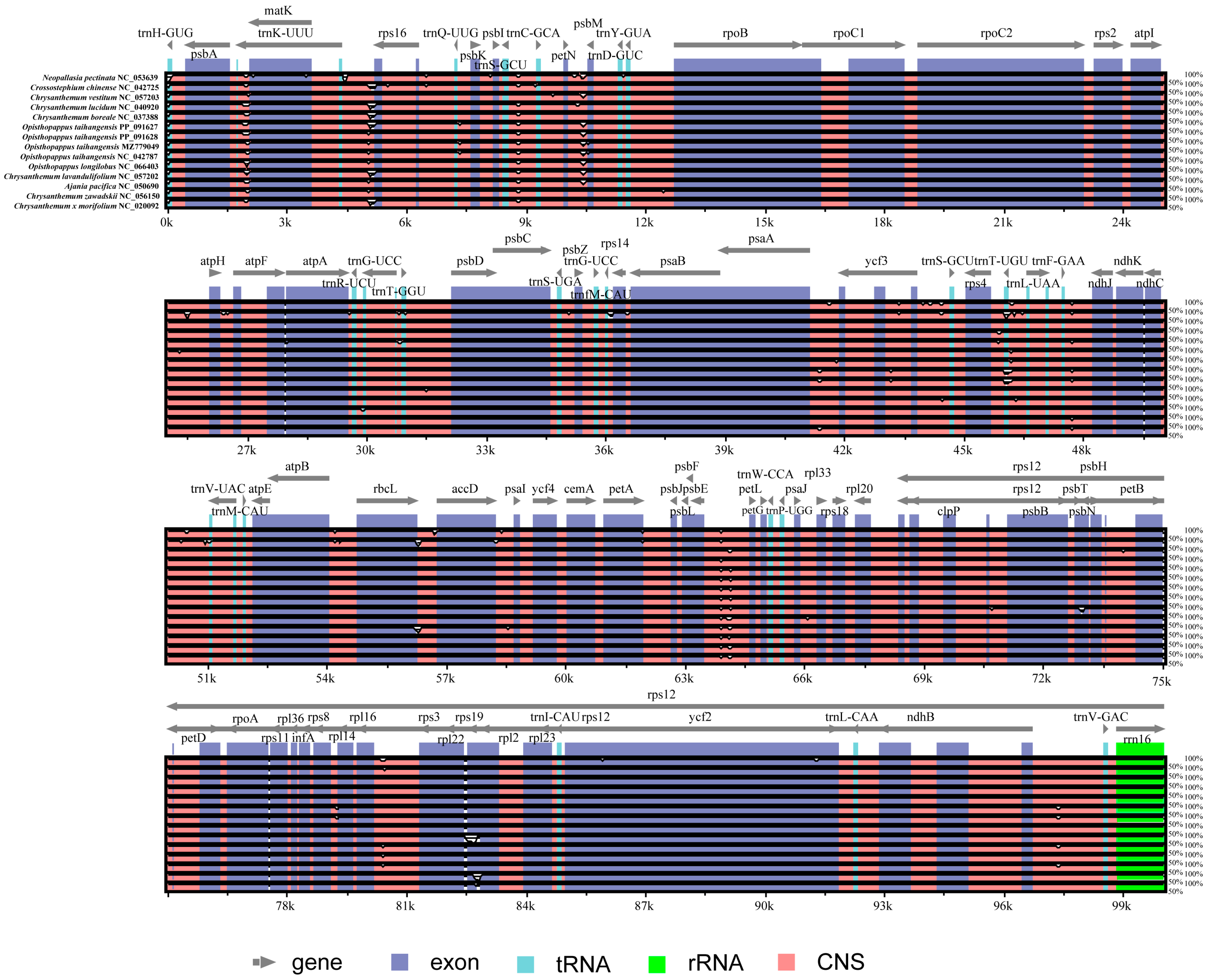

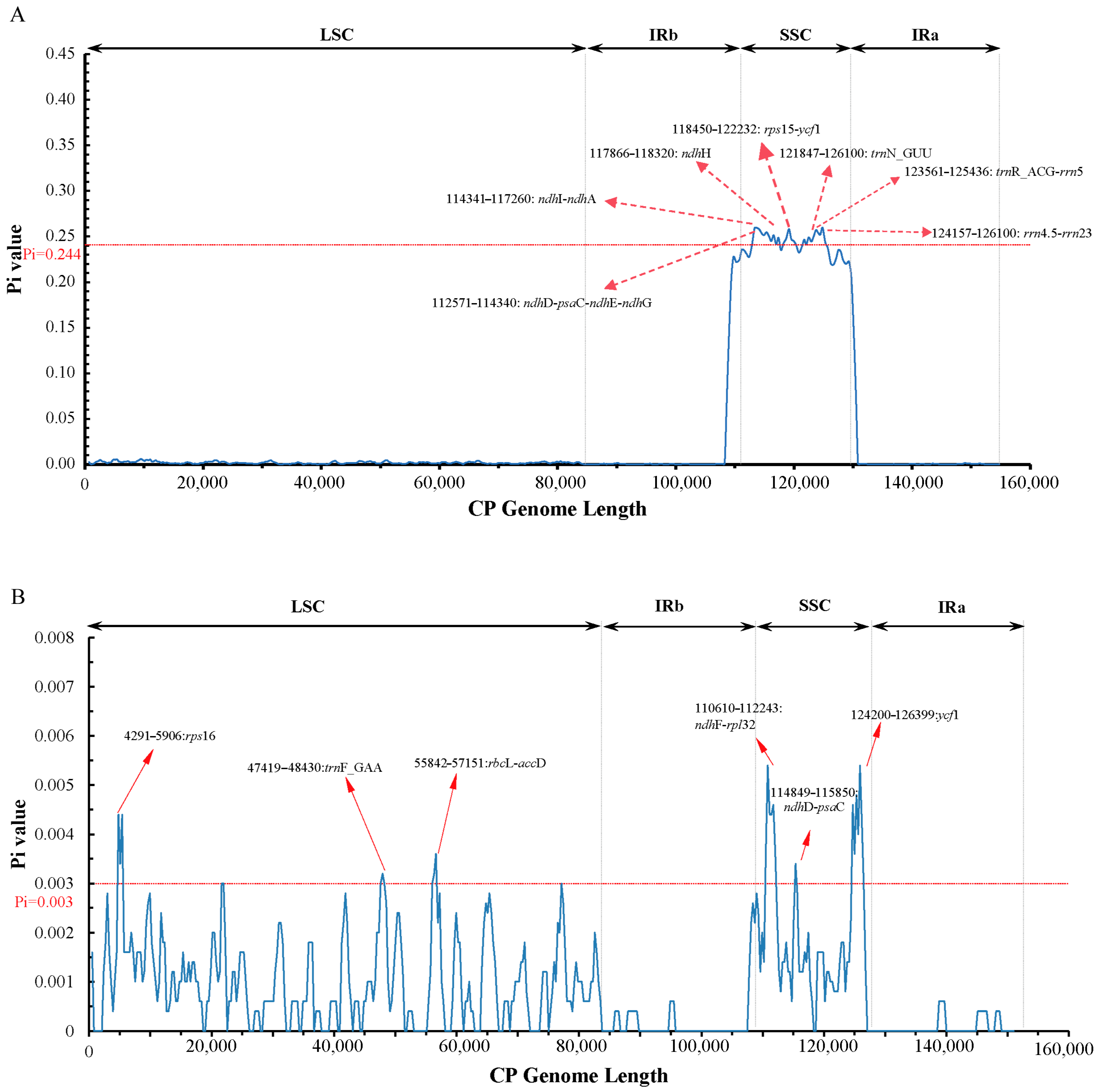

| Gene Category | Gene Group | Gene Name |
|---|---|---|
| rRNA | rRNA genes | rrn16S (×2), rrn23S (×2), rrn5S (×2), rrn4.5S (×2) |
| tRNA | tRNA genes | trnA-UGC (×2), trnC-GCA, trnD-GUC, trnE-UUC (×3), |
| trnF-GAA, trnM-CAU (×4), trnG-GCC, trnH-GUG, trnKUUU, trnL-CAA (×2), trnL-UAA, trnL-UAG, trnM-CAU, | ||
| trnN-GUU (×2), trnP-UGG, trnQ-UUG, trnR-ACG (×2), | ||
| trnR-UCU, trnS-CGA, trnS-GCU, trnS-GGA (×2), trnTGGU, trnT-UGU, trnV-GAC (×2), trnW-CCA, trnY-GUA | ||
| Self-replication | Small subunit of ribosome | rps11, rps12 (×2), rps14, rps15 (×2), rps16, rps18, rps19, rps2, rps3, rps4, rps7 (×2), rps8 |
| Large subunit of ribosome | rpl14, rpl16, rpl2 (×2), rpl20, rpl22, rpl23 (×2), rpl32, rpl33, rpl36 | |
| DNA dependent RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| Photosynthesis | Subunits of NADH-dehydrogenase | ndhA, ndhB (×2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbI, psbJ, psbK, | |
| psbM, psbN, psbT, psbZ, ycf3 | ||
| Subunits of cytochrome b/f complex | petA, petB, petD, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI | |
| Large subunit of rubisco | rbcL | |
| Other genes | Maturase | matK |
| Protease | clpP | |
| Envelope membrane protein | cemA | |
| Subunit of Acetyl-CoA-carboxylase | accD | |
| c-type cytochrome synthesis gene | ccsA | |
| Translational initiation factor | infA | |
| Unknown | Conserved open reading frames | ycf1, ycf15(×2), ycf2 (×2), ycf4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, L.; Ma, B.; Han, M.; Feng, X.; Dan, H.; Wang, T.; Han, J.; Yang, M.; Liu, L.; Sun, G.; et al. Comparative Chloroplast Genomes to Gain Insights into the Phylogenetic Relationships and Evolution of Opisthopappus Species. Horticulturae 2025, 11, 1209. https://doi.org/10.3390/horticulturae11101209
Liang L, Ma B, Han M, Feng X, Dan H, Wang T, Han J, Yang M, Liu L, Sun G, et al. Comparative Chloroplast Genomes to Gain Insights into the Phylogenetic Relationships and Evolution of Opisthopappus Species. Horticulturae. 2025; 11(10):1209. https://doi.org/10.3390/horticulturae11101209
Chicago/Turabian StyleLiang, Liqin, Bingui Ma, Mian Han, Xiaolong Feng, Haoyuan Dan, Tingyu Wang, Jinghui Han, Minghui Yang, Li Liu, Genlou Sun, and et al. 2025. "Comparative Chloroplast Genomes to Gain Insights into the Phylogenetic Relationships and Evolution of Opisthopappus Species" Horticulturae 11, no. 10: 1209. https://doi.org/10.3390/horticulturae11101209
APA StyleLiang, L., Ma, B., Han, M., Feng, X., Dan, H., Wang, T., Han, J., Yang, M., Liu, L., Sun, G., & Wang, Y. (2025). Comparative Chloroplast Genomes to Gain Insights into the Phylogenetic Relationships and Evolution of Opisthopappus Species. Horticulturae, 11(10), 1209. https://doi.org/10.3390/horticulturae11101209







