Tree Species Overcome Edaphic Heterogeneity in Shaping the Urban Orchard Soil Microbiome and Metabolome
Abstract
1. Introduction
2. Materials and Methods
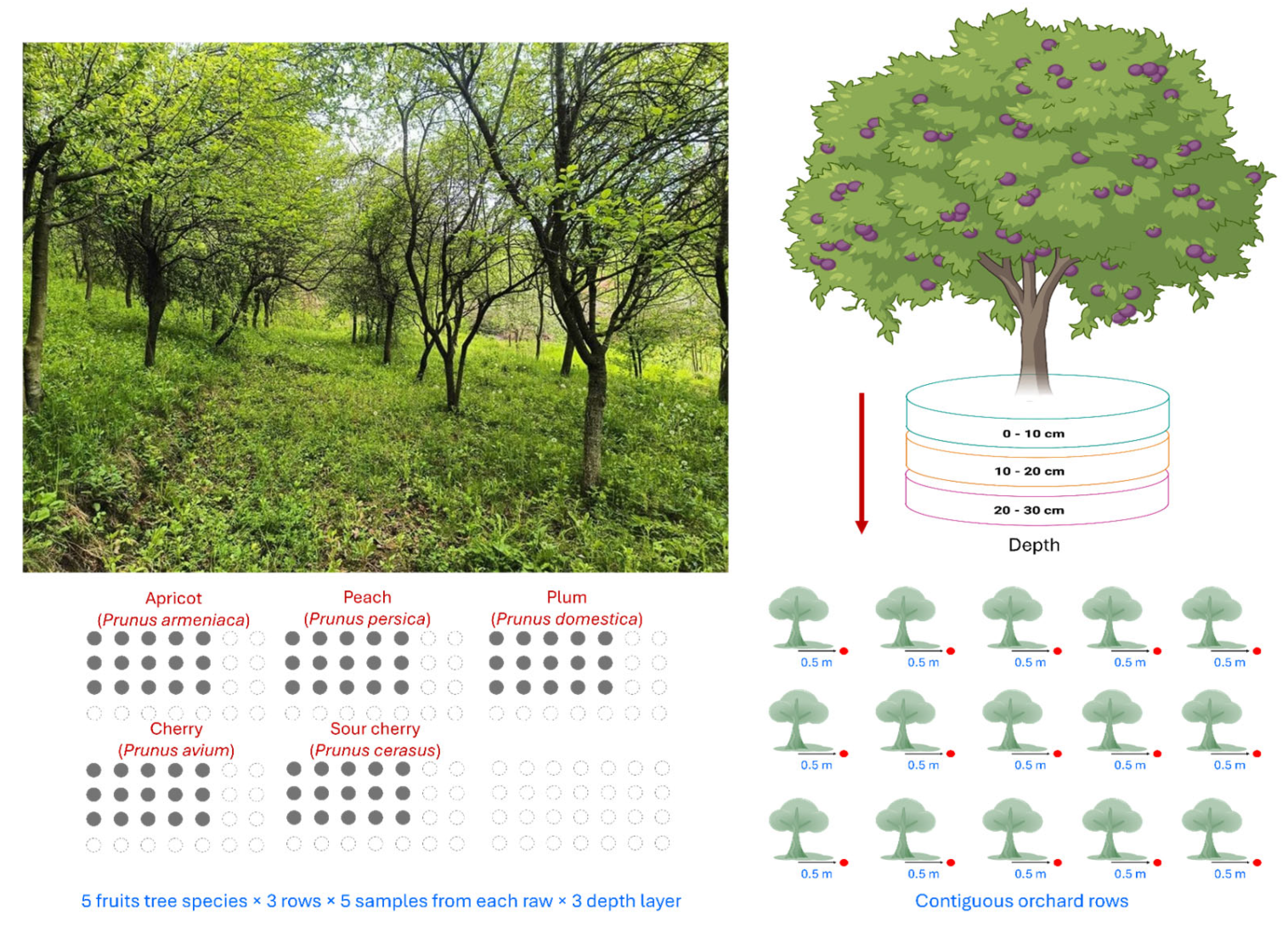
2.1. Sampling Design
2.2. Assessment of Soil Physicochemical Properties
2.3. PLFA Approach
2.4. Mass Spectrometric Assessment of Untargeted Soil Metabolites
2.5. Statistical Analysis
3. Results
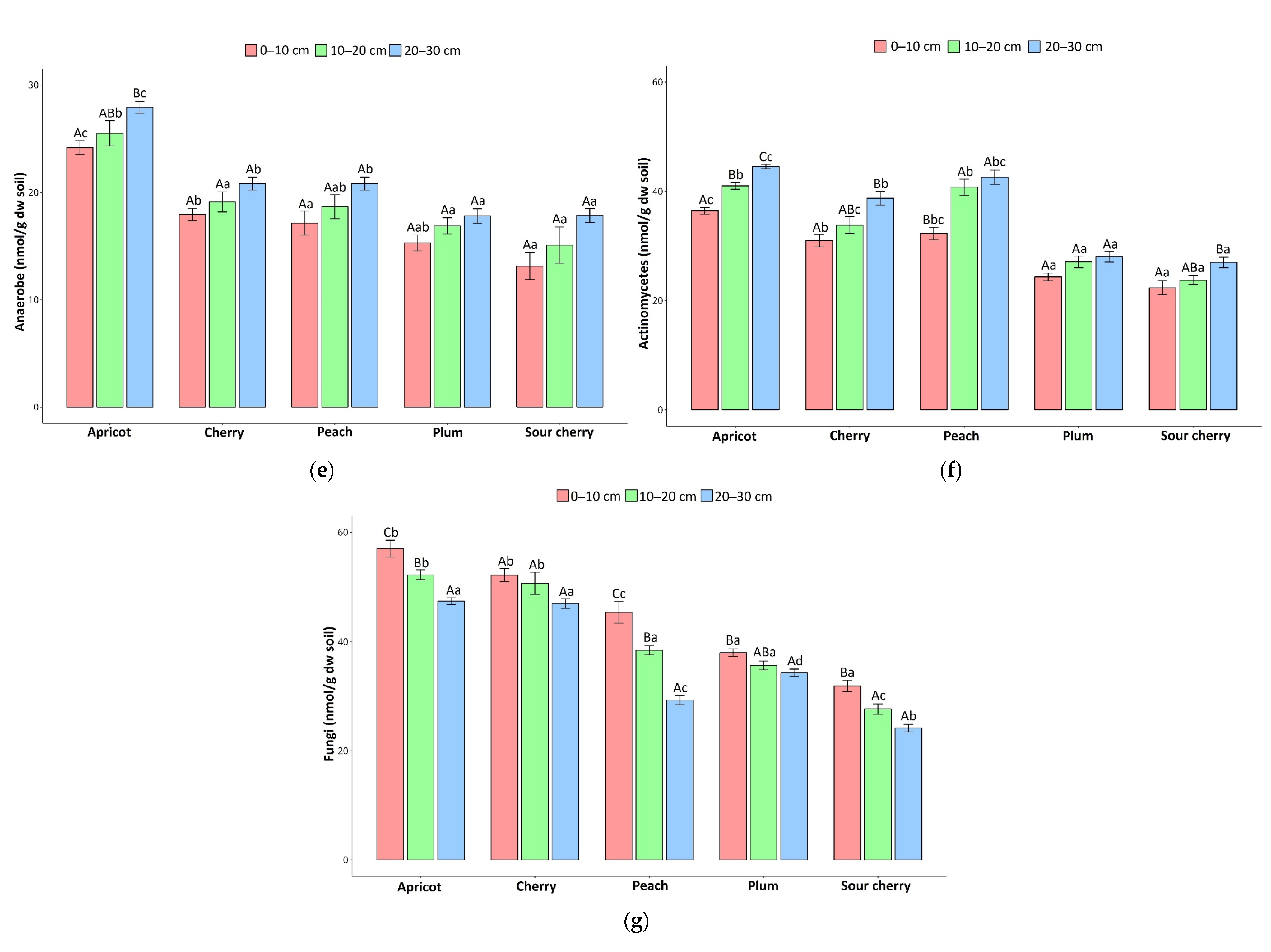
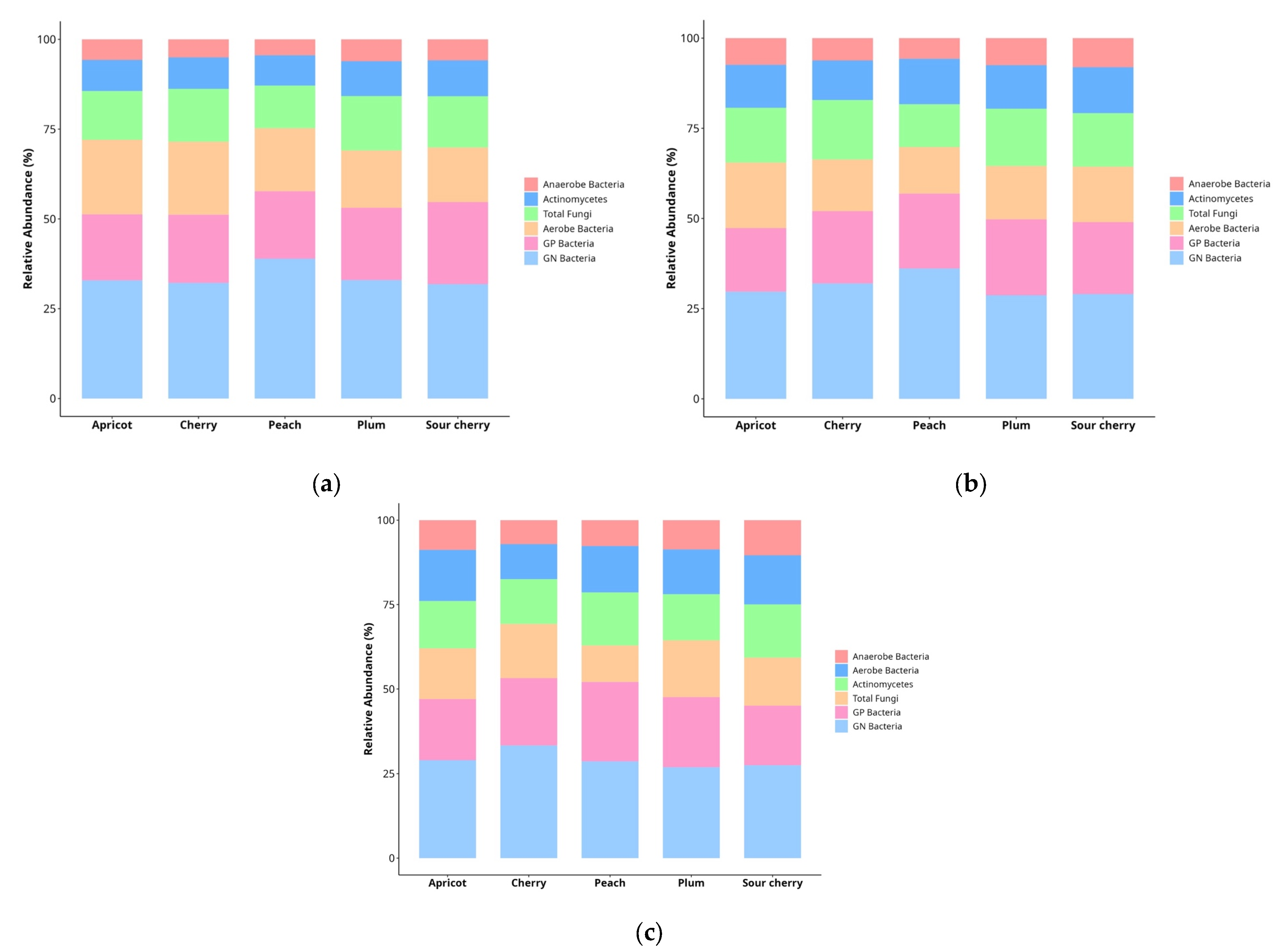
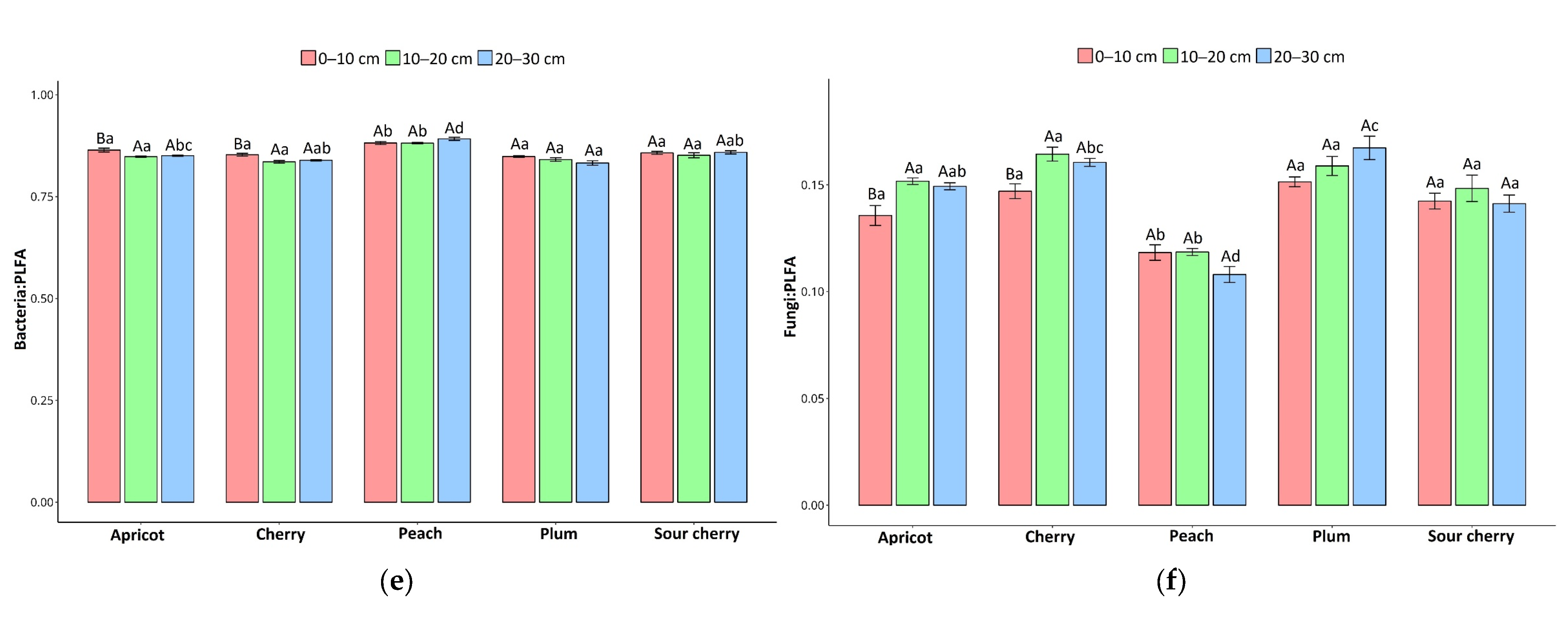
3.1. Soil Microbial Community Composition
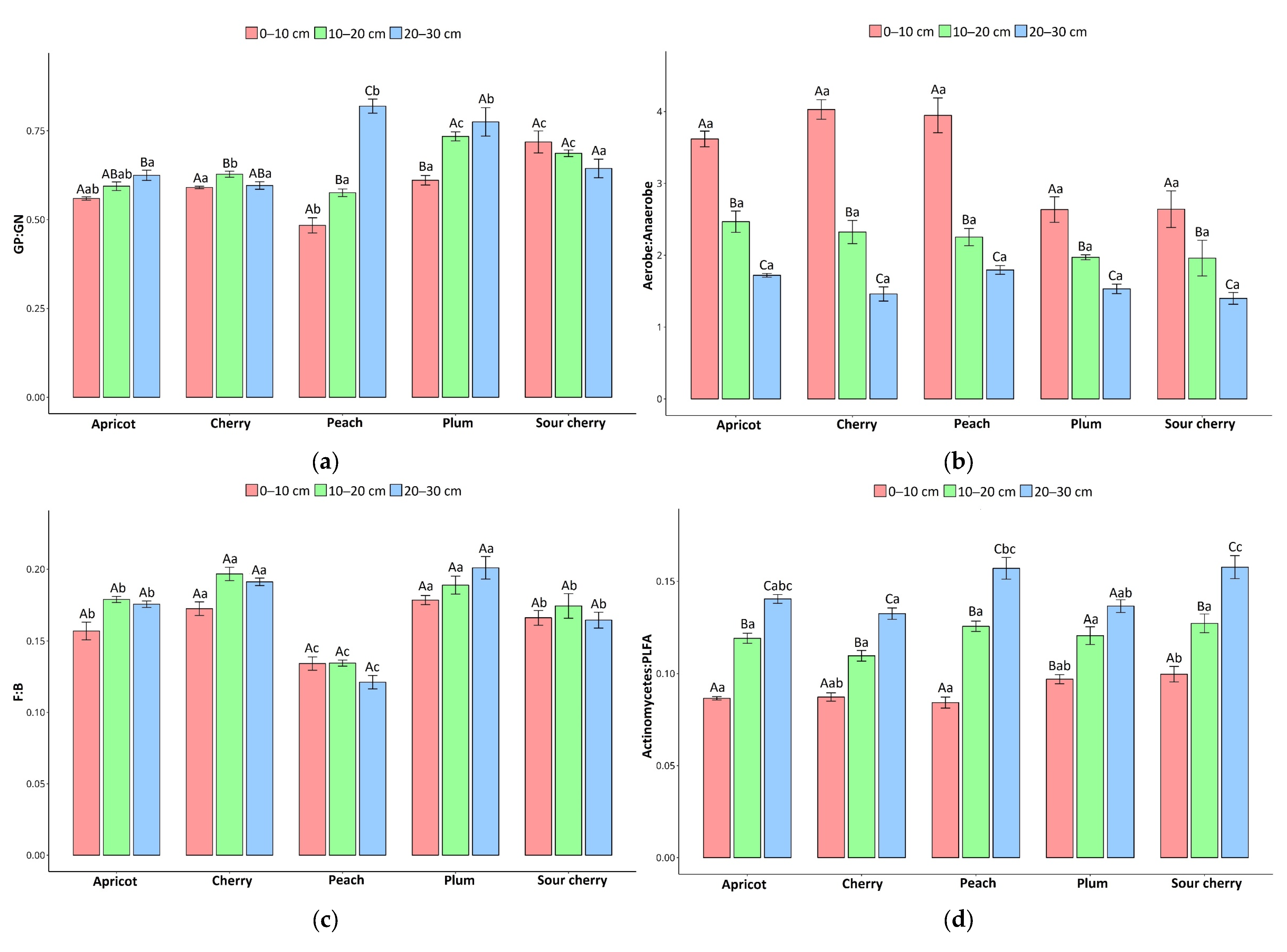
3.2. Correlations Between Soil Microbial Communities and Abiotic Parameters
3.3. Principal Component Analysis of Abiotic/Biotic Parameter Variation in Urban Orchards
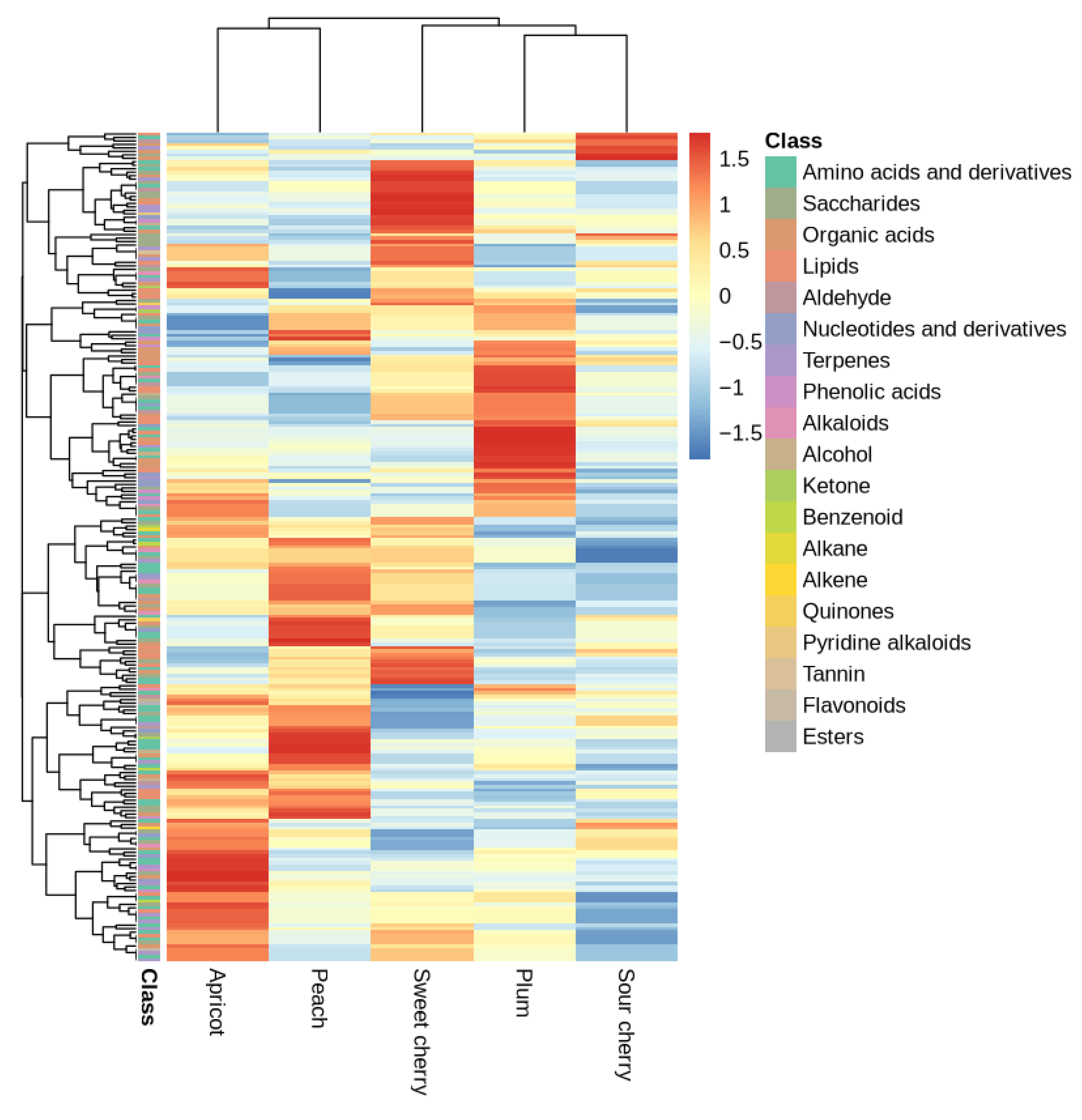
3.4. Soil Metabolites Variation in Urban Orchards
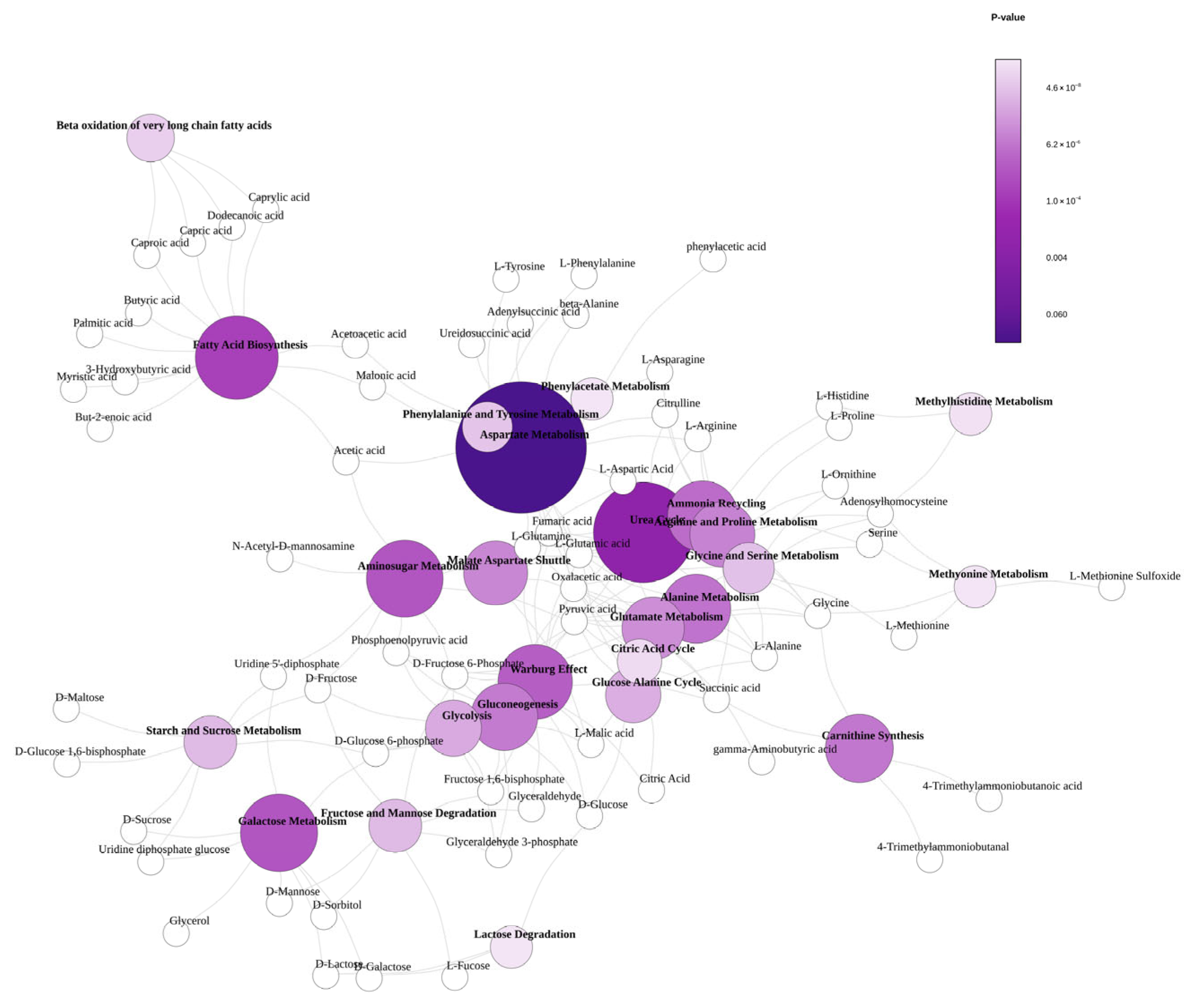
3.5. Metabolic Network Topology and Functional Organization Analysis of Urban Orchard Soils
4. Discussion
4.1. Differences in Soil Microbial Communities Among Orchard Tree Species
4.2. Factors Influencing Microbial Dominance in Urban Orchards
4.3. Urban Orchard Soil Metabolome Profile
4.4. The Ecological Implications
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviation
| Abbreviation | Description |
| PLFA | Phospholipid derived fatty acid |
| GN | Gram-negative bacteria |
| GP | Gram-positive bacteria |
| F:B | Fungal-Bacterial ratio |
| GP:GN | Gram-negative bacteria–Gram-positive bacteria |
| GC-FID | Gas chromatograph with Flame ionization Detector |
| GC-MS/MS | Gas Chromatography-Tandem Mass Spectrometer |
| SSL | Split/Splitless Injector |
| MALDI TOF/TOF IMS | Matrix-Assisted Laser Desorption/Ionization Time-of-Flight/Time-of-Flight Imagistic Mass Spectrometry |
| MSTFA | N-Methyl-N-(trimethylsilyl)trifluoroacetamide |
| TFA | Trifluoroacetic acid |
| AMP | Adenosine Monophosphate |
| ATP | Adenosine Triphosphate |
| CoA | Coenzyme A |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| BD | Bulk density |
| SOC | Soil organic carbon |
| Calcium ion | |
| Chloride ion | |
| Potassium ion | |
| Ammonium ion | |
| Nitrate ion | |
| Magnesium ion | |
| PCA | Principal Component Analysis |
References
- Pascual, U.; Balvanera, P.; Anderson, C.B.; Chaplin-Kramer, R.; Christie, M.; González-Jiménez, D.; Martin, A.; Raymond, C.M.; Termansen, M.; Vatn, A.; et al. Diverse values of nature for sustainability. Nature 2023, 620, 813–823. [Google Scholar] [CrossRef]
- Bull, J.W.; Milner-Gulland, E.J.; Addison, P.F.E.; Arlidge, W.N.S.; Baker, J.; Brooks, T.M.; Burgass, M.J.; Hinsley, A.; Maron, M.; Robinson, J.G.; et al. Net positive outcomes for nature. Nat. Ecol. Evol. 2020, 4, 4–7. [Google Scholar] [CrossRef]
- McPhearson, T.; Cook, E.M.; Berbés-Blázquez, M.; Cheng, C.; Grimm, N.B.; Andersson, E.; Barbosa, O.; Chandler, D.G.; Chang, H.; Chester, M.V.; et al. A social-ecological-technological systems framework for urban ecosystem services. One Earth 2022, 5, 505–518. [Google Scholar] [CrossRef]
- Frac, M.; Hannula, E.S.; Bełka, M.; Salles, J.F.; Jedryczka, M. Soil mycobiome in sustainable agriculture. Front. Microbiol. 2022, 13, 1033824. [Google Scholar] [CrossRef] [PubMed]
- Wankhade, A.; Wilkinson, E.; Britt, D.W.; Kaundal, A. A review of plant–microbe interactions in the rhizosphere and the role of root exudates in microbiome engineering. Appl. Sci. 2025, 15, 7127. [Google Scholar] [CrossRef]
- Marcos-Romero, J.C.; Poveda, J.; Diez, J.J. Relation of the soil microbiota of cork oak groves and surrounding grasslands to tree decline. Appl. Soil Ecol. 2025, 212, 106165. [Google Scholar] [CrossRef]
- Finney, D.M.; Buyer, J.S.; Kaye, J.P. Living cover crops have immediate impacts on soil microbial community structure and function. J. Soil Water Conserv. 2017, 72, 361–373. [Google Scholar] [CrossRef]
- Tosi, M.; Drummelsmith, J.; Obregón, D.; Chahal, I.; Van Eerd, L.L.; Dunfield, K.E. Cover crop-driven shifts in soil microbial communities could modulate early tomato biomass via plant-soil feedbacks. Sci. Rep. 2022, 12, 9140. [Google Scholar] [CrossRef] [PubMed]
- Lasa, A.V.; Lopez-Hinojosa, M.; Villadas, P.J.; Fernandez-Gonzalez, A.J.; Cervera, M.T.; Fernandez-Lopez, M. Unravelling the shifts in the belowground microbiota and metabolome of Pinus pinaster trees affected by forest decline. Sci. Total Environ. 2025, 963, 178486. [Google Scholar] [CrossRef]
- Chauhan, P.; Sharma, N.; Tapwal, A.; Kumar, A.; Verma, G.S.; Meena, M.; Seth, C.S.; Swapnil, P. Soil microbiome: Diversity, benefits and interactions with plants. Sustainability 2023, 15, 14643. [Google Scholar] [CrossRef]
- Dip, D.P.; Sannazzaro, A.I.; Otondo, J.; Pistorio, M.; Estrella, M.J. Exploring Phosphate Solubilizing Bacterial Communities in Rhizospheres of Native and Exotic Forage Grasses in Alkaline-Sodic Soils of the Flooding Pampa. Curr. Microbiol. 2024, 81, 189. [Google Scholar] [CrossRef]
- Jiang, H.; Chen, Y.; Hu, Y.; Wang, Z.; Lu, X. Soil Bacterial Communities and Diversity in Alpine Grasslands on the Tibetan Plateau Based on 16S rRNA Gene Sequencing. Front. Ecol. Evol. 2021, 9, 630722. [Google Scholar] [CrossRef]
- Dasgupta, D.; Brahmaprakash, G.P. Soil Microbes are Shaped by Soil Physico-chemical Properties: A Brief Review of Existing Literature. Int. J. Plant Sci. 2021, 33, 59–71. [Google Scholar] [CrossRef]
- Dukunde, A.; Schneider, D.; Schmidt, M.; Veldkamp, E.; Daniel, R. Tree species shape soil bacterial community structure and function in temperate deciduous forests. Front. Microbiol. 2019, 10, 1519. [Google Scholar] [CrossRef]
- Strukelj, M.; Parker, W.; Corcket, E.; Augusto, L.; Khlifa, R.; Jactel, H.; Munson, A.D. Tree species richness and water availability interact to affect soil microbial processes. Soil Biol. Biochem. 2021, 155, 108180. [Google Scholar] [CrossRef]
- Sridhar, B.; Lawrence, G.B.; Debenport, S.J.; Fahey, T.J.; Buckley, D.H.; Wilhelm, R.C.; Goodale, C.L. Watershed-scale liming reveals the short- and long-term effects of pH on the forest soil microbiome and carbon cycling. Environ. Microbiol. 2022, 24, 6184–6199. [Google Scholar] [CrossRef] [PubMed]
- Wan, W.; Hao, X.; Xing, Y.; Liu, S.; Zhang, X.; Li, X.; Chen, W.; Huang, Q. Spatial differences in soil microbial diversity caused by pH-driven organic phosphorus mineralization. Land Degrad. Dev. 2021, 32, 766–776. [Google Scholar] [CrossRef]
- Luan, L.; Jiang, Y.; Dini-Andreote, F.; Crowther, T.W.; Li, P.; Bahram, M.; Zheng, J.; Xu, Q.; Zhang, X.X.; Sun, B. Integrating pH into the metabolic theory of ecology to predict bacterial diversity in soil. Proc. Natl. Acad. Sci. USA 2023, 120, e2207832120. [Google Scholar] [CrossRef]
- Kaur, M.; Li, J.; Zhang, P.; Yang, H.F.; Wang, L.; Xu, M. Agricultural soil physico-chemical parameters and microbial abundance and diversity under long-run farming practices: A greenhouse study. Front. Ecol. Evol. 2022, 10, 1026771. [Google Scholar] [CrossRef]
- Lori, M.; Kundel, D.; Mader, P.; Singh, A.; Patel, D.; Sisodia, B.S.; Riar, A.; Krause, H.M. Organic farming systems improve soil quality and shape microbial communities across a cotton-based crop rotation in an Indian Vertisol. FEMS Microbiol. Ecol. 2024, 100, fiae127. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Du, Y.; Zhihao, Z.; Islam, W.; Zeng, F. Variation in root-associated microbial communities among three different plant species in natural desert ecosystem. Plants 2024, 13, 2468. [Google Scholar] [CrossRef] [PubMed]
- Nugent, A.; Allison, S.D. A framework for soil microbial ecology in urban ecosystems. Ecosphere 2022, 13, e3968. [Google Scholar] [CrossRef]
- Li, H.; Tang, B.; Lehmann, A.; Rongstock, R.; Zhu, Y.; Rillig, M.C. The dissimilarity between multiple management practices drives the impact on soil properties and functions. Soil Ecol. Lett. 2025, 7, 240278. [Google Scholar] [CrossRef]
- Rezultate Definitive ale Recensamantului Populatiei si Locuintelor-2011-Analiza. Cluj County Regional Statistics Directorate. 2013. Available online: https://www.recensamantromania.ro/rpl-2011/rezultate-2011/ (accessed on 18 February 2025).
- Morar, I.M.; Stefan, R.; Dan, C.; Sestras, R.E.; Truta, P.; Medeleanu, M.; Ranga, F.; Sestras, P.; Truta, A.M.; Sestras, A.F. FT-IR and HPLC analysis of silver fir (Abies alba Mill.) bark compounds from different geographical provenances. Helyon 2024, 10, e26820. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, E.D.; Kovacs, M.H.; Barcelo, D.; Pereira, P. Nonsteroidal anti-inflammatory drugs impact the microbial community in three different soil types—A laboratory experiment. Case Stud. Chem. Environ. Eng. 2024, 10, 100833. [Google Scholar] [CrossRef]
- Kovacs, E.D.; Silaghi-Dumitrescu, L.; Roman, C.; Tian, D. Structural and metabolic profiling of Lycopersicon esculentum rhizosphere microbiota artificially exposed at commonly used non-steroidal anti-inflammatory drugs. Microorganisms 2022, 10, 254. [Google Scholar] [CrossRef]
- Blight, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Lai, Z.; Tsugawa, H.; Wohlgemuth, G.; Mehta, S.; Mueller, M.; Zheng, Y.; Ogiwara, A.; Meissen, J.; Showalter, M.; Takeuchi, K.; et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat. Methods 2018, 15, 53–56. [Google Scholar] [CrossRef]
- Kovacs, E.D.; Rusu, T.; Kovacs, M.H. Sustainable soil volatilome: Discrimination of land uses through GC–MS-identified volatile organic compounds. Separations 2025, 12, 92. [Google Scholar] [CrossRef]
- Baquer, G.; Semente, L.; Rafols, P.; Martin-Saiz, L.; Bookmeyer, C.; Fernandez, J.A.; Correig, X.; Garcia-Altares, M. rMSIfragment: Improving MALDI-MSI lipidomics through automated in-source fragment annotation. J. Cheminform. 2023, 15, 80. [Google Scholar] [CrossRef]
- Peris-Diaz, M.D.; Sweeney, S.R.; Rodak, O.; Sentandreu, E.; Tiziani, S. R-MetaboList 2: A flexible tool for metabolite annotation from high-resolution data-independent acquisition mass spectrometry analysis. Metabolites 2019, 9, 187. [Google Scholar] [CrossRef]
- Luo, X.; Xie, Y.; Yue, S.; Yang, M.; Han, C.; Zhao, Y.; Zhao, Y.; Li, J. Plant species richness enhances aboveground primary productivity via net biodiversity effects and bacterial community interactions. Appl. Soil Ecol. 2025, 209, 106052. [Google Scholar] [CrossRef]
- Sharma, I.; Kashyap, S.; Agarwala, N. Biotic stress-induced changes in root exudation confer plant stress tolerance by altering rhizospheric microbial community. Front. Plant Sci. 2023, 14, 1132824. [Google Scholar] [CrossRef] [PubMed]
- Molefe, R.R.; Amoo, A.E.; Babalola, O.O. Communication between plant roots and the soil microbiome; involvement in plant growth and development. Symbiosis 2023, 90, 231–239. [Google Scholar] [CrossRef]
- Shi, Z.; Fan, C.; Zhao, S.; Xiao, R.; Miao, R.; Yang, Z.; Wan, S. Effect of litter changes on soil microbial community and respiration in a coniferous—Broadleaf mixed forest. Ecosystems 2025, 28, 26. [Google Scholar] [CrossRef]
- Ma, Z.; Wu, J.; Li, L.; Zhou, Q.; Hou, F. Litter-Induced Reduction in Ecosystem Multifunctionality Is Mediated by Plant Diversity and Cover in an Alpine Meadow. Front. Plant Sci. 2021, 12, 773804. [Google Scholar] [CrossRef]
- Prescott, C.E.; Vesterdal, L. Decomposition and transformations along the continuum from litter to soil organic matter in forest soils. For. Ecol. Manag. 2021, 498, 119522. [Google Scholar] [CrossRef]
- Ortuno-Hernandez, G.; Silva, M.; Toledo, R.; Ramos, H.; Reis-Mendes, A.; Ruiz, D.; Martinez-Gomez, P.; Ferreira, I.M.P.L.V.O.; Salazar, J.A. Nutraceutical Profile Characterization in Apricot (Prunus armeniaca L.) Fruits. Plants 2025, 14, 1000. [Google Scholar] [CrossRef]
- Keiluweit, M.; Wanzek, T.; Kleber, M.; Nico, P.; Fendorf, S. Anaerobic microsites have an unaccounted role in soil carbon stabilization. Nat. Commun. 2017, 8, 1771. [Google Scholar] [CrossRef]
- Rumpel, C.; Kögel-Knabner, I. Deep soil organic matter-a key but poorly understood component of terrestrial C cycle. Plant Soil 2011, 338, 143–158. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, C.; Li, X.; Wang, X.; Dai, X.; Xu, Y. Microbial community and metabolic pathways in anaerobic digestion of organic solid wastes: Progress, challenges and prospects. Fermentation 2025, 11, 457. [Google Scholar] [CrossRef]
- Pei, J.; Li, J.; Luo, Y.; Rillig, M.C.; Smith, P.; Gao, W.; Li, B.; Fang, C.; Nie, M. Patterns and drivers of soil microbial carbon use efficiency across soil depths in forest ecosystems. Nat. Commun. 2025, 16, 5218. [Google Scholar] [CrossRef]
- Zhou, W.; Han, G.; Liu, M.; Zeng, J.; Liang, B.; Liu, J.; Qu, R. Determining the distribution and interaction of soil organic carbon, nitrogen, pH and texture in soil profiles: A case study in the Lancangjiang river basin, Southwest China. Forests 2020, 11, 532. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, J.; Wang, L.; Liu, K.; Tian, S.; Zhou, W. Alterations in litter chemical traits and soil environmental properties limit the litter decomposition of near-mature Robinia pseudoacacia plantations. Geoderma 2023, 439, 116668. [Google Scholar] [CrossRef]
- Ma, W.; Tang, S.; Dengzeng, Z.; Zhang, D.; Zhang, T.; Ma, X. Root exudates contribute to belowground ecosystem hotspots: A review. Front. Microbiol. 2022, 13, 937940. [Google Scholar] [CrossRef]
- Naylor, D.; McClure, R.; Jansson, J. Trends in microbial community composition and function by soil depth. Microorganisms 2022, 10, 540. [Google Scholar] [CrossRef]
- D’Entremont, T.W.; Kivlin, S.N. Specificity in plant-mycorrhizal fungal relationships: Prevalence, parameterization, and prospects. Front. Plant Sci. 2023, 14, 1260286. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, M.; Zhao, Y.; Hu, N.; Qin, L.; Ren, Z.; Wang, G.; Jiang, M. Soil microbial abundance was more affected by soil depth than the altitude in peatlands. Front. Microbiol. 2022, 13, 1068540. [Google Scholar] [CrossRef]
- Cui, H.; Mo, C.; Chen, P.; Lan, R.; He, C.; Lin, J.; Jiang, Z.; Yang, J. Impact of rhizosphere priming on soil organic carbon dynamics: Insights from the perspective of carbon fractions. Appl. Soil Ecol. 2023, 189, 104982. [Google Scholar] [CrossRef]
- Das, S.; Beegum, S.; Acharya, B.S.; Panday, D. Soil carbon sequestration: A mechanistic perspective on limitations and future possibilities. Sustainability 2025, 17, 6015. [Google Scholar] [CrossRef]
- Gao, G.; Li, G.; Liu, M.; Liu, J.; Ma, S.; Li, D.; Liang, X.; Wu, M.; Li, Z. Microbial carbon metabolic activity and bacterial cross-profile network in paddy soils of different fertility. Appl. Soil Ecol. 2024, 195, 105233. [Google Scholar] [CrossRef]
- Yang, Y.; Xie, H.; Mao, Z.; Bao, X.; He, H.; Zhang, X.; Liang, C. Fungi determine increased soil organic carbon more than bacteria through their necromass inputs in conservation tillage croplands. Soil Biol. Biochem. 2022, 167, 108587. [Google Scholar] [CrossRef]
- Ramirez, P.B.; Fuentes-Alburquenque, S.; Diez, B.; Vargas, I.; Bonilla, C.A. Soil microbial community responses to labile organic carbon fractions in relation to soil type and land use along a climate gradient. Soil Biol. Biochem. 2020, 141, 107692. [Google Scholar] [CrossRef]
- Sun, H.; Sun, F.; Deng, X.; Storn, N.; Wan, S. Unfolding the roles of particulate- and mineral-associated organic carbon in soil microbial communities. Forests 2025, 16, 27. [Google Scholar] [CrossRef]
- Wang, S.; Yan, X.; Li, T.; Yang, J.; Zhao, L.; Yuan, D.; Guo, Z.; Liu, C.; Duan, C. Cahnges in soil microbe-mediated carbon, nitrogen and phosphorus cycling during spontaneous succession in abandoned Pb–Zn mining areas. Sci. Total Environ. 2024, 920, 171018. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chai, Y.; Xie, H.; Zhang, L.; Zhang, Z.; Yang, X.; Hao, S.; Gai, J.; Chen, Y. Responses of soil microbial diversity, network complexity and multifunctionality to three land-use changes. Sci. Total Environ. 2023, 859, 160255. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Zhang, X.; Chen, W.; Liang, Z.; Wang, K.; Zhang, Y.; Song, Y. Differential responses of bacterial and fungal community structure in soil to nitrogen deposition in two planted forests in Southwest China in relation to pH. Forests 2024, 15, 1112. [Google Scholar] [CrossRef]
- Muneer, M.A.; Hou, W.; Li, J.; Huang, X.; ur Rehman Kayani, M.; Cai, Y.; Yang, W.; Wu, L.; Ji, B.; Zheng, C. Soil pH: A key edaphic factor regulating distribution and functions of bacterial community along vertical soil profiles in red soil of pomelo orchard. BMC Microbiol. 2022, 22, 38. [Google Scholar] [CrossRef]


| Soil Microbiota | pH | BD | Ca2+ | Cl− | K+ | NH4+ | NO3− | Mg2+ | SOC |
|---|---|---|---|---|---|---|---|---|---|
| GP | 0.787 *** | −0.648 ** | −0.229 | −0.218 | −0.19 | −0.272 | 0.046 | 0.266 | 0.759 ** |
| GN | 0.79 *** | −0.785 *** | −0.399 | 0.055 | −0.255 | −0.345 | 0.318 | −0.029 | 0.704 ** |
| Aerobe | 0.87 *** | −0.513 | −0.253 | −0.307 | −0.297 | −0.159 | 0.037 | 0.332 | 0.799 ** |
| Anaerobe | 0.714 ** | −0.513 | −0.358 | −0.205 | −0.522 * | −0.118 | −0.024 | 0.545 | 0.509 |
| Actinomycetes | 0.837 *** | −0.623 * | −0.322 | −0.177 | −0.318 | −0.25 | 0.138 | 0.259 | 0.719 ** |
| Fungi | 0.855 *** | −0.377 | −0.277 | −0.361 | −0.36 | 0.032 | 0.07 | 0.325 | 0.777 *** |
| Total PLFA | 0.857 *** | −0.657 ** | −0.333 | −0.157 | −0.3 | −0.238 | 0.16 | 0.196 | 0.772 *** |
| GP:GN | −0.756 ** | 0.734 ** | 0.618 * | −0.325 | 0.394 | 0.25 | −0.658 ** | 0.383 | −0.534 * |
| Aerobe–Anaerobe | 0.741 ** | −0.346 | −0.064 | −0.27 | 0.082 | −0.129 | 0.141 | 0.001 | 0.848 *** |
| F:B | −0.294 | 0.761 *** | 0.151 | −0.303 | −0.098 | 0.65 ** | −0.173 | 0.143 | −0.308 |
| Actinomycetes–PLFA | −0.698 ** | 0.515 * | 0.281 | 0.085 | 0.174 | 0.087 | −0.195 | 0.039 | −0.691 ** |
| Bacteria–PLFA | 0.293 | −0.763 *** | −0.156 | 0.308 | 0.096 | −0.644 ** | 0.181 | −0.152 | 0.306 |
| Soil Microbiota | pH | BD | Ca2+ | Cl− | K+ | NH4+ | NO3− | Mg2+ | SOC |
|---|---|---|---|---|---|---|---|---|---|
| GP | 0.913 *** | −0.49 | −0.348 | −0.169 | −0.177 | −0.008 | 0.377 | −0.28 | 0.634 * |
| GN | 0.864 *** | −0.529 * | −0.305 | −0.193 | −0.086 | −0.127 | 0.249 | −0.206 | 0.646 ** |
| Aerobe | 0.643 ** | −0.547 * | −0.489 | −0.546 * | −0.241 | −0.109 | −0.047 | 0.333 | 0.498 |
| Anaerobe | 0.555 * | −0.583 * | −0.494 | −0.428 | −0.261 | −0.064 | 0.005 | 0.3 | 0.316 |
| Actinomycetes | 0.826 *** | −0.711 ** | −0.475 | −0.21 | −0.173 | −0.168 | 0.205 | −0.108 | 0.507 |
| Fungi | 0.735 ** | −0.201 | −0.273 | −0.609 * | −0.163 | 0.252 | 0.054 | 0.276 | 0.725 ** |
| Total PLFA | 0.872 *** | −0.552 * | −0.405 | −0.357 | −0.174 | −0.057 | 0.189 | −0.018 | 0.654 ** |
| GP:GN | −0.636 * | 0.479 | 0.106 | 0.345 | −0.187 | 0.318 | 0.125 | −0.088 | −0.644 ** |
| Aerobe–Anaerobe | 0.48 | −0.195 | −0.201 | −0.466 | −0.059 | −0.099 | −0.103 | 0.188 | 0.584 * |
| F:B | −0.175 | 0.59 * | 0.181 | −0.412 | −0.042 | 0.597 * | −0.18 | 0.46 | 0.148 |
| Actinomycetes–PLFA | −0.321 | −0.354 | −0.088 | 0.424 | 0.094 | −0.366 | −0.077 | −0.179 | −0.508 |
| Bacteria–PLFA | 0.185 | −0.59 * | −0.179 | 0.417 | 0.043 | −0.591 * | 0.187 | −0.468 | −0.14 |
| Soil Microbiota | pH | BD | Ca2+ | Cl− | K+ | NH4+ | NO3− | Mg2+ | SOC |
|---|---|---|---|---|---|---|---|---|---|
| GP | 0.75 ** | −0.535 * | −0.241 | −0.195 | −0.109 | −0.276 | 0.226 | −0.333 | 0.585 * |
| GN | 0.935 *** | −0.207 | −0.001 | −0.6 * | 0.038 | −0.037 | −0.054 | 0.112 | 0.82 *** |
| Aerobe | 0.627 * | −0.769 *** | −0.495 | −0.463 | −0.3 | −0.561 * | −0.208 | 0.2 | 0.362 |
| Anaerobe | 0.644 ** | −0.558 * | −0.356 | −0.698 ** | −0.213 | −0.395 | −0.353 | 0.409 | 0.459 |
| Actinomycetes | 0.729 ** | −0.627 * | −0.198 | −0.413 | −0.011 | −0.436 | −0.133 | 0.007 | 0.59 * |
| Fungi | 0.908 *** | 0.019 | −0.181 | −0.742 ** | −0.275 | 0.211 | 0.007 | 0.267 | 0.748 ** |
| Total PLFA | 0.905 *** | −0.434 | −0.213 | −0.566 * | −0.12 | −0.212 | −0.042 | 0.072 | 0.726 ** |
| GP:GN | −0.416 | −0.483 | −0.477 | 0.766 *** | −0.37 | −0.287 | 0.592 * | −0.831 *** | −0.502 |
| Aerobe–Anaerobe | 0.371 | −0.794 *** | −0.507 | 0.134 | −0.315 | −0.583 * | 0.164 | −0.28 | 0.084 |
| F:B | 0.19 | 0.623 * | −0.092 | −0.346 | −0.385 | 0.642 ** | 0.189 | 0.234 | 0.145 |
| Actinomycetes–PLFA | −0.593 * | −0.318 | 0.199 | 0.377 | 0.406 | −0.47 | −0.355 | −0.029 | −0.401 |
| Bacteria–PLFA | −0.192 | −0.626 * | 0.084 | 0.359 | 0.376 | −0.641 ** | −0.17 | −0.255 | −0.15 |
| Metabolites | No. of Comp.(a.) | Apricot | Peach | Plum | Cherry | Sour Cherry | Significance |
|---|---|---|---|---|---|---|---|
| Organic acids | 37 | 14.31 ± 0.92 | 16.23 ± 0.23 | 13.29 ± 0.39 | 15.50 ± 0.71 | 23.46 ± 2.88 | *** |
| Nucleotides and derivatives | 15 | 5.51 ± 0.35 | 3.86 ± 0.16 | 4.35 ± 0.40 | 2.68 ± 0.46 | 4.03 ± 0.28 | *** |
| Saccharides | 39 | 10.06 ± 0.16 | 10.19 ± 1.45 | 7.99 ± 0.25 | 12.12 ± 1.64 | 9.25 ± 1.01 | * |
| Amino acids and derivatives | 59 | 31.40 ± 0.77 | 32.41 ± 1.64 | 29.38 ± 0.86 | 25.13 ± 1.36 | 24.54 ± 2.04 | *** |
| Lipids | 26 | 10.88 ± 0.34 | 11.66 ± 1.6 | 19.45 ± 1.67 | 15.67 ± 0.69 | 14.55 ± 1.07 | *** |
| Terpenes | 21 | 12.69 ± 0.59 | 9.62 ± 0.63 | 11.57 ± 1.65 | 14.12 ± 1.15 | 11.02 ± 0.22 | ** |
| Phenolic acids | 9 | 3.28 ± 0.28 | 4.17 ± 0.26 | 2.97 ± 0.21 | 1.58 ± 0.04 | 1.99 ± 0.15 | *** |
| Ketone | 4 | 1.33 ± 0.11 | 1.36 ± 0.08 | 1.05 ± 0.07 | 0.98 ± 0.08 | 1.07 ± 0.13 | ** |
| Alkaloids | 8 | 3.05 ± 0.21 | 3.14 ± 0.28 | 2.87 ± 0.36 | 2.49 ± 0.28 | 2.44 ± 0.20 | * |
| Aldehyde | 9 | 1.91 ± 0.08 | 2.49 ± 0.20 | 2.33 ± 0.39 | 2.26 ± 0.32 | 2.70 ± 0.55 | ns |
| Alkene | 2 | 0.73 ± 0.04 | 0.33 ± 0.21 | 0.14 ± 0.10 | 0.40 ± 0.07 | 0.74 ± 0.11 | *** |
| Alcohol | 6 | 0.93 ± 0.05 | 1.03 ± 0.20 | 1.01 ± 0.13 | 0.49 ± 0.03 | 0.74 ± 0.21 | ** |
| Flavonoids | 1 | 0.88 ± 0.11 | 0.48 ± 0.07 | 0.62 ± 0.05 | 0.93 ± 0.08 | 0.47 ± 0.07 | *** |
| Esters | 1 | 1.16 ± 0.15 | 0.80 ± 0.07 | 0.53 ± 0.04 | 0.23 ± 0.10 | 0.89 ± 0.17 | *** |
| Benzenoid | 3 | 1.18 ± 0.21 | 1.20 ± 0.09 | 0.97 ± 0.04 | 0.73 ± 0.03 | 0.65 ± 0.08 | *** |
| Quinones | 5 | 0.17 ± 0.04 | 0.40 ± 0.03 | 0.46 ± 0.05 | 0.60 ± 0.10 | 0.27 ± 0.06 | *** |
| Tannin | 1 | 0.22 ± 0.03 | 0.15 ± 0.04 | 0.08 ± 0.01 | 0.24 ± 0.03 | 0.10 ± 0.02 | *** |
| Pyridine alkaloids | 1 | 0.32 ± 0.04 | 0.49 ± 0.06 | 0.95 ± 0.1 | 3.84 ± 0.21 | 1.07 ± 0.18 | *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kovacs, E.D.; Kovacs, M.H. Tree Species Overcome Edaphic Heterogeneity in Shaping the Urban Orchard Soil Microbiome and Metabolome. Horticulturae 2025, 11, 1163. https://doi.org/10.3390/horticulturae11101163
Kovacs ED, Kovacs MH. Tree Species Overcome Edaphic Heterogeneity in Shaping the Urban Orchard Soil Microbiome and Metabolome. Horticulturae. 2025; 11(10):1163. https://doi.org/10.3390/horticulturae11101163
Chicago/Turabian StyleKovacs, Emoke Dalma, and Melinda Haydee Kovacs. 2025. "Tree Species Overcome Edaphic Heterogeneity in Shaping the Urban Orchard Soil Microbiome and Metabolome" Horticulturae 11, no. 10: 1163. https://doi.org/10.3390/horticulturae11101163
APA StyleKovacs, E. D., & Kovacs, M. H. (2025). Tree Species Overcome Edaphic Heterogeneity in Shaping the Urban Orchard Soil Microbiome and Metabolome. Horticulturae, 11(10), 1163. https://doi.org/10.3390/horticulturae11101163







