Transcriptome Analysis Identified PyNAC42 as a Positive Regulator of Anthocyanin Biosynthesis Induced by Nitrogen Deficiency in Pear (Pyrus spp.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Media Components
2.3. Total Anthocyanin Measurement
2.4. Genomic DNA and Total RNA Extraction and cDNA Synthesis
2.5. Library Construction and RNA Sequencing
2.6. Analysis of Transcriptome Data
2.7. Real-Time Quantitative PCR Assay
2.8. Phylogenetic Analysis and Sequence Alignment
2.9. Subcellular Localization Analysis
2.10. Transient Transformation Assay of “Zaosu” Pear Peels
2.11. Dual Luciferase and Yeast One-Hybrid (Y1H) Assays
2.12. Yeast Two-Hybrid (Y2H) Assay
2.13. Pull-Down Assay
2.14. Statistical Analysis
3. Results
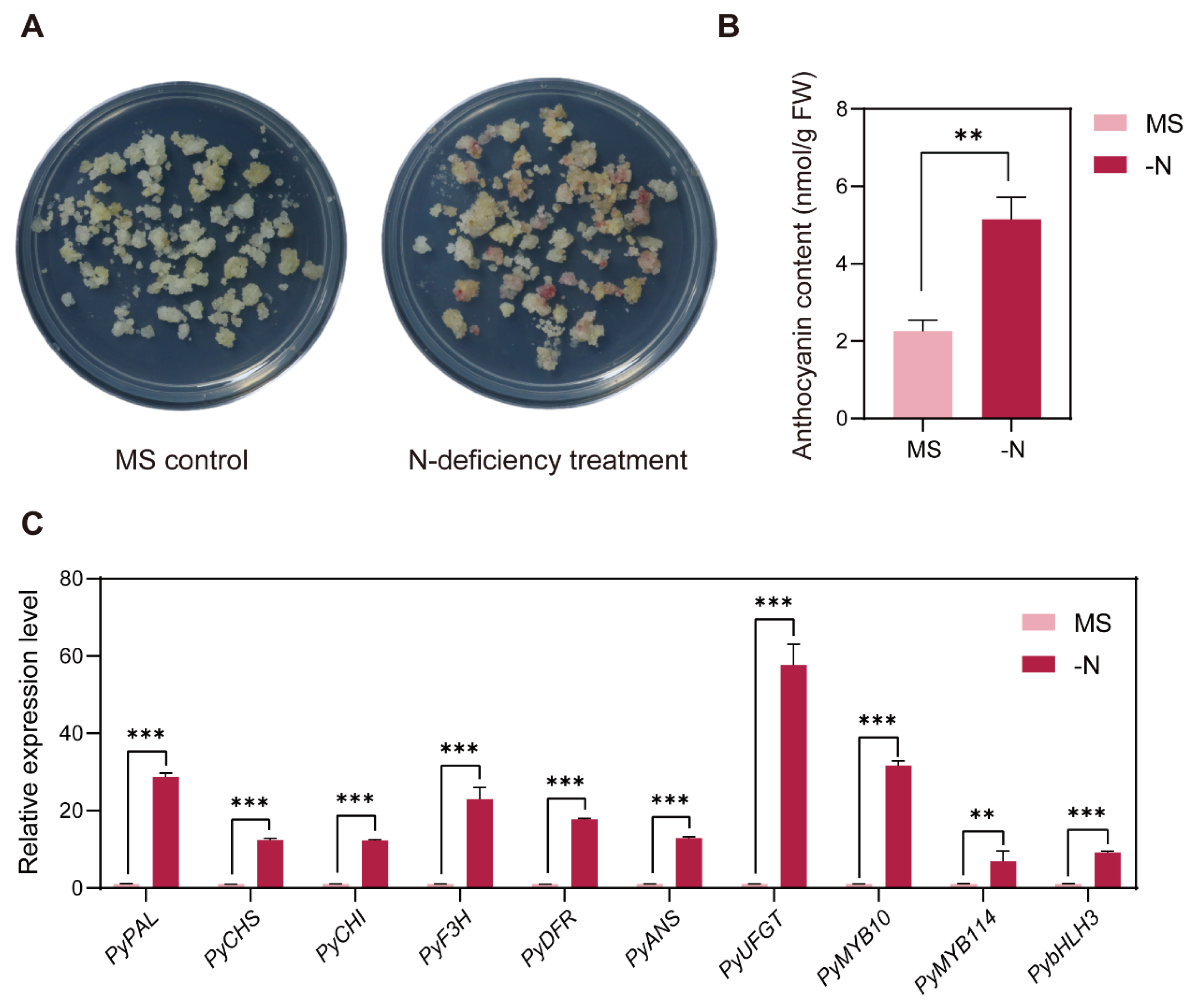
3.1. N-Deficiency Treatment Induced Anthocyanin Accumulation in Pear Callus
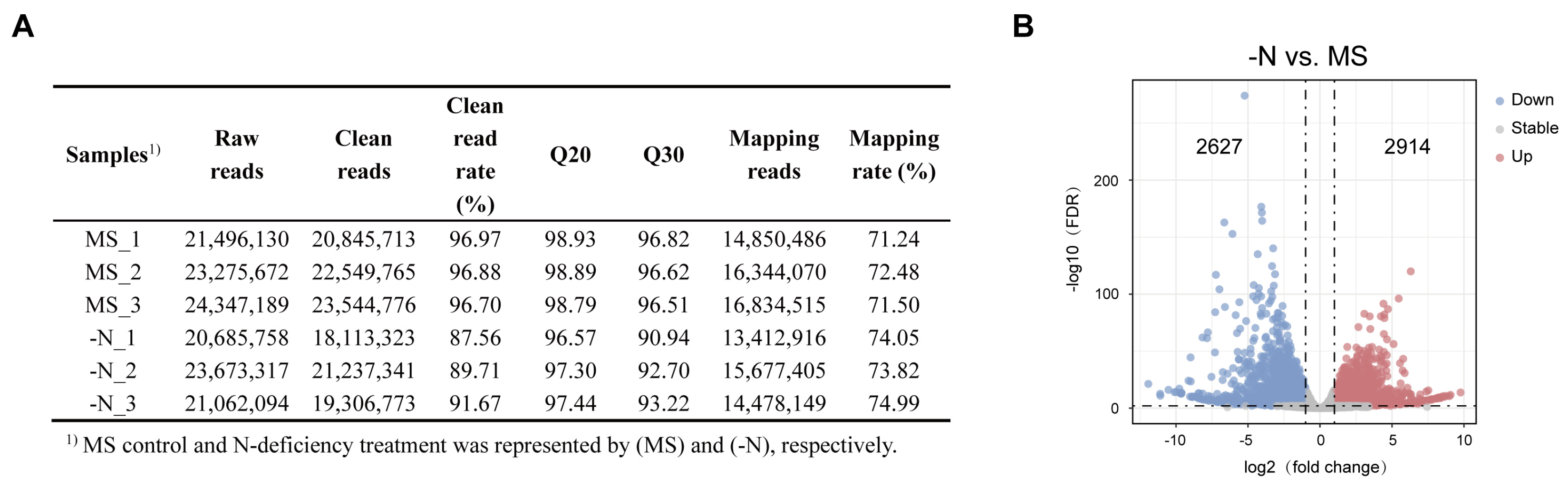
3.2. Analysis of RNA-seq Data and Identification of DEGs
3.3. Functional Enrichment Analysis of DEGs
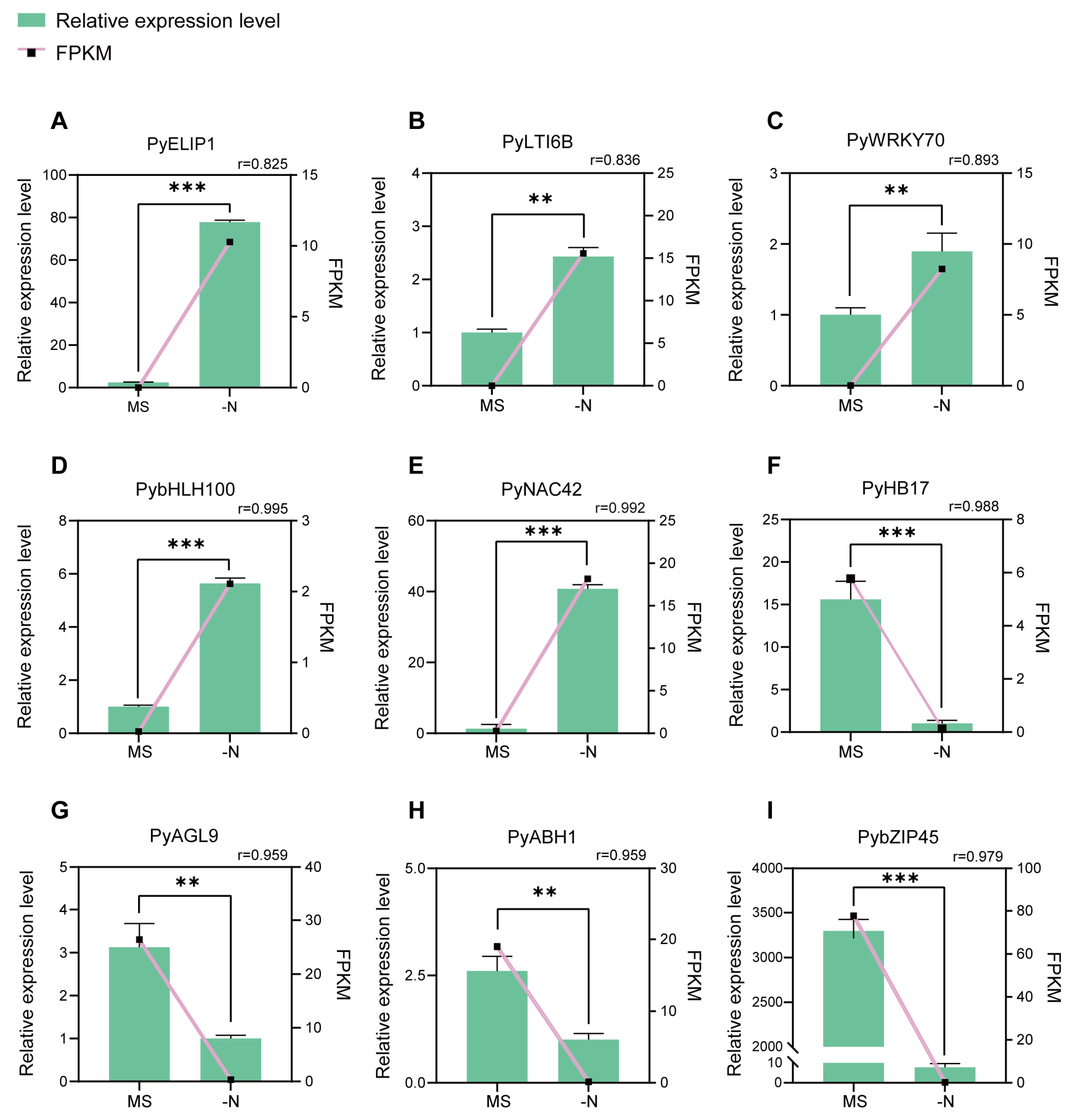
3.4. Validation of RNA-seq Data and Screening of Candidate DEGs
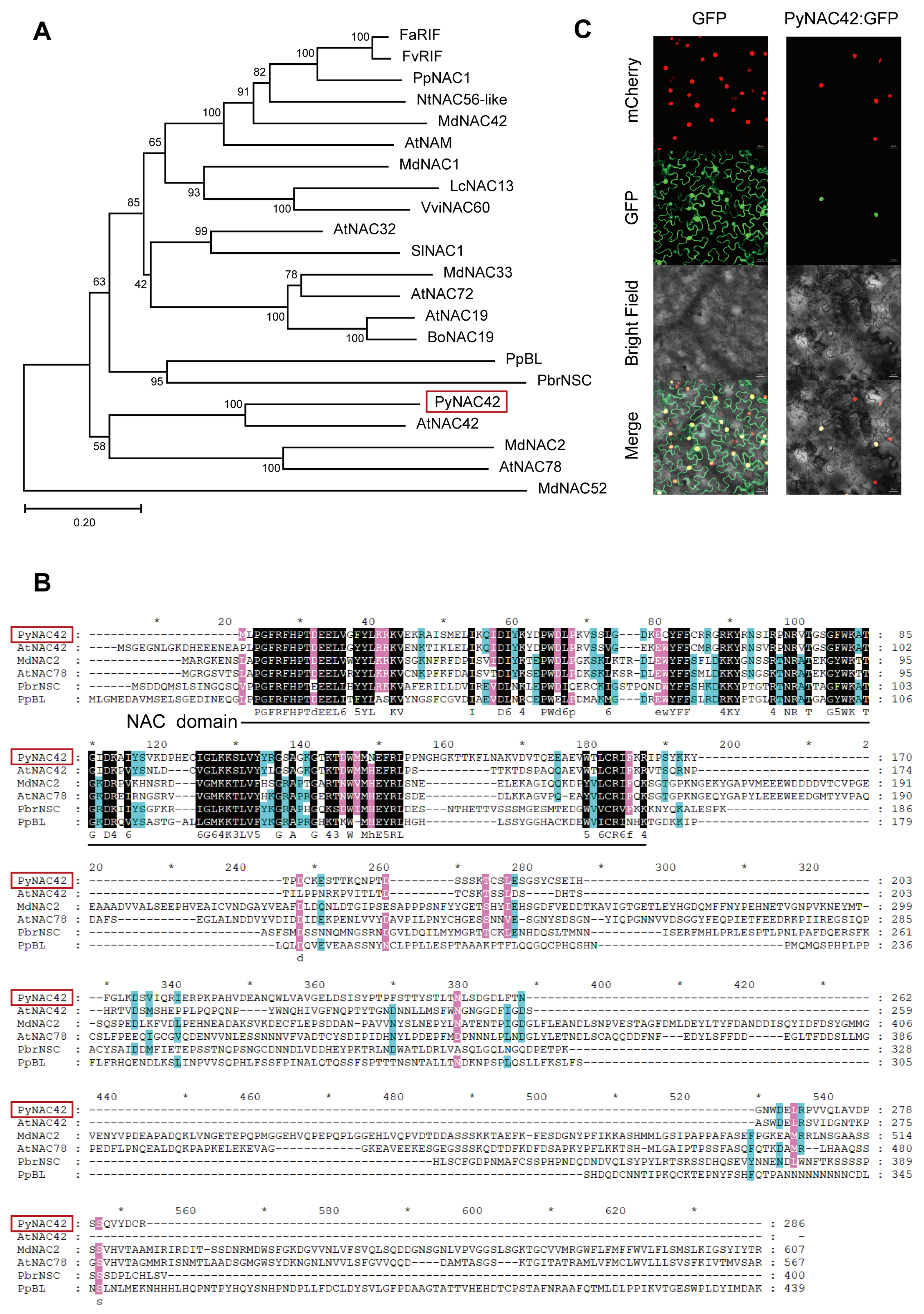
3.5. Phylogenetic Analysis and Subcellular Localization of PyNAC42
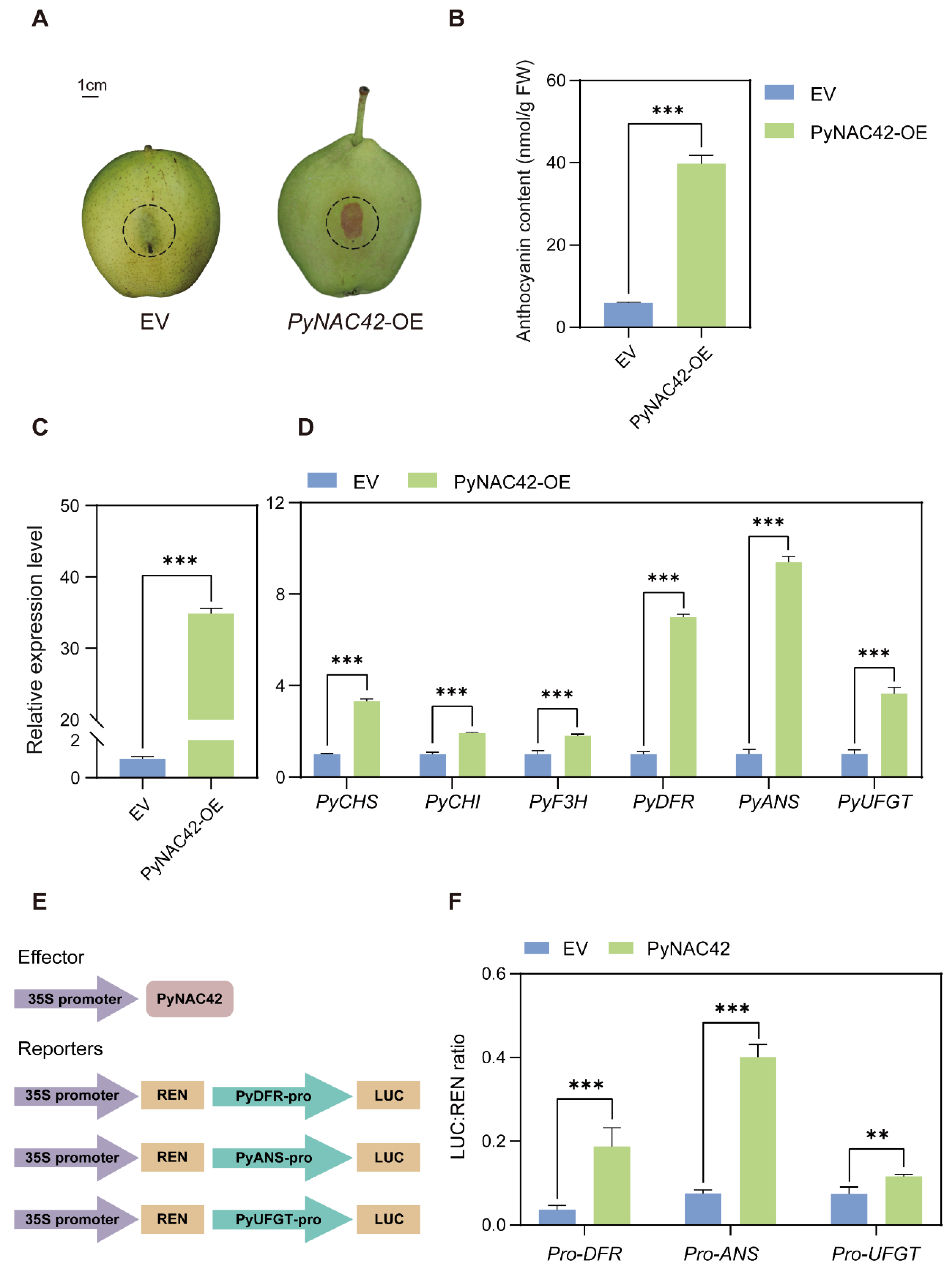
3.6. Overexpression of PyNAC42 Induced Anthocyanin Accumulation in “Zaosu” Pear Peels
3.7. PyNAC42 Activated the Expression of PyDFR, PyANS and PyUFGT
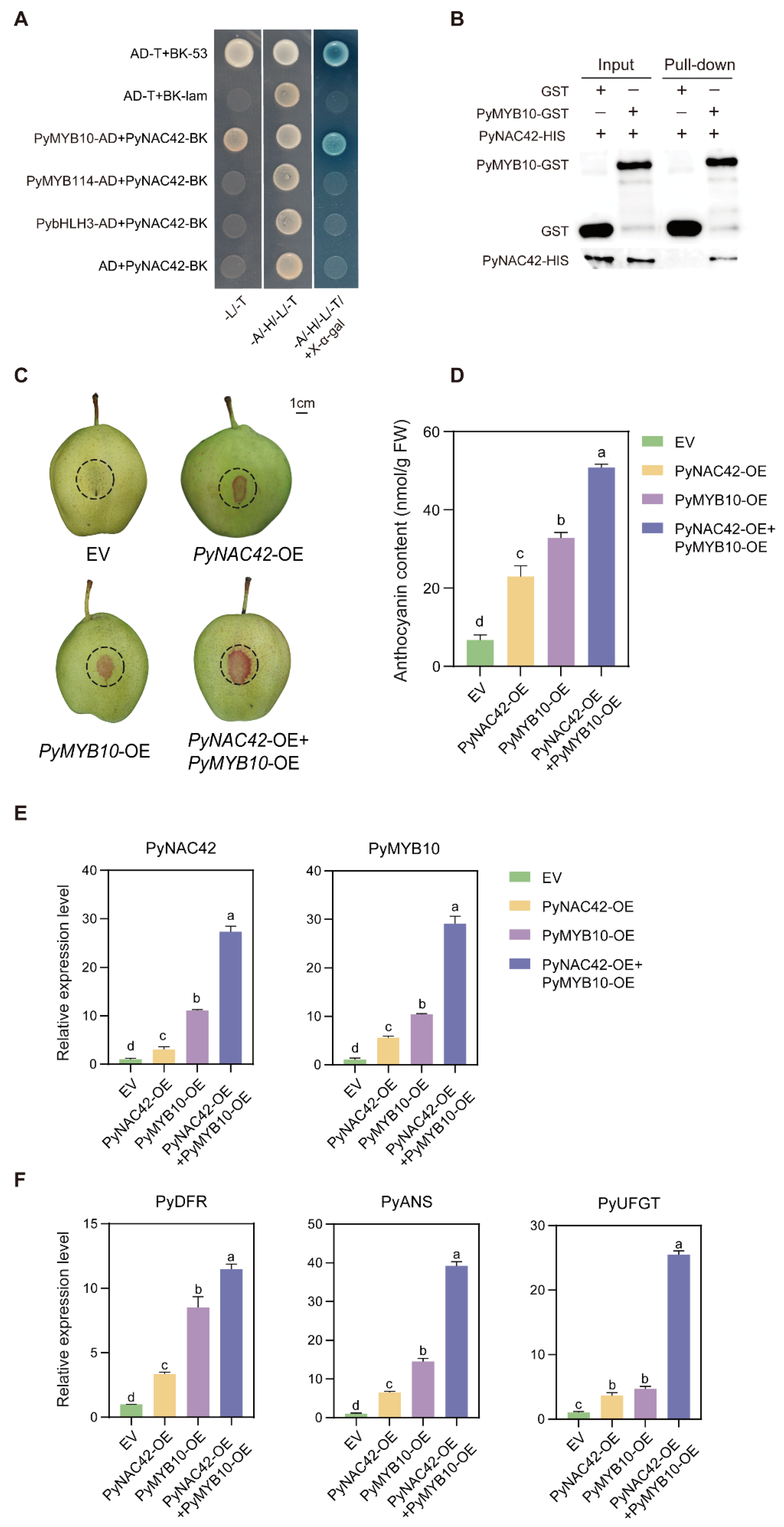
3.8. PyNAC42 Interacts with PyMYB10
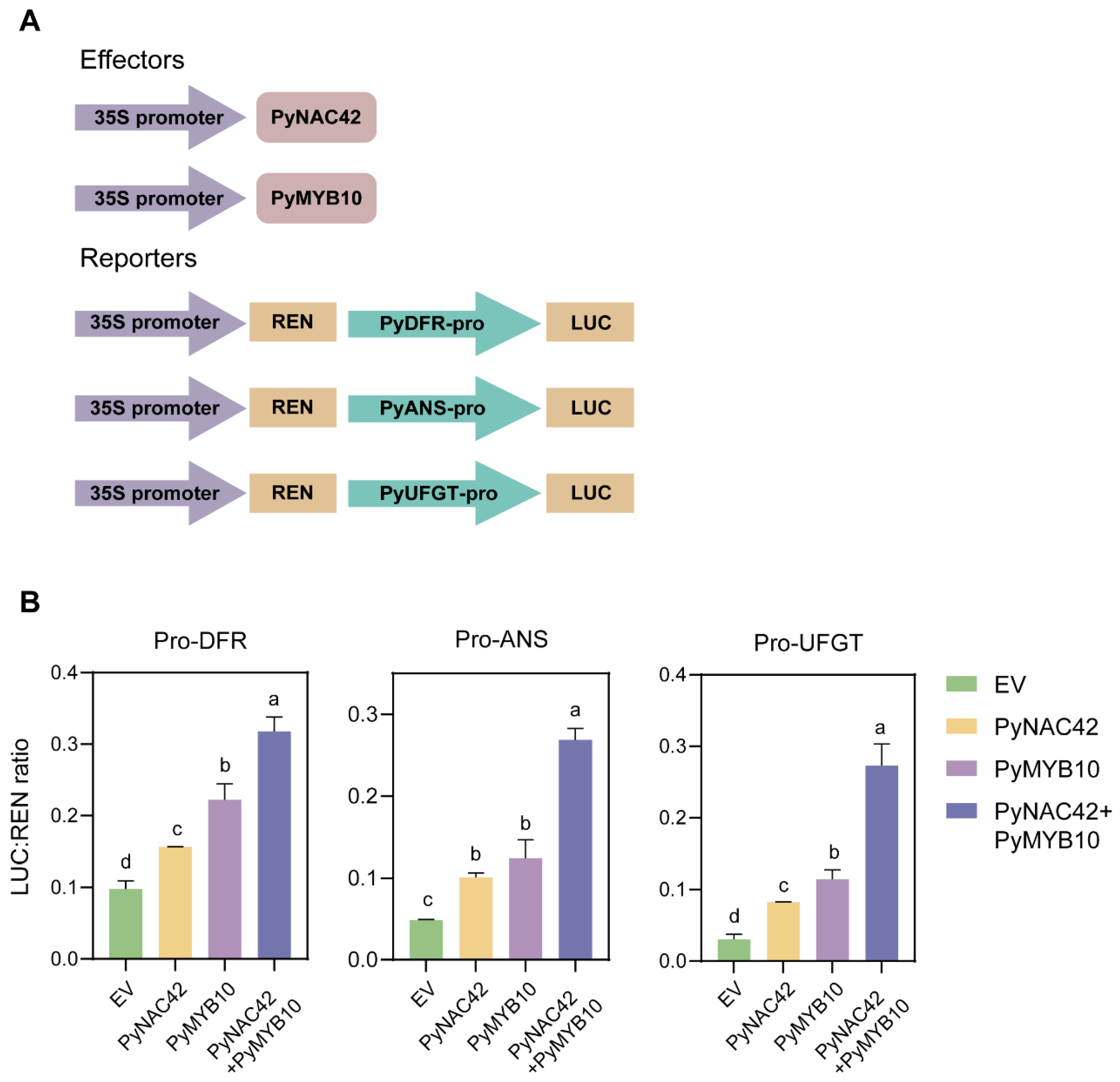
3.9. PyNAC42 Enhanced the Activation of PyMYB10 to PyDFR, PyANS and PyUFGT
4. Discussion
4.1. N Deficiency Promoted Anthocyanin Accumulation in Pear Callus
4.2. Effects of N Deficiency on Anthocyanin Biosynthesis and Other Metabolisms Based on the Analysis of Transcriptome Data
4.3. Regulation of Nutrient Limitation-Mediated Anthocyanin Biosynthesis in Plants
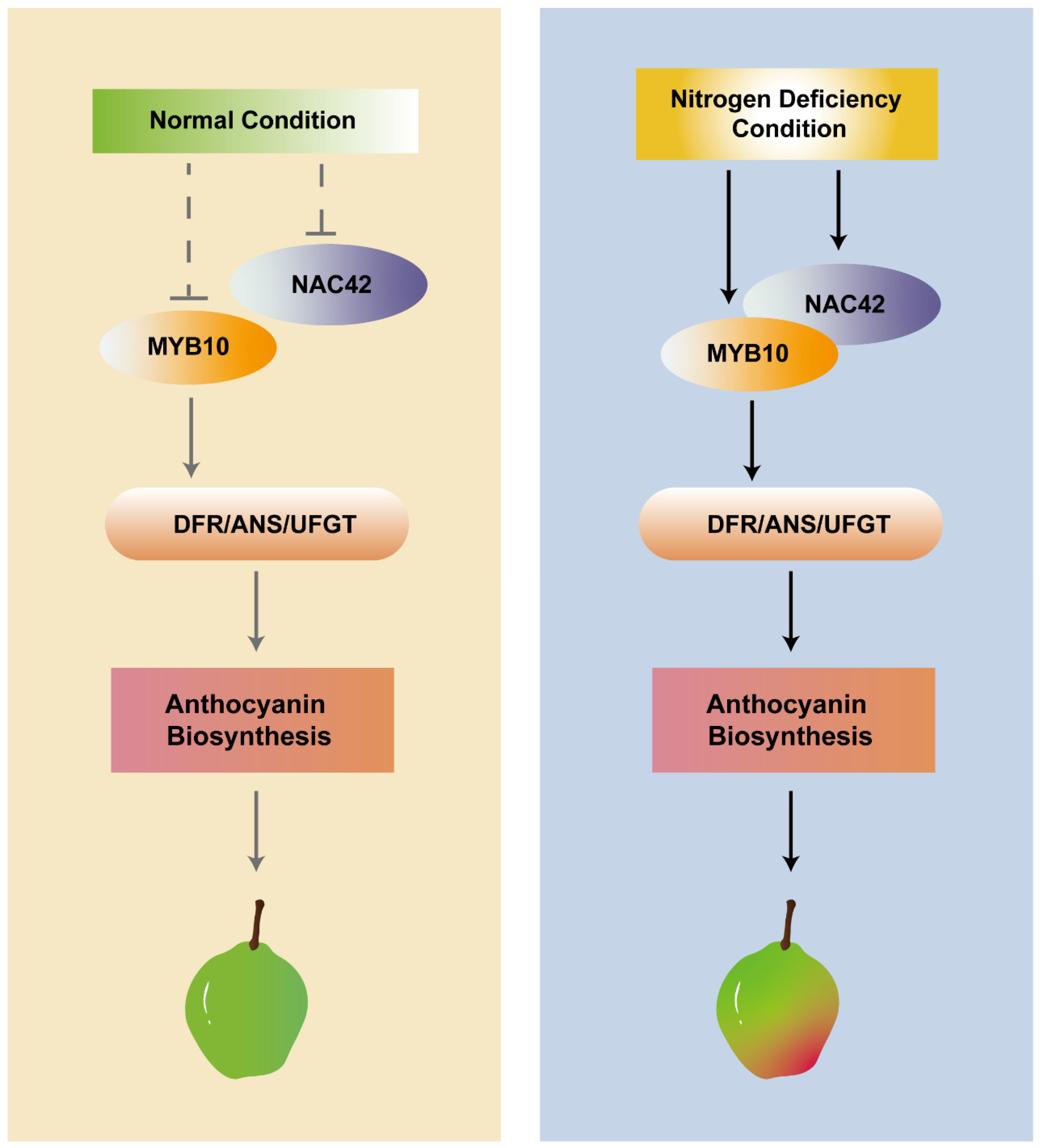
4.4. PyNAC42 Acted as a Positive Regulator Induced by N-Deficiency
PyNAC42 Enhanced the Activation of PyDFR, PyANS, and PyUFGT by Interacting with PyMYB10
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, P.; Wu, X.; Shi, Z.; Tao, S.; Liu, Z.; Qi, K.; Xie, Z.; Qiao, X.; Gu, C.; Yin, H.; et al. A large-scale proteogenomic atlas of pear. Mol. Plant 2023, 16, 599–615. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Gou, S.; Zhong, T.; Wei, S.; An, X.; Sun, H.; Sun, C.; Hu, K.; Zhang, H. Persulfidation of transcription factor MYB10 inhibits anthocyanin synthesis in red-skinned pear. Plant Physiol. 2023, 192, 2185–2202. [Google Scholar] [CrossRef] [PubMed]
- Alappat, B.; Alappat, J. Anthocyanin pigments: Beyond aesthetics. Molecules 2020, 25, 5500. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tian, C.; Zhang, Y.; Li, C.; Li, X.; Yu, Q.; Wang, S.; Wang, X.; Chen, X.; Feng, S. Transcriptomic and metabolomic analysis provides insights into anthocyanin and procyanidin accumulation in pear. BMC Plant Biol. 2020, 20, 129. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Gao, Y.; Han, M.; Liu, P.; Yang, C.; Shen, T.; Li, H. In vitro anthocyanin induction and metabolite analysis in Malus spectabilis leaves under low nitrogen conditions. Hortic. Plant J. 2020, 6, 284–292. [Google Scholar] [CrossRef]
- Steyn, W.J.; Wand, S.J.; Jacobs, G.; Rosecrance, R.C.; Roberts, S.C. Evidence for a photoprotective function of low-temperature-induced anthocyanin accumulation in apple and pear peel. Physiol. Plant 2009, 136, 461–472. [Google Scholar] [CrossRef]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef]
- Mattioli, R.; Francioso, A.; Mosca, L.; Silva, P. Anthocyanins: A comprehensive review of their chemical properties and health effects on cardiovascular and neurodegenerative diseases. Molecules 2020, 25, 3809. [Google Scholar] [CrossRef]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, Y.; Long, T.; Wang, S.; Yang, J. Regulation mechanism of plant pigments biosynthesis: Anthocyanins, carotenoids, and betalains. Metabolites 2022, 12, 871. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Wang, S.; Yu, W.; Liao, Y.; Pan, C.; Zhang, M.; Tao, R.; Wei, J.; Gao, Y.; Wang, D.; et al. The ethylene-responsive transcription factor PpERF9 represses PpRAP2.4 and PpMYB114 via histone deacetylation to inhibit anthocyanin biosynthesis in pear. Plant Cell 2023, 35, 2271–2292. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Jia, X.; Huo, L.; Che, R.; Gong, X.; Wang, P.; Ma, F. MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 2018, 41, 469–480. [Google Scholar] [CrossRef]
- Yang, L.; Li, W.C.; Fu, F.L.; Qu, J.; Sun, F.; Yu, H.; Zhang, J. Characterization of phenylalanine ammonia-lyase genes facilitating flavonoid biosynthesis from two species of medicinal plant Anoectochilus. PeerJ 2022, 10, e13614. [Google Scholar] [CrossRef]
- Guo, N.; Cheng, F.; Wu, J.; Liu, B.; Zheng, S.; Liang, J.; Wang, X. Anthocyanin biosynthetic genes in Brassica rapa. BMC Genom. 2014, 15, 426. [Google Scholar] [CrossRef]
- Ni, J.; Premathilake, A.T.; Gao, Y.; Yu, W.; Tao, R.; Teng, Y.; Bai, S. Ethylene-activated PpERF105 induces the expression of the repressor-type R2R3-MYB gene PpMYB140 to inhibit anthocyanin biosynthesis in red pear fruit. Plant J. 2021, 105, 167–181. [Google Scholar] [CrossRef]
- Goswami, G.; Nath, U.K.; Park, J.I.; Hossain, M.R.; Biswas, M.K.; Kim, H.T.; Kim, H.R.; Nou, I.S. Transcriptional regulation of anthocyanin biosynthesis in a high-anthocyanin resynthesized Brassica napus cultivar. J. Biol. Res. 2018, 25, 19. [Google Scholar] [CrossRef]
- LaFountain, A.M.; Yuan, Y.W. Repressors of anthocyanin biosynthesis. New Phytol. 2021, 231, 933–949. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Premathilake, A.T.; Ni, J.; Bai, S.; Tao, R.; Ahmad, M.; Teng, Y. R2R3-MYB transcription factor PpMYB17 positively regulates flavonoid biosynthesis in pear fruit. Planta 2020, 252, 59. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Wang, Y.; Yang, S.; Xu, Y.; Chen, X. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 2010, 232, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.; Sun, Y.; Allan, A.C.; Teng, Y.; Zhang, D. The red sport of ‘Zaosu’ pear and its red-striped pigmentation pattern are associated with demethylation of the PyMYB10 promoter. Phytochemistry 2014, 107, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ban, Z.J.; Li, X.H.; Wu, M.Y.; Wang, A.L.; Jiang, Y.Q.; Jiang, Y.H. Differential expression of anthocyanin biosynthetic genes and transcription factor PcMYB10 in pears (Pyrus communis L.). PLoS ONE 2012, 7, e46070. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Bai, S.; Zhao, Y.; Qian, M.; Tao, R.; Yin, L.; Gao, L.; Teng, Y. Ethylene response factors Pp4ERF24 and Pp12ERF96 regulate blue light-induced anthocyanin biosynthesis in ‘Red Zaosu’ pear fruits by interacting with MYB114. Plant Mol. Biol. 2019, 99, 67–78. [Google Scholar] [CrossRef]
- Yao, G.; Ming, M.; Allan, A.C.; Gu, C.; Li, L.; Wu, X.; Wang, R.; Chang, Y.; Qi, K.; Zhang, S.; et al. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017, 92, 437–451. [Google Scholar] [CrossRef]
- Bai, S.; Tao, R.; Tang, Y.; Yin, L.; Ma, Y.; Ni, J.; Yan, X.; Yang, Q.; Wu, Z.; Zeng, Y.; et al. BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol. J. 2019, 17, 1985–1997. [Google Scholar] [CrossRef]
- Li, Z.; Ahammed, G.J. Plant stress response and adaptation via anthocyanins: A review. Plant Stress 2023, 10, 100230. [Google Scholar] [CrossRef]
- Shi, L.; Li, X.; Fu, Y.; Li, C. Environmental stimuli and phytohormones in anthocyanin biosynthesis: A comprehensive review. Int. J. Mol. Sci. 2023, 24, 16415. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Zhang, X.W.; Xu, H.F.; Bi, S.Q.; You, C.X.; Hao, Y.J. An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol. J. 2020, 18, 337–353. [Google Scholar] [CrossRef]
- Shin, D.H.; Choi, M.; Kim, K.; Bang, G.; Cho, M.; Choi, S.B.; Choi, G.; Park, Y.I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013, 587, 1543–1547. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Wang, X.F.; Hao, Y.J. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 2020, 101, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Alabd, A.; Ahmad, M.; Zhang, X.; Gao, Y.; Peng, L.; Zhang, L.; Ni, J.; Bai, S.; Teng, Y. Light-responsive transcription factor PpWRKY44 induces anthocyanin accumulation by regulating PpMYB10 expression in pear. Hortic. Res. 2022, 9, uhac199. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Lei, D.; Yao, W.; Li, S.; Wang, H.; Lu, J.; Zhang, Y.; Lin, Y.; Wang, Y.; He, W.; et al. A novel R2R3-MYB transcription factor PbMYB1L of Pyrus bretschneideri regulates cold tolerance and anthocyanin accumulation. Plant Cell Rep. 2024, 43, 34. [Google Scholar] [CrossRef]
- Ren, Y.R.; Zhao, Q.; Yang, Y.Y.; Zhang, T.E.; Wang, X.F.; You, C.X.; Hao, Y.J. The apple 14-3-3 protein MdGRF11 interacts with the BTB protein MdBT2 to regulate nitrate deficiency-induced anthocyanin accumulation. Hortic. Res. 2021, 8, 22. [Google Scholar] [CrossRef]
- Lee, Y.J.; Lee, W.J.; Le, Q.T.; Hong, S.W.; Lee, H. Growth performance can be increased under high nitrate and high salt stress through enhanced nitrate reductase activity in Arabidopsis anthocyanin over-producing mutant plants. Front. Plant Sci. 2021, 12, 644455. [Google Scholar] [CrossRef]
- Liu, K.H.; Liu, M.; Lin, Z.; Wang, Z.F.; Chen, B.; Liu, C.; Guo, A.; Konishi, M.; Yanagisawa, S.; Wagner, G.; et al. NIN-like protein 7 transcription factor is a plant nitrate sensor. Science 2022, 377, 1419–1425. [Google Scholar] [CrossRef]
- Wang, R.; Cresswell, T.; Johansen, M.P.; Harrison, J.J.; Jiang, Y.; Keitel, C.; Cavagnaro, T.R.; Dijkstra, F.A. Reallocation of nitrogen and phosphorus from roots drives regrowth of grasses and sedges after defoliation under deficit irrigation and nitrogen enrichment. J. Ecol. 2021, 109, 4071–4080. [Google Scholar] [CrossRef]
- Liang, J.; He, J. Protective role of anthocyanins in plants under low nitrogen stress. Biochem. Biophys. Res. Commun. 2018, 498, 946–953. [Google Scholar] [CrossRef]
- Peng, M.; Hudson, D.; Schofield, A.; Tsao, R.; Yang, R.; Gu, H.; Bi, Y.M.; Rothstein, S.J. Adaptation of Arabidopsis to nitrogen limitation involves induction of anthocyanin synthesis which is controlled by the NLA gene. J. Exp. Bot. 2008, 59, 2933–2944. [Google Scholar] [CrossRef]
- Wang, X.F.; An, J.P.; Liu, X.; Su, L.; You, C.X.; Hao, Y.J. The nitrate-responsive protein MdBT2 regulates anthocyanin biosynthesis by interacting with the MdMYB1 transcription factor. Plant Physiol. 2018, 178, 890–906. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Yang, K.; Kan, Z.; Dang, H.; Feng, S.; Yang, Y.; Li, L.; Hou, N.; Xu, L.; Wang, X.; et al. The regulatory module MdBT2-MdMYB88/MdMYB124-MdNRTs regulates nitrogen usage in apple. Plant Physiol. 2021, 185, 1924–1942. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Li, X.; Wang, Y.; Xu, J.; Jiang, S.; Zhang, Y. MdMKK9-mediated the regulation of anthocyanin synthesis in red-fleshed apple in response to different nitrogen signals. Int. J. Mol. Sci. 2022, 23, 7755. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Sun, M.; Brewer, L.; Tang, Z.; Nieuwenhuizen, N.; Cooney, J.; Xu, S.; Sheng, J.; Andre, C.; Xue, C.; et al. Allelic variation of BBX24 is a dominant determinant controlling red coloration and dwarfism in pear. Plant Biotechnol. J. 2024, 22, 1468–1490. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Andrews, S. FastQC a quality control tool for high throughput sequence data. In Babraham Bioinformatics; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Wilkinson, L. ggplot2: Elegant graphics for data analysis by WICKHAM, H. Biometrics 2011, 67, 678–679. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Song, B.; Li, B.; Zhang, S.; Liu, Y.; Chen, G.; Zhang, J.; Li, J.; Wu, J. Genome-wide identification and expression analysis of fifteen gene families involved in anthocyanin synthesis in pear. Horticulturae 2024, 10, 335. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimian-Motlagh, S.; Ribone, P.A.; Thirumalaikumar, V.P.; Allu, A.D.; Chan, R.L.; Mueller-Roeber, B.; Balazadeh, S. JUNGBRUNNEN1 confers drought tolerance downstream of the HD-Zip I transcription factor AtHB13. Front. Plant Sci. 2017, 8, 2118. [Google Scholar] [CrossRef] [PubMed]
- Shahnejat-Bushehri, S.; Mueller-Roeber, B.; Balazadeh, S. Arabidopsis NAC transcription factor JUNGBRUNNEN1 affects thermomemory-associated genes and enhances heat stress tolerance in primed and unprimed conditions. Plant Signal. Behav. 2012, 7, 1518–1521. [Google Scholar] [CrossRef]
- Alshareef, N.O.; Wang, J.Y.; Ali, S.; Al-Babili, S.; Tester, M.; Schmöckel, S.M. Overexpression of the NAC transcription factor JUNGBRUNNEN1 (JUB1) increases salinity tolerance in tomato. Plant Physiol. Biochem. 2019, 140, 113–121. [Google Scholar] [CrossRef]
- Diao, P.; Chen, C.; Zhang, Y.; Meng, Q.; Lv, W.; Ma, N. The role of NAC transcription factor in plant cold response. Plant Signal Behav. 2020, 15, 1785668. [Google Scholar] [CrossRef]
- Su, H.; Zhang, S.; Yuan, X.; Chen, C.; Wang, X.F.; Hao, Y.J. Genome-wide analysis and identification of stress-responsive genes of the NAM-ATAF1,2-CUC2 transcription factor family in apple. Plant Physiol. Biochem. 2013, 71, 11–21. [Google Scholar] [CrossRef]
- Liu, H.; Su, J.; Zhu, Y.; Yao, G.; Allan, A.C.; Ampomah-Dwamena, C.; Shu, Q.; Lin-Wang, K.; Zhang, S.; Wu, J. The involvement of PybZIPa in light-induced anthocyanin accumulation via the activation of PyUFGT through binding to tandem G-boxes in its promoter. Hortic. Res. 2019, 6, 134. [Google Scholar] [CrossRef]
- Cong, L.; Qu, Y.; Sha, G.; Zhang, S.; Ma, Y.; Chen, M.; Zhai, R.; Yang, C.; Xu, L.; Wang, Z. PbWRKY75 promotes anthocyanin synthesis by activating PbDFR, PbUFGT, and PbMYB10b in pear. Physiol. Plant 2021, 173, 1841–1849. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, Y.; Zhao, L.; Li, C.; Yu, J.; Li, T.; Yang, W.; Zhang, S.; Su, H.; Wang, L. A novel NAC transcription factor, MdNAC42, regulates anthocyanin accumulation in red-fleshed apple by interacting with MdMYB10. Tree Physiol. 2020, 40, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Li, Z.; Song, Y.; Zhu, H.; Lin, S.; Huang, R.; Jiang, Y.; Duan, X. LcNAC13 physically interacts with LcR1MYB1 to coregulate anthocyanin biosynthesis-related genes during litchi fruit ripening. Biomolecules 2019, 9, 135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, J.; Wei, B.; Li, Y.; Fang, X.; Zhong, Y.; Wang, L. Transcription factor MdNAC33 is involved in ALA-induced anthocyanin accumulation in apples. Plant Sci. 2024, 339, 111949. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Shu, Q.; Lin-Wang, K.; Allan, A.C.; Espley, R.V.; Su, J.; Pei, M.; Wu, J. The PyPIF5-PymiR156a-PySPL9-PyMYB114/MYB10 module regulates light-induced anthocyanin biosynthesis in red pear. Mol. Hortic. 2021, 1, 14. [Google Scholar] [CrossRef] [PubMed]
- Jezek, M.; Zörb, C.; Merkt, N.; Geilfus, C.M. Anthocyanin management in fruits by fertilization. J. Agric. Food Chem. 2018, 66, 753–764. [Google Scholar] [CrossRef]
- Xu, X.; Qin, H.; Liu, C.; Liu, J.; Lyu, M.; Wang, F.; Xing, Y.; Tian, G.; Zhu, Z.; Jiang, Y.; et al. Transcriptome and metabolome analysis reveals the effect of nitrogen-potassium on anthocyanin biosynthesis in “Fuji” apple. J. Agric. Food Chem. 2022, 70, 15057–15068. [Google Scholar] [CrossRef]
- Li, Z.; Ahammed, G.J. Hormonal regulation of anthocyanin biosynthesis for improved stress tolerance in plants. Plant Physiol. Biochem. 2023, 201, 107835. [Google Scholar] [CrossRef]
- Peng, Z.; Tian, J.; Luo, R.; Kang, Y.; Lu, Y.; Hu, Y.; Liu, N.; Zhang, J.; Cheng, H.; Niu, S.; et al. MiR399d and epigenetic modification comodulate anthocyanin accumulation in Malus leaves suffering from phosphorus deficiency. Plant Cell Environ. 2020, 43, 1148–1159. [Google Scholar] [CrossRef]
- Liu, Z.; Wu, X.; Wang, E.; Liu, Y.; Wang, Y.; Zheng, Q.; Han, Y.; Chen, Z.; Zhang, Y. PHR1 positively regulates phosphate starvation-induced anthocyanin accumulation through direct upregulation of genes F3′H and LDOX in Arabidopsis. Planta 2022, 256, 42. [Google Scholar] [CrossRef]
- Wu, X.; Liu, Z.; Liu, Y.; Wang, E.; Zhang, D.; Huang, S.; Li, C.; Zhang, Y.; Chen, Z.; Zhang, Y. SlPHL1 is involved in low phosphate stress promoting anthocyanin biosynthesis by directly upregulation of genes SlF3H, SlF3′H, and SlLDOX in tomato. Plant Physiol. Biochem. 2023, 200, 107801. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Zhao, L.; Song, X.; Lin, Z.; Gu, B.; Yan, J.; Zhang, S.; Tao, S.; Huang, X. Genome-wide analyses and expression patterns under abiotic stress of NAC transcription factors in white pear (Pyrus bretschneideri). BMC Plant Biol. 2019, 19, 161. [Google Scholar] [CrossRef] [PubMed]
- Morishita, T.; Kojima, Y.; Maruta, T.; Nishizawa-Yokoi, A.; Yabuta, Y.; Shigeoka, S. Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol. 2009, 50, 2210–2222. [Google Scholar] [CrossRef]
- Liu, W.; Mei, Z.; Yu, L.; Gu, T.; Li, Z.; Zou, Q.; Zhang, S.; Fang, H.; Wang, Y.; Zhang, Z.; et al. The ABA-induced NAC transcription factor MdNAC1 interacts with a bZIP-type transcription factor to promote anthocyanin synthesis in red-fleshed apples. Hortic. Res. 2023, 10, uhad049. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Lin-Wang, K.; Wang, H.; Gu, C.; Dare, A.P.; Espley, R.V.; He, H.; Allan, A.C.; Han, Y. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015, 82, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Martín-Pizarro, C.; Zhou, L.; Hou, B.; Wang, Y.; Shen, Y.; Li, B.; Posé, D.; Qin, G. Deciphering the regulatory network of the NAC transcription factor FvRIF, a key regulator of strawberry (Fragaria vesca) fruit ripening. Plant Cell 2023, 35, 4020–4045. [Google Scholar] [CrossRef]
- Xu, H.; Wang, N.; Liu, J.; Qu, C.; Wang, Y.; Jiang, S.; Lu, N.; Wang, D.; Zhang, Z.; Chen, X. The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol. Biol. 2017, 94, 149–165. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Zhao, Y.; Sun, J.; Cherono, S.; An, J.P.; Allan, A.C.; Han, Y. Colorful hues: Insight into the mechanisms of anthocyanin pigmentation in fruit. Plant Physiol. 2023, 192, 1718–1732. [Google Scholar] [CrossRef]

| Gene ID | Gene Name | Relative Expression Level | ||
|---|---|---|---|---|
| Log2 (Fold-Change) | p-Value (Adjusted) | Expression Trend | ||
| Pbr032667.1 | PyELIP1 | 8.98 | 3.28 × 10−11 | Up |
| Pbr005987.1 | PyLTI6B | 7.75 | 4.88 × 10−08 | Up |
| Pbr001238.1 | PyWRKY70 | 8.55 | 2.97 × 10−11 | Up |
| Pbr005190.1 | PybHLH100 | 6.00 | 4.08 × 10−05 | Up |
| Pbr002372.1 | PyNAC42 | 5.74 | 1.02 × 10−33 | Up |
| Pbr028712.1 | PyHB17 | −5.76 | 7.80 × 10−11 | Down |
| Pbr022183.1 | PyAGL9 | −6.56 | 4.23 × 10−12 | Down |
| Pbr006840.1 | PyABH1 | −7.30 | 1.37 × 10−49 | Down |
| Pbr030279.1 | PybZIP45 | −8.99 | 2.88 × 10−45 | Down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Song, B.; Chen, G.; Yang, G.; Ming, M.; Zhang, S.; Xue, Z.; Han, C.; Li, J.; Wu, J. Transcriptome Analysis Identified PyNAC42 as a Positive Regulator of Anthocyanin Biosynthesis Induced by Nitrogen Deficiency in Pear (Pyrus spp.). Horticulturae 2024, 10, 980. https://doi.org/10.3390/horticulturae10090980
Zhang J, Song B, Chen G, Yang G, Ming M, Zhang S, Xue Z, Han C, Li J, Wu J. Transcriptome Analysis Identified PyNAC42 as a Positive Regulator of Anthocyanin Biosynthesis Induced by Nitrogen Deficiency in Pear (Pyrus spp.). Horticulturae. 2024; 10(9):980. https://doi.org/10.3390/horticulturae10090980
Chicago/Turabian StyleZhang, Jianhui, Bobo Song, Guosong Chen, Guangyan Yang, Meiling Ming, Shiqiang Zhang, Zhaolong Xue, Chenhui Han, Jiaming Li, and Jun Wu. 2024. "Transcriptome Analysis Identified PyNAC42 as a Positive Regulator of Anthocyanin Biosynthesis Induced by Nitrogen Deficiency in Pear (Pyrus spp.)" Horticulturae 10, no. 9: 980. https://doi.org/10.3390/horticulturae10090980
APA StyleZhang, J., Song, B., Chen, G., Yang, G., Ming, M., Zhang, S., Xue, Z., Han, C., Li, J., & Wu, J. (2024). Transcriptome Analysis Identified PyNAC42 as a Positive Regulator of Anthocyanin Biosynthesis Induced by Nitrogen Deficiency in Pear (Pyrus spp.). Horticulturae, 10(9), 980. https://doi.org/10.3390/horticulturae10090980







