Exploring Sampling Strategies and Genetic Diversity Analysis of Red Beet Germplasm Resources Using SSR Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Material
2.1.1. Plant Materials
2.1.2. SSR Primers
2.1.3. Experimental Instruments and Reagents
2.2. Test Methods
2.2.1. Extraction of Sugar Beet Genomic DNA
2.2.2. Sampling Gradient Division in Sampling Strategy Research
2.2.3. PCR Amplification and Electrophoresis Detection
2.2.4. Data Analysis
3. Result and Analysis
3.1. Analysis of Sampling Strategies for Red Beet Germplasm Resources
3.1.1. Verification of Feasibility of Mixed Sampling
3.1.2. Analysis of Genetic Diversity within Six Test Material Populations
3.1.3. Comparison of Amplification Polymorphism between Red Sweet Menu Strain Samples (Group A) and Fixed Mixed Samples (Group B)
3.1.4. Analysis of Amplification Parameters for Randomly Numbered (Group C) Mixed DNA Samples
3.2. Genetic Diversity Analysis
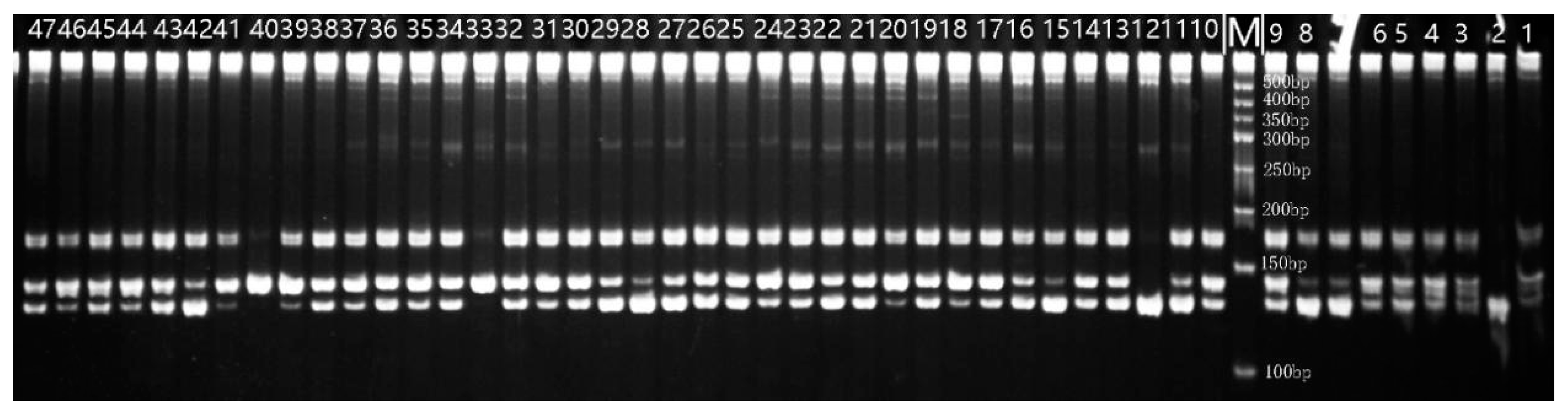
3.2.1. Amplification Results of 21 SSR Primer Pairs and Polymorphism of Loci
3.2.2. Genetic Diversity Analysis of 47 Red Beet Germplasm Resources
3.2.3. Genetic Distance and Cluster Analysis of Red Beet Germplasm Resources
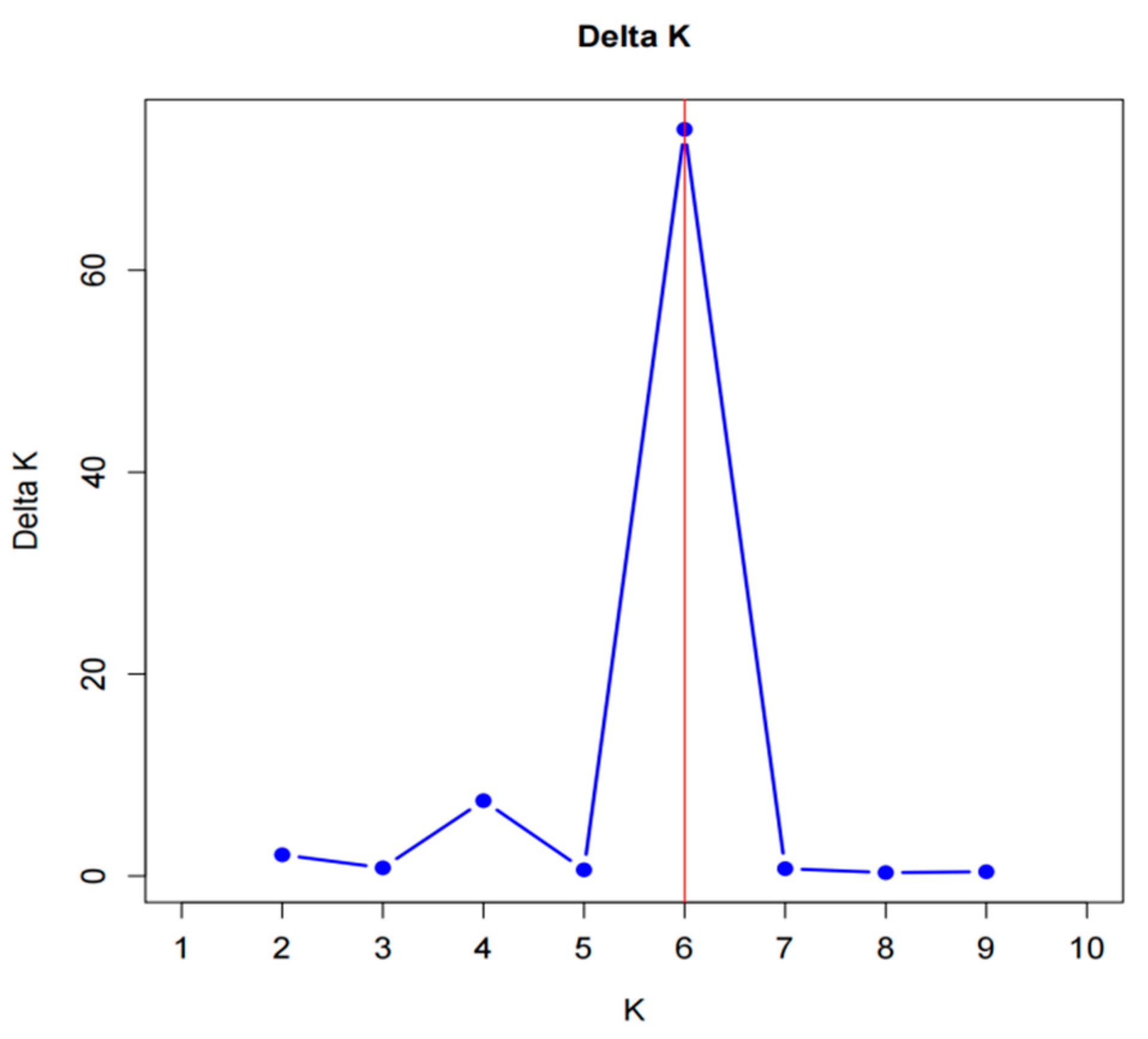
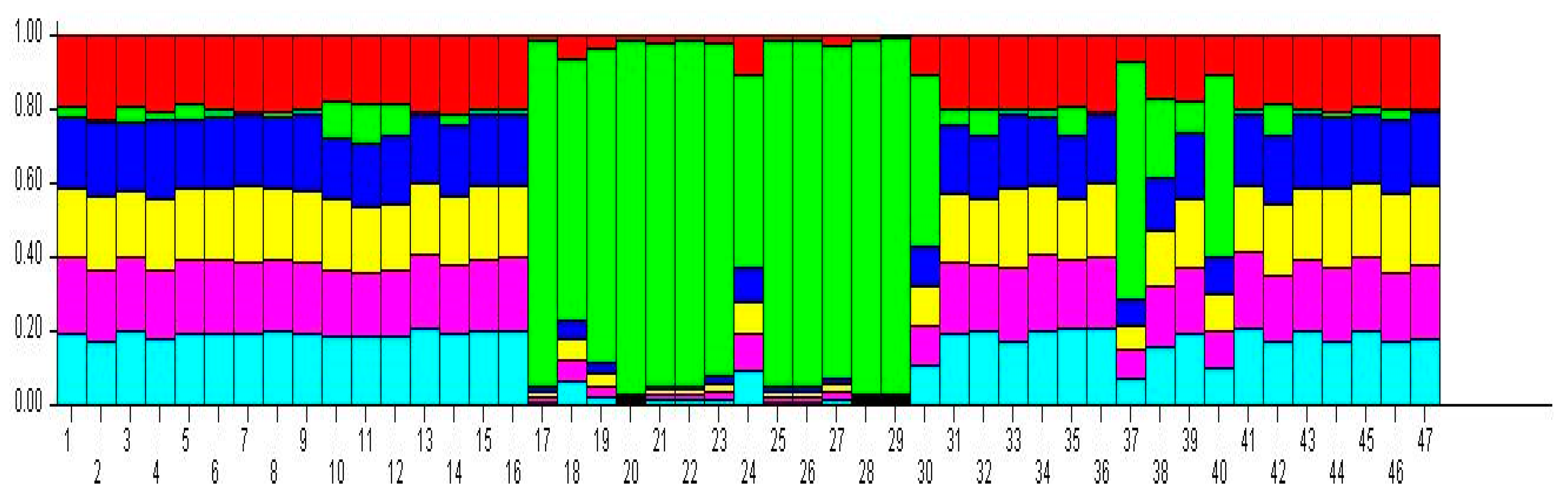
3.2.4. Genetic Structure of Red Beet Germplasm Resources
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wu, X.; Pi, Z.; Li, S.; Wu, Z. Identification of the Fertility Types of Red Beet Varieties (Lines) Using Molecular-Marker Technology. Sugar Tech 2024, 1–9. [Google Scholar] [CrossRef]
- Wiczkowski, W. Thematic Issue on “Red Beetroot as a Source of Nutrients, Bioactive Compounds and Pigments”. Pol. J. Food Nutr. Sci. 2020, 70, 5–6. [Google Scholar] [CrossRef]
- Ćurčić, Z.; Taški-Ajduković, K.; Nagl, N. Relationship between hybrid performance and genetic variation in self-fertile and self-sterile sugar beet pollinators as estimated by SSR markers. Euphytica 2017, 213, 1–16. [Google Scholar] [CrossRef]
- Kornienko, A.V.; Podvigina, O.A.; Zhuzhzhalova, T.P.; Fedulova, T.P.; Bogomolov, M.A.; Oshevnev, V.P.; Butorina, A.K. High-priority research directions in genetics, and the breeding of the sugar beet (Beta vulgaris L.) in the 21st century. Genetika 2014, 50, 1137–1148. [Google Scholar] [CrossRef]
- Xiong, H.; Chen, Y.; Gao, S.-J.; Pan, Y.-B.; Shi, A. Population Structure and Genetic Diversity Analysis in Sugarcane (Saccharum spp. hybrids) and Six Related Saccharum Species. Agronomy 2022, 12, 412. [Google Scholar] [CrossRef]
- Khanlou, K.M.; Vandepitte, K.; Asl, L.K.; Van Bockstaele, E. Towards an optimal sampling strategy for assessing genetic variation within and among white clover (Trifolium repens L.) cultivars using AFLP. Genet. Mol. Biol. 2011, 34, 252–258. [Google Scholar] [CrossRef]
- Lu, B.-R.; Yan, J. Sampling strategy for genetic diversity. Biodivers. Sci. 2003, 11, 155–161. [Google Scholar] [CrossRef]
- Wu, Y.; He, R.; Lu, Y.; Zhang, Z.; Yang, L.; Guan, X.; Zhang, R.; Zheng, J. Development and evaluation of EST-SSR markers in Sorbus pohuashanensis (Hance) Hedl. and their application to other Sorbus species. Trees-Struct. Funct. 2020, 34, 455–467. [Google Scholar] [CrossRef]
- Jin, Y.; Zhang, W.; Fu, D.; Lu, B. Sampling Strategy Within a Wild Soybean Population Based on Its Genetic Variation Detected by ISSR Markers. Acta Bot. Sin. 2003, 8, 995–1002. [Google Scholar]
- Liu, W.; Li, L.; Liu, W.; Zhang, Z.; Wu, Z.; Wang, C. SSR Analysis on the Sampling Strategy of Psathyrostachys Huashanic Keng Population. J. Triticeae Crops 2006, 26, 16–20. [Google Scholar]
- Zhang, Q.; Guo, R.; Chen, X.; Xu, Y.; Guo, M.; Guo, H.; Yang, M. Sampling strategy for genetic diversity ISSR analysis of wild Cannabis sativa L. population. J. South. Agric. 2017, 48, 973–978. [Google Scholar] [CrossRef]
- Ober, E.S.; Luterbacher, M.C. Genotypic variation for drought tolerance in Beta vulgaris. Ann. Bot. 2002, 89, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yuan, Q.; Meng, Y.; Li, X.; Nan, Z.B.; Wang, Y.; Zhang, W. A genetic diversity analysis of wild Lespedeza populations based on morphological characters, allozyme and RAPD methods. Plant Breed. 2007, 126, 89–94. [Google Scholar] [CrossRef]
- Wang, P.; Bai, Y.; Wang, M.; Hu, B.; Pu, Z.; Zhang, Z.; Zhang, Q.; Xu, D.; Luo, W.; Chen, Z. Breeding of CMS maintainer lines through anther culture assisted by high-resolution melting-based markers. J. Integr. Agric. 2020, 19, 2965–2973. [Google Scholar] [CrossRef]
- McGrath, J.M. Assisted Breeding in Sugar Beets. Sugar Tech 2010, 12, 187–193. [Google Scholar] [CrossRef]
- Cruz, V.M.V.; Kilian, A.; Dierig, D.A. Development of DArT Marker Platforms and Genetic Diversity Assessment of the U.S. Collection of the New Oilseed Crop Lesquerella and Related Species. PLoS ONE 2013, 8, e64062. [Google Scholar] [CrossRef]
- Peleman, J.D.; van der Voort, J.R. Breeding by design. Trends Plant Sci. 2003, 8, 330–334. [Google Scholar] [CrossRef]
- Van Inghelandt, D.; Melchinger, A.E.; Lebreton, C.; Stich, B. Population structure and genetic diversity in a commercial maize breeding program assessed with SSR and SNP markers. Theor. Appl. Genet. 2010, 120, 1289–1299. [Google Scholar] [CrossRef]
- Bini, L.; Gori, M.; Novello, M.A.; Biricolti, S.; Giordani, E.; Lara, M.V.; Niella, F.; Nunziata, A.; Rocha, P.; Filippi, J.M.; et al. Assessing the Genetic Diversity of Wild and Commercial Feijoa sellowiana Accessions Using AFLPs. Horticulturae 2024, 10, 366. [Google Scholar] [CrossRef]
- Choe, Y.-I.; Song, S.-R.; Ho, U.-H.; Ho, T.-S.; Sin, S.-J.; Pak, I.-C.; Choe, M.-B. Sequence-related amplified polymorphism (SRAP) markers reveal genetic variation of rice strains obtained by millet DNA injection through coleoptile. Genet. Resour. Crop. Evol. 2024, 1–8. [Google Scholar] [CrossRef]
- Bahmankar, M.; Rahnama, H.; Salehi, M.; Noori, S.A.S. Somatic embryogenesis and genetic fidelity in camelina by RAPD markers and flow cytometry. Plant Cell Tissue Organ Cult. 2024, 156, 67. [Google Scholar] [CrossRef]
- Zeljković, M.K.; Bosančić, B.; Đurić, G.; Flachowsky, H.; Garkava-Gustavsson, L. Genetic diversity of pear germplasm in Bosnia and Herzegovina, as revealed by SSR markers. Zemdirb. -Agric. 2021, 108, 71–78. [Google Scholar] [CrossRef]
- Bagheri, M.; Heidari, B.; Dadkhodaie, A.; Heidari, Z.; Daneshnia, N.; Richards, C.M. Analysis of genetic diversity in a collection of Plantago species: Application of ISSR markers. J. Crop. Sci. Biotechnol. 2022, 25, 1–8. [Google Scholar] [CrossRef]
- Carranza, J.; Pérez-González, J.; Anaya, G.; de Jong, M.; Broggini, C.; Zachos, F.E.; McDevitt, A.D.; Niedziałkowska, M.; Sykut, M.; Csányi, S.; et al. Genome-wide SNP assessment of contemporary European red deer genetic structure highlights the distinction of peripheral populations and the main admixture zones in Europe. Mol. Ecol. 2024, e17508. [Google Scholar] [CrossRef] [PubMed]
- Igwe, D.O.; Ihearahu, O.C.; Osano, A.A.; Acquaah, G.; Ude, G.N. Assessment of genetic diversity of Musa species accessions with variable genomes using ISSR and SCoT markers. Genet. Resour. Crop. Evol. 2022, 69, 49–70. [Google Scholar] [CrossRef]
- Srivastava, S.; Pathak, A.D.; Kumar, R.; Joshi, B.B. Genetic diversity of sugar beet genotypes evaluated by microsatellite DNA markers. J. Environ. Biol. 2017, 38, 777–783. [Google Scholar] [CrossRef]
- Li, Q.; Wang, Y.; Pi, Z.; Wu, Z. Genetic diversity analysis of sugar beet germplasm resources based on SSR molecular markers. Sugar Crops China 2023, 45, 1–7. [Google Scholar] [CrossRef]
- Fugate, K.K.; Fajardo, D.; Schlautman, B.; Ferrareze, J.P.; Bolton, M.D.; Campbell, L.G.; Wiesman, E.; Zalapa, J. Generation and Characterization of a Sugarbeet Transcriptome and Transcript-Based SSR Markers. Plant Genome 2014, 7, plantgenome2013-11. [Google Scholar] [CrossRef]
- Yan, C.; Zou, Y.; Wu, Z.; Xing, W.; Li, H. A method suitable for large-scale and rapid extraction of DNA from sugar beets. Sugar Crops China 2018, 40, 44–46. [Google Scholar] [CrossRef]
- Zhang, H.; Li, X.; Chen, Z.; Hu, W.; Li, W.; Huang, M.; Yu, F.; Yang, L. Genetic diversity analysis and DNA fingerprinting construction of naked flower purple pearl germplasm resources based on SSR molecular markers. Chin. Tradit. Herb. Drugs 2023, 54, 3971–3982. [Google Scholar]
- Chen, S.; Li, J.; Chen, W.; Luo, T.; Chen, S. Genetic diversity analysis of Pinus kesiya var.langbianensis germplasm resources based on SSR molecular markers. Southwest China J. Agric. Sci. 2024, 37, 532–541. [Google Scholar] [CrossRef]
- Lin, T.; Zhu, G.; Zhang, J.; Xu, X.; Yu, Q.; Zheng, Z.; Zhang, Z.; Lun, Y.; Li, S.; Wang, X.; et al. Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 2014, 46, 1220–1226. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. The genetical structure of populations. Ann. Eugen. 1951, 15, 323–354. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Ji, P.; Wang, J.; He, Y.; Zhang, Y.; Zhang, F.; Yun, Y.; Zhang, G. Estimation of Genetic Diversity between and within Biparental Clones and Full-Sib Families of the Chinese Pine Using SSR Markers. Horticulturae 2023, 9, 1205. [Google Scholar] [CrossRef]
- Zhu, Y.; An, W.; Peng, J.; Li, J.; Gu, Y.; Jiang, B.; Chen, L.; Zhu, P.; Yang, H. Genetic Diversity of Nanmu (Phoebe zhennan S. Lee. et F. N. Wei) Breeding Population and Extraction of Core Collection Using nSSR, cpSSR and Phenotypic Markers. Forests 2022, 13, 1320. [Google Scholar] [CrossRef]
- Peng, F.; Pi, Z.; Li, S.; Wu, Z. Genetic Diversity and Population Structure Analysis of Excellent Sugar Beet (Beta vulgaris L.) Germplasm Resources. Horticulturae 2024, 10, 120. [Google Scholar] [CrossRef]
- Liang, X.-M.; Pi, Z.; Wu, Z.-D.; Li, S.-N. Constructing DNA Fingerprinting and Evaluating Genetic Diversity Among Sugar Beet (Beta vulgaris L.) Varieties Based on Four Molecular Markers. Sugar Tech 2023, 25, 1361–1373. [Google Scholar] [CrossRef]
- Liu, D.; Wang, X.; Li, W.; Li, J.; Tan, W.; Xing, W. Genetic Diversity Analysis of the Phenotypic Traits of 215 Sugar Beet Germplasm Resources. Sugar Tech 2022, 24, 1790–1800. [Google Scholar] [CrossRef]
- Sandell, F.L.; Stralis-Pavese, N.; McGrath, J.M.; Schulz, B.; Himmelbauer, H.; Dohm, J.C. Genomic distances reveal relationships of wild and cultivated beets. Nat. Commun. 2024, 15, 1078. [Google Scholar] [CrossRef]
- Brown, A.H.D. Isozymes, plant population genetic structure and genetic conservation. TAG Theor. Appl. Genet. Theor. Und Angew. Genet. 1978, 52, 145–157. [Google Scholar] [CrossRef]
- Li, L.; Guo, R.; Du, G.; Guo, H.; Lü, P.; Xu, Y.; Guo, M.; Zhang, Y.; Chen, X.; Zhang, Q.; et al. Sampling Strategy for Genetic Diversity Analysis and Variety Identification of Hemp (Cannabis sativa L.) Based on SSR Markers. Mol. Plant Breed. 2023. Available online: https://link.cnki.net/urlid/46.1068.S.20231211.1122.002 (accessed on 11 September 2024).
- Li, Q.; Wang, Y.; Liu, R.; Pi, Z.; Wu, Z. Exploring The Sampling Strategy of Sugar Beet Germplasm by SSR Molecular Markers. Mol. Plant Breed. 2023. Available online: https://kns.cnki.net/kcms/detail/46.1068.S.20220520.1754.017.html (accessed on 11 September 2024).
- Cardon, L.R.; Palmer, L.J. Population stratification and spurious allelic association. Lancet 2003, 361, 598–604. [Google Scholar] [CrossRef] [PubMed]

| Number | Name of Germplasm Resource | Number | Name of Germplasm Resource | Number | Name of Germplasm Resource |
|---|---|---|---|---|---|
| 1 | Yulu Red Beet | 17 | S214 | 33 | GOLDEN |
| 2 | Tianyong Big Red Ball | 18 | HTC-95 | 34 | CYCLHDAR |
| 3 | Red New three | 19 | JHTC 1601 | 35 | ChioGGiA |
| 4 | Marunouchi mating | 20 | JHTC 1504 | 36 | Geralt Yi |
| 5 | Hua Yu | 21 | JHTC 1602 | 37 | Red sugar beet in the north |
| 6 | Jilin Qingfeng | 22 | HTC 2005-2 | 38 | HTC 2403 |
| 7 | 96001-2/1-2 | 23 | HTC 2006-1 | 39 | HTC 2401 |
| 8 | 96001/2 | 24 | BOLDOR-ORI:FR | 40 | HTC 2406 |
| 9 | Red New two | 25 | WODAN-F1-ORI:AU | 41 | HTC 2402 |
| 10 | Red stem sugar beet | 26 | SUBETO-F1-ORI:FR | 42 | HTC 2405 |
| 11 | Shan Yan Hong Sweet Cabbage Head | 27 | ACTION-F1-ORI:FR | 43 | HTC 2404 |
| 12 | Inner Mongolia Red | 28 | PABLO-F1-ORI:AU | 44 | Yien |
| 13 | 357357 | 29 | Red New one | 45 | Seed Agriculture |
| 14 | American Hybrid | 30 | Heida Red No.30 | 46 | JinTai |
| 15 | rootstock | 31 | Heida Red No.5 | 47 | Guilan |
| 16 | Striped Red Beet | 32 | Gongda Shitian No.1 |
| Processing Groups | Processing | Numbers | Sample Size |
|---|---|---|---|
| Individual plant (Group A) | Individual | 1–50 | 50 |
| Fixed number, single-plant mixed (Group B) | DNA mixture with numbers 1–5 | 51 | 1 |
| DNA mixture with numbers 1–10 | 52 | 1 | |
| DNA mixture with numbers 1–20 | 53 | 1 | |
| DNA mixture with numbers 1–30 | 54 | 1 | |
| DNA mixture with numbers 1–40 | 55 | 1 | |
| DNA mixture with numbers 1–50 | 56 | 1 | |
| Single plant, random sample mixture (Group C) | Randomly mix 5 individual plants | 57–66 | 10 |
| Randomly mix 10 individual plants | 67–76 | 10 | |
| Randomly mix 20 individual plants | 77–81 | 5 | |
| Randomly mix 30 individual plants | 82–86 | 5 | |
| Randomly mix 40 individual plants | 87–88 | 2 | |
| Randomly mix 50 individual plants | 89 | 1 |
| Single Plant and Fixed Number Gradient Processing | Name of Red Beet Variety (Series) | |||||
|---|---|---|---|---|---|---|
| Marunouchi Mating | Inner Mongolia Red | ACTION-F1-ORI:FR | Yulu Red Beet | Heida Red No.30 | CYCLHDAR | |
| Single plant 1–5 | 28 | 28 | 35 | 31 | 30 | 32 |
| Fixed numbers 1–5 | 31 | 28 | 35 | 31 | 34 | 41 |
| Single plant 1–10 | 34 | 32 | 35 | 42 | 36 | 39 |
| Fixed numbers 1–10 | 36 | 33 | 42 | 42 | 34 | 37 |
| Single plant 1–20 | 36 | 35 | 36 | 42 | 39 | 41 |
| Fixed numbers 1–20 | 37 | 35 | 43 | 41 | 38 | 43 |
| Single plant 1–30 | 38 | 37 | 35 | 42 | 35 | 41 |
| Fixed numbers 1–30 | 41 | 36 | 44 | 40 | 42 | 47 |
| Single plant 1–40 | 37 | 36 | 36 | 44 | 37 | 42 |
| Fixed numbers 1–40 | 41 | 39 | 45 | 35 | 44 | 43 |
| Single plant 1–50 | 33 | 30 | 34 | 39 | 29 | 35 |
| Fixed numbers 1–50 | 44 | 41 | 45 | 38 | 47 | 46 |
| Amplification Parameters | Name of Red Beet Variety (Series) | |||||
|---|---|---|---|---|---|---|
| Marunouchi Mating | Inner Mongolia Red | ACTION-F1-ORI:FR | Heida Red No.30 | Yulu Red Beet | CYCLHDAR | |
| Total number of amplified bands | 52 | 43 | 46 | 52 | 50 | 55 |
| Number of polymorphic bands | 48 | 38 | 41 | 45 | 46 | 45 |
| Heterogeneity ratio (%) | 92% | 88% | 89% | 87% | 92% | 82% |
| Na | 2 | 2 | 2 | 2 | 2 | 2 |
| Ne | 1.6784 | 1.6882 | 1.7526 | 1.8121 | 1.8175 | 1.8308 |
| I | 0.5696 | 0.5674 | 0.5981 | 0.6196 | 0.6329 | 0.6414 |
| Genetic distance within the population | 0.0417~0.6875 | 0.0357~0.3889 | 0.0385~0.6250 | 0.0357~0.4444 | 0.0455~0.6000 | 0.0384~0.5500 |
| Sampling Method | Name of Red Beet Variety (Series) | ||||||
|---|---|---|---|---|---|---|---|
| Marunouchi Mating | Inner Mongolia Red | ACTION-F1-ORI:FR | Heida Red No.30 | Yulu Red Beet | CYCLHDAR | ||
| Individual | A | 34.92 | 33.36 | 35.14 | 40.58 | 34.58 | 38.7 |
| Fixed number single plant mixed sample | B1 | 31 | 28 | 35 | 31 | 34 | 41 |
| B2 | 36 | 33 | 42 | 42 | 34 | 37 | |
| B3 | 37 | 35 | 43 | 41 | 38 | 43 | |
| B4 | 41 | 36 | 44 | 41 | 42 | 47 | |
| B5 | 41 | 39 | 45 | 35 | 44 | 43 | |
| B6 | 44 | 41 | 45 | 38 | 47 | 46 | |
| Amplification Parameters | Variety (Series) Name | Randomly Numbered Sample Mix Number | |||||
|---|---|---|---|---|---|---|---|
| 5 | 10 | 20 | 30 | 40 | 50 | ||
| Expand the number of sites | 27 | 43.7 | 44.5 | 44.8 | 45.4 | 42.5 | 44 |
| 4 | 45.3 | 48.9 | 49.2 | 50 | 49.5 | 46 | |
| 12 | 38.8 | 40.9 | 39.6 | 40.6 | 40.5 | 39 | |
| 30 | 44.7 | 45.9 | 36.8 | 41.6 | 40 | 50 | |
| 1 | 43.4 | 46.2 | 46 | 46.2 | 45.5 | 47 | |
| 34 | 44.8 | 46 | 41.8 | 46.6 | 44 | 45 | |
| Ne | 27 | 1.9105 | 1.9921 | 1.9193 | 1.9376 | 1.9000 | 1.9333 |
| 4 | 1.9411 | 1.9718 | 1.9425 | 1.9890 | 1.8714 | 1.7143 | |
| 12 | 1.8563 | 1.9461 | 1.9806 | 1.8867 | 1.8333 | 1.7500 | |
| 30 | 1.8654 | 1.9375 | 1.8857 | 1.8805 | 1.6857 | 1.8462 | |
| 1 | 1.9400 | 1.9890 | 1.9714 | 2.0000 | 1.9714 | 2.0000 | |
| 34 | 1.9451 | 1.9868 | 1.9748 | 1.9945 | 1.9667 | 1.8571 | |
| I | 27 | 0.6652 | 0.6911 | 0.6604 | 0.6755 | 0.6343 | 0.6469 |
| 4 | 0.6767 | 0.6855 | 0.6735 | 0.6903 | 0.6249 | 0.4951 | |
| 12 | 0.6330 | 0.6768 | 0.6880 | 0.6472 | 0.5776 | 0.5199 | |
| 30 | 0.6409 | 0.6738 | 0.6302 | 0.6498 | 0.4858 | 0.5865 | |
| 1 | 0.6658 | 0.6903 | 0.6838 | 0.6931 | 0.6838 | 0.6931 | |
| 34 | 0.6778 | 0.6895 | 0.6858 | 0.6917 | 0.6822 | 0.5941 | |
| Nei’s | 27 | 0.4731 | 0.4980 | 0.4707 | 0.4827 | 0.4554 | 0.4667 |
| 4 | 0.4838 | 0.4924 | 0.4814 | 0.4971 | 0.4464 | 0.3571 | |
| 12 | 0.4460 | 0.4842 | 0.4949 | 0.4586 | 0.4167 | 0.3750 | |
| 30 | 0.4525 | 0.4813 | 0.4514 | 0.4599 | 0.3482 | 0.4231 | |
| 1 | 0.4761 | 0.4971 | 0.4911 | 0.5000 | 0.4911 | 0.5000 | |
| 34 | 0.4849 | 0.4964 | 0.4929 | 0.4986 | 0.4896 | 0.4268 | |
| Primers | Total Loci | Polymorphic Loci | PIC |
|---|---|---|---|
| W21 | 2 | 1 | 0.0782 |
| TC94 | 3 | 3 | 0.4326 |
| TC55 | 6 | 6 | 0.7045 |
| SB06 | 3 | 2 | 0.2856 |
| L70 | 6 | 6 | 0.7202 |
| L59 | 3 | 2 | 0.6718 |
| L48 | 4 | 3 | 0.6194 |
| L16 | 3 | 2 | 0.4435 |
| 77067 | 3 | 2 | 0.5213 |
| 57236 | 4 | 3 | 0.5940 |
| 27906 | 5 | 3 | 0.6951 |
| 27374 | 3 | 2 | 0.5335 |
| 26391 | 6 | 5 | 0.7298 |
| 24552 | 4 | 3 | 0.6675 |
| 18963 | 4 | 3 | 0.5663 |
| 17923 | 5 | 5 | 0.6872 |
| 17623 | 4 | 2 | 0.6313 |
| 16898 | 4 | 3 | 0.5909 |
| 14118 | 7 | 5 | 0.5574 |
| 11965 | 6 | 5 | 0.7066 |
| 2305 | 4 | 3 | 0.6131 |
| Total | 89 | 69 | __ |
| Average | 4.24 | 3.29 | 0.5738 |
| Mean Value of Genetic Diversity Parameters | Numerical Value |
|---|---|
| Observing the number of alleles (Na) | 4.1905 |
| Effective number of alleles (Ne) | 2.8962 |
| Shannon’s information index (I) | 1.1299 |
| Expected heterozygosity rate (He) | 0.6127 |
| Observation of heterozygosity rate (Ho) | 0.3806 |
| Expected heterozygosity (Nei’s) | 0.6127 |
| Gene flow between germplasms (Nm) | 0.43 |
| Genetic differentiation index between populations (Fst) | 0.3677 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Pi, Z.; Li, S.; Wu, Z. Exploring Sampling Strategies and Genetic Diversity Analysis of Red Beet Germplasm Resources Using SSR Markers. Horticulturae 2024, 10, 1008. https://doi.org/10.3390/horticulturae10091008
Wu X, Pi Z, Li S, Wu Z. Exploring Sampling Strategies and Genetic Diversity Analysis of Red Beet Germplasm Resources Using SSR Markers. Horticulturae. 2024; 10(9):1008. https://doi.org/10.3390/horticulturae10091008
Chicago/Turabian StyleWu, Xiangjia, Zhi Pi, Shengnan Li, and Zedong Wu. 2024. "Exploring Sampling Strategies and Genetic Diversity Analysis of Red Beet Germplasm Resources Using SSR Markers" Horticulturae 10, no. 9: 1008. https://doi.org/10.3390/horticulturae10091008
APA StyleWu, X., Pi, Z., Li, S., & Wu, Z. (2024). Exploring Sampling Strategies and Genetic Diversity Analysis of Red Beet Germplasm Resources Using SSR Markers. Horticulturae, 10(9), 1008. https://doi.org/10.3390/horticulturae10091008






