Abstract
The cystathionine-β-synthase (CBS) domain is present in the proteins of all living organisms and functions as an energy sensor, regulating protein activity through the binding capacity of its adenosine ligands. The role of the CBS gene in plant growth and development, as well as tolerance to abiotic stresses, remains largely unknown, especially in grapevine. In our study, 32 members of the CBS gene family were obtained that were distributed on 15 chromosomes. The results of the structural and evolutionary tree analyses indicated that the VvCBS gene family exhibits diverse intron-exon patterns and highly conserved motifs. Furthermore, the phylogenetic classification of the VvCBS genes revealed the presence of three subfamilies. Subcellular localization analyses showed that the VvCBS genes are mainly located in the plasma membrane region. The secondary structure of the VvCBS protein mainly consists of α-helices, extended strands, β-turns, and random coils. The VvCBS gene family exhibited four co-linear gene pairs, while the numbers for Arabidopsis thaliana and rice were 21 and 7, respectively. The promoter cis-acting element analysis revealed the presence of light-responsive, hormone-responsive, stress-responsive, and growth- and development-related elements in the VvCBS family. The expression characterization demonstrated that 12 VvCBS genes exhibited high expression levels in all grape tissues. Additionally, the RT-qPCR expression analyses showed that the 32 VvCBS exhibited different responses to a variety of abiotic stresses (cold, drought, salt), suggesting that they were functionally differentiated. VvCBS27 was cloned from ‘Pinot Noir’ of grapevine with a coding sequence of 624 bp. Subcellular localization showed that VvCBS27 protein was mainly located in the cytoplasm, cell membrane, and nucleus. This study lays a foundation for elucidating the function of grape CBS protein.
1. Introduction
The CBS (cystathionine-β-synthase) structural domain was originally found as a conserved structural domain in archaeal proteins [1]. It was subsequently discovered in human cystathionine-β-synthase [2]. A typical CBS structural domain consists of approximately 60 amino acid residues forming two α and three β, i.e., a β-α-β-β-α secondary structure composed of β-α-β-β-α, and further folded into a globular tertiary structure containing a three-sided antiparallel β-folding, with two α-helices on one side [3]. CBS domains are usually found in pairs in protein sequences, and each pair of these domains is tightly bound in a pseudo-dimer arrangement by their β-folding, forming CBS pairs or Bat Batman domains, which are a functional unit. Moreover, two Bateman modules are usually combined into a disk—such as structure (CBS module) formed in 95% of the known proteins by the “head-to-head” type—the contacts making up homonymous CBS domains of two Bateman modules. It is usually found as tandem repeat sequences forming CBS pairs or so-called Bateman structural domains, which define the family of CBS structural domain-containing proteins (CDCP) [4,5,6].
The CBS structural domain is known to have affinity for a wide range of ligands (mainly adenosine nucleotides) and has a regulatory function based on ligand-induced conformational changes [7,8]. The CBS structural domain has been shown to bind adenosine compounds such as adenosine monophosphate (AMP), adenosine diphosphate (ADP), adenosine triphosphate (ATP), and S-adenosylmethionine and to function in cellular energy sensing [9,10,11]. In addition to CBS, CBS domains are present in other proteins and regulate biological processes by binding to other functional regions of CBS domain proteins (CDCPs). For example, the inosine 5′-monophosphate dehydrogenase (IMPDH) structural domain, the AMP-activated protein kinase (AMPK) structural domain, and the CBS structural domain are present in other proteins and regulate biological processes by binding to other functional regions of CBS domain proteins (CDCPs)—activated protein kinase (AMPK) and voltage-gated chloride channels (CLC) [12,13,14]. The CBS structural domain is not only widely distributed among archaea, bacteria, and yeast but has also been identified in animal and plant proteins [1,15]. Previous studies have identified point mutations in the CBS structural domain as a causal factor in several hereditary diseases in humans, including homocystinuria, retinitis pigmentosa, Bartter syndrome, osteoporosis, and others [2,16,17,18,19]. In mammals, the binding of AMP/ADP to the CBS structural domain of AMPK induces conformational changes that stabilize the protein activation loop phosphorylated by upstream kinases [20,21]. This suggests that the CBS structural domain or a wide range of CDCPs are indispensable for this process. Despite the importance of CBS or CDCP studies in human clinical and animal directions, however, genome-wide identification and expression analysis of grape CBS or CDCP have not been reported.
Previously, 34, 59, 66, 71, and 95 CDCPs have been identified from Arabidopsis thaliana, rice (Oryza sativa L.), wheat (Triticum aestivum L.), soybean (Glycine max (L.) Merr.), and sugarcane (Saccharum officinarum) by genome-wide analyses [22,23,24], and 37 members of CBSs have been identified in maize (Zea mays L.), respectively [25]. Some of these members have been demonstrated to perform crucial functions in plant growth and development, as well as in response to biotic and abiotic stresses. In Arabidopsis, AtCBSX1 and AtCBSX2 were found to be highly expressed in anthers, which directly activate thioredoxins (Trx) in the thioredoxin-Trx system in chloroplasts. This results in the control of the cellular level of H2O2, which in turn regulates the development and growth of plants [10,26]. AtCBSX3 is localized to the mitochondria, where it activates o-type thioredoxins (Trx-o2). These play an important role in the regulation of plant development and the redox system, modulating the production of reactive oxygen species (ROS) in the mitochondria [27]. In rice, OsBi1 is a CBS-containing gene that has been linked to resistance to the brown plant hopper (Nilaparvata lugens Stal.). The overexpression of OsBi1 has been demonstrated to enhance plant resistance to the herbivorous brown plant hopper [28]. A number of rice CDCP genes have been identified as playing a role in the plant’s response to a range of environmental stresses, including drought, salinity, and injury [15]. Rice inoculation with Botrytis cinerea, exogenous application of salicylic acid (SA) or methyl jasmonate (MeJA) resulted in up-regulation of the OsCBSX3 transcript, and overexpression of OsCBSX3 conferred resistance to Botrytis cinerea [29]. Furthermore, the overexpression of OsCBSX4 in tobacco plants resulted in a notable increase in resistance to salt stress [30].
Additionally, OsCBSX9 and OsCBSX4 exhibited considerably elevated expression levels in rice plants subjected to both salt and drought stress conditions [31]. In soybean, GmCBS14 is specifically expressed in nodule-infected regions and vascular bundles, where it regulates the biological nitrogen fixation capacity of soybean nodules. Furthermore, the overexpression of GmCBS21 and GmCBSDUF3 in Arabidopsis thaliana demonstrated an enhanced tolerance of transgenic plants to low nitrogen stress, drought, and salt stress [32,33]. In wheat, the expression of CBS structural domain-containing proteins was observed to be upregulated in response to flooding [34]. In maize, the overexpression of CBS structural domain-containing proteins has been demonstrated to enhance maize tolerance to a range of abiotic stresses [35]. It has been demonstrated that the expression levels of ZmCBS3 and ZmCBS14 are significantly elevated in response to cold stress, while the expression levels of ZmCBS22 and ZmCBS14 are significantly enhanced in the presence of drought stress. Nevertheless, to date, only a limited number of plant proteins containing CBS structural domains have been functionally characterized, with the majority of members within this family remaining unstudied, particularly in relation to their roles in abiotic stresses.
In this experiment, a genome-wide identification of the grape CBS gene family was carried out using bioinformatic analysis, and its protein physicochemical properties, chromosomal localization, and promoter cis-acting elements were analyzed. In addition, based on the classification of the gene structure and the phylogenetic tree, we focused on the expression analysis of the members of the gene family under different abiotic stresses, aiming at the identification of the key members of the CBS gene family in grapes and providing a theoretical basis for further functional studies.
2. Materials and Methods
2.1. Plant Materials and Stress Treatment
The experimental material was grape ‘Pinot Noir’ test tube plantlets. The stem segments of single bud were cultured in solid GS (modified B5 solid medium) medium and white light LED lamp (PHILIPS, Eindhoven, The Netherlands) in incubator for 16 h, dark for 8 h, and for 30 d, and then the test tube plantlets with strong growth and no pollution were selected for abiotic stress treatment. The grape test tube plantlets used for treatment were completely removed from the culture medium in the ultra-clean workbench, the agar from the roots was washed off with sterile water, and then transferred to GS liquid medium for culture by paper bridge culture [36]. In the treatment group, 400 mM NaCl and 10% PEG were added to GS liquid medium, and the same amount of distilled water was used as control. The grape plantlets were treated at 4 °C in a low-temperature plant incubator (Percival LT36VL, Perry, IA, USA). All treatment groups were cultured for 24 h [37,38]. Three biological repeats were set for each process. All materials were collected from grape test tube plantlets and stored at −80 °C after rapid freezing in liquid nitrogen for RNA extraction and gene expression analysis.
2.2. Identification and Characterization of Grape CBS Genes
The full length, CDS, and protein sequences of the grape CBS gene family were obtained from the plant genome website Phytozome v13 (https://phytozome-next.jgi.doe.gov/, accessed on 16 December 2023) [39]. The grape CBS domains were compared and screened by HMMER online software (https://www.ebi.ac.uk/Tools/hmmer/, accessed on 20 December 2023) to remove the sequences that did not contain CBS-specific domains. The molecular weight (MW), theoretical isoelectric point (pI), amino acid (aa), instability index, aliphatic index, and grand average of hydropathicity (GRAVY) of the VvCBS gene were obtained from the online software ExPASy [40] (https://web.expasy.org/protparam/, accessed on 24 December 2023). Basic chromosome information is obtained from the plant genome website Phytozome v13 (https://phytozome-next.jgi.doe.gov/, accessed on 26 December 2023) and visualized using software TBtools-Ⅱ (Toolbox for Biologists) v2.086.
2.3. Construction and Phylogenetic Analysis of the CBS Gene Family in Grape
The gene structure map was constructed by online software GSDS2.0 (https://gsds.gao-lab.org/index.php, accessed on 29 December 2023). The amino acid sequences of grape, apple, Arabidopsis, and rice were obtained from the plant genome website Phytozome v13 (https://phytozome-next.jgi.doe.gov/, accessed on 16 December 2023). Multiple amino acid sequence alignments were performed using the ClustalX 1.83 software default settings; the phylogenetic tree was constructed using the neighbor-joining method (NJ) in MEGA 5.0 software, and the bootstraping value was set to 1000 [41]. The evolution tree was embellished with ChiPolt online software (https://www.chiplot.online/, accessed on 4 April 2024).
2.4. Collinearity and Protein Interaction Analysis of the CBS Gene Family in Grape
The data were obtained from the plant genome website, and the collinear relationship of the VvCBS gene family was drawn by using the Multiple Synteny Plot tool of TBtools-Ⅱ (Toolbox for Biologists) v2.086. For intergroup covariance analysis, gene density files were obtained using the TBtools software Gene Density Profile tool, and the covariance genes between VvCBS and Arabidopsis AtCBS were visualized by the One Step MCScanX function. The grape chromosome ID and length information was obtained using the TBtools software Fasat Stats tool, and the covariance file between grape CBS genes was obtained using the plug-in One Step MCScanX-Super Fast and visualized in the Advanced Circos tool, specific parameter data reference Wang et al. [42]. Select Arabidopsis as the model plant and set the highest confidence to 0.900. Protein interaction network prediction is performed by the STRING Version 11 (https://cn.string-db.org/, accessed on 10 April 2024) [43].
2.5. Subcellular Localization and Protein Secondary Structure Analysis of CBS Gene in Grape
The online software WoLF PSORT [44] (https://wolfpsort.hgc.jp/, accessed on 29 December 2023) was used to predict the distribution and quantity of VvCBS in each organelle, and TBtools-Ⅱ (Toolbox for Biologists) v2.086 was used for visualization. The secondary structure of VvCBS gene proteins was analyzed by the online tool Prabi (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html, accessed on 15 April 2024).
2.6. Prediction of cis-Acting Elements and Analysis of Tissue Differential Expression Pattern of CBS Genes in Grape
The cis-acting elements of the upstream 2000 bp sequence of the VvCBS gene were obtained by PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 19 April 2024). The obtained VvCBS gene ID was submitted to the grape tissue expression database (https://bar.utoronto.ca/, accessed on 25 April 2024) to obtain the expression amount of the CBS gene in different tissues of grape. The data were further analyzed using Excel 2021 software, and finally, the heatmap was then generated using clustering in the Heml 1.0.3.7 software.
2.7. Gene Expression Analysis by RT-qPCR
The RNA was extracted from grapes by using the E.Z.N.A Plant RNA Kit (OMEGA BIO-TEK, Beijing, China) following the operating instructions. The Reverse Transcriptase M-MLV (RNase H) kit (TaKaRa, Tokyo, Japan) was used to synthesize cDNA. All primers were designed and synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) using CDS sequences. The GAPDH gene (GenBank number, CB973647) was used as an internal reference gene (Table S1). The total reaction volume was 20 μL, including ddH2O 6.5 μL, cDNA 1.5 μL, and 0.2 μM of each primer, TaKaRa SYBR Premix Ex Taq (2x) 10 μL (TaKaRa). The cycling parameters were 95 °C for 30 s, 95 °C for 5 s, and 60 °C for 34 s for a total of 40 cycles. The qPCR was performed using a LightCycle® 96 (Roche, Switzerland) instrument. qPCR amplification and lysis curves were confirmed at the end of the reaction [45]. All reactions were implemented in three biological replicates, and each replicate was measured in triplicate, and representative data are shown as the mean values ± SE. The relative expression profile was calculated using the 2−∆∆CT method [46], relatively to WT set to 1.
2.8. Cloning and Vector Construction of the VvCBS27 Gene
The total RNA from the leaves of ‘Pinot Noir’ was reverse-transcribed into cDNA, and the primers of the cloning vector were designed in SnapGene 1.1.3 software according to the sequence and cleavage site of the target gene (upstream primer: 5′-ATGCAAGGAATTGGCCGAG-3′; downstream primer: 5′-TCAATAGTAATCTCCTTTAATGAACTCGT-3′). Using cDNA as a template, PCR amplification was performed. The amplified target fragment was recovered and purified and connected to the pART-CAM-eGFP cloning vector with the linking system of 5 μL: template (DNA) 4 μL and pART-CAM-eGFP vector 1 μL. The connected product was transformed into a DH5α strain, and a single colony was selected and sent to Sangong Bioengineering (Shanghai, China) Co., Ltd. for sequencing.
2.9. Subcellular Localization of VvCBS27 Protein
Tobacco transient expression transformation refers to Che et al. [47]. The purified cDNA fragment was combined with the pART-CAM-GFP vector to form a fusion protein vector. The constructed expression vector plasmid was transferred into Agrobacterium GV3101 by heat-excitation method, and Agrobacterium tumefaciens infiltration method was used to inject the N. benthamiana leaves, and the positions of tobacco fusion proteins VvCBS27-GFP and GFP that produced fluorescence inside the cell were observed under a laser confocal microscope (Olympus FV1000 Viewer, Tokyo, Japan) to determine the subcellular localization of the VvCBS27 protein.
3. Results
3.1. Analysis of Physicochemical Properties and Chromosome Mapping of the CBS Gene Family in Grape
In this study, a total of 32 non-redundant VvCBS genes were identified from the whole grape genome, and their amino acid sequences all contained CBS-specific structural domains. According to their position on the chromosome, the VvCBS genes were named VvCBS1-VvCBS32 (Table S2). Through the analysis of the physical and chemical characteristics of the VvCBS gene family, it is found that VvCBS genes are clustered on 15 chromosomes, of which the number of VvCBS genes on chromosome 7 is the largest, VvCBS13, VvCBS14, VvCBS15, VvCBS16, and VvCBS17, respectively. VvCBS31 and VvCBS32 genes are distributed on unknown chromosomes. With the exception of VvCBS20 and VvCBS30, the other genes were located in regions with high gene density (Figure 1). The length of the CDS sequence of VvCBS is between 624 bp (VvCBS27) and 2443 bp (VvCBS19). The VvCBS19 protein sequence is the longest (811 amino acids), and the VvCBS27 protein sequence is the shortest (208 amino acids). The molecular weight is between 23,133.56 (VvCBS27) and 89,201.44 Da (VvCBS19), indicating that the molecular weight was quite different. The isoelectric point (pI) indicates that 12 of VvCBS are alkaline proteins and the rest are acidic proteins. The large average values of 18 VvCBS grand average of hydropathicity (GRAVY) are all greater than zero, indicating that they are hydrophobic in nature, and the rest of VvCBS are hydrophilic proteins. The adipose index was between 87.05 (VvCBS27) and 111.12 (VvCBS6). The instability index showed that 18 VvCBS were unstable and 14 VvCBS were stable (Table S2).
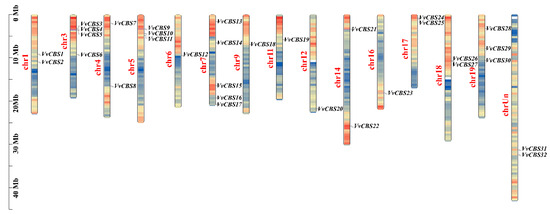
Figure 1.
Distribution of the VvCBS gene on different grape chromosomes. Note: The left scale indicates the chromosome length (Mb), with VvCBS gene markers on the right side of each chromosome. Different chromosomal colors indicate different gene densities, with red indicating the highest density and blue the lowest density.
3.2. Gene Structure and Phylogenetic Analysis of the CBS Gene Family in Grape
The genetic structure was constructed by using the DNA sequence of the VvCBS gene family. The results showed that the number of exons of the VvCBS gene was between 2 and 23, that of VvCBS8, VvCBS22, and VvCBS28 genes was only 2 exons, and that of VvCBS11 was 23 exons. The more similar the genetic structure between the VvCBS gene structures, the closer the theoretical relationship, such as VvCBS and VvCBS32 (Figure 2).
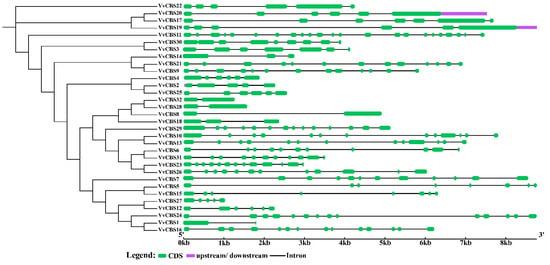
Figure 2.
Gene structure analysis of the CBS gene family in grape. Note: The exon-intron structure of VvCBS genes. Exon CDS, upstream/downstream, and intron were denoted by the green boxes, purple boxes, and black lines, respectively. The scale bar represented 1 kb (right).
In order to better understand the evolutionary relationship between grape and other representative species of CBS gene families, phylogenetic trees were constructed using amino acid sequences of Arabidopsis, rice, apple, and grape (Text S1). The results showed that CBS was obviously divided into 3 subfamilies, including 14 members of the VvCBS family in subfamily Ⅰ, 9 members of the VvCBS family in Ⅱ subfamily, and 9 members of the VvCBS family in Ⅲ subfamily. In this study, it was observed that there were 40 pairs of sideline homologous genes in grape, such as VvCBS20 and AT5G49890.1 (Figure 3).
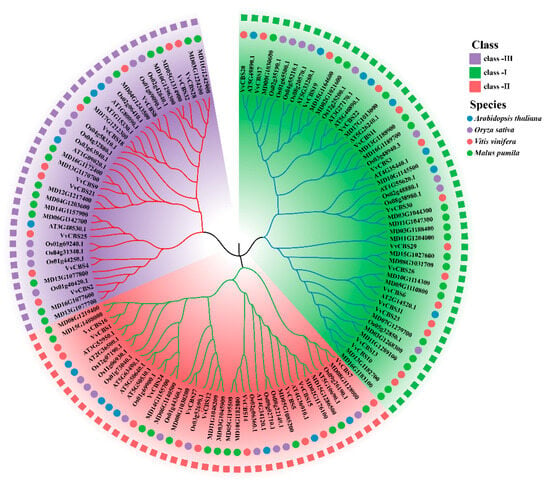
Figure 3.
Evolutionary analysis of VvCBS protein in Arabidopsis thaliana, rice, apple, and grape.
3.3. Subcellular Localization and Secondary Structure Analysis of the CBS Gene Family in Grape
Subcellular localization analysis showed that the VvCBS gene was mainly located in the plasma membrane region, indicating that the gene family may be involved in material transport, cell recognition and information transmission, and other functions (Table S3). Among them, VvCBS25 was mainly located in the peroxisome, VvCBS8 and VvCBS27 were mainly located in the chloroplast, VvCBS32 was mainly located in the nucleus, and VvCBS1, VvCBS10, VvCBS14, VvCBS15, and VvCBS18 were mainly located in the cytoplasm. These results indicate that this gene family plays an important role in plant growth and development (Figure 4A).
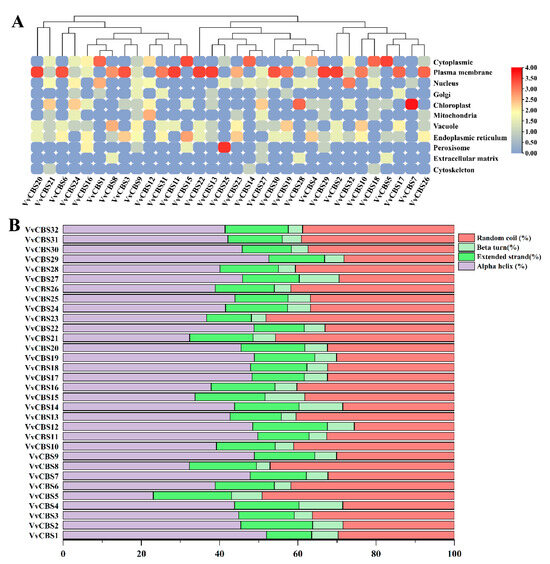
Figure 4.
Prediction of secondary structure and subcellular location of the VvCBS protein. (A) subcellular location prediction of VvCBS protein. Blue and orange colors represent low and high quantities, respectively. (B) the secondary structure of VvCBS protein. Different colors represent different secondary structures.
VvCBS protein secondary structure (Figure 4B) showed that the secondary structure mainly consisted of alpha helix, extended strand, beta turn, and random coil. Among them, the proportion of alpha helix structure was the largest (23.04–52.62%), followed by random coil structure (25.57–49.13%) and extended strand structure (11.48–20.00%), and the proportion of beta turn (3.51–11.18%) was the smallest.
3.4. Collinearity and Protein Interaction Analysis of the CBS Gene Family in Grape
The analysis of covariate relationships (Figure 5A) showed that there were 4 pairs of covariate relationships in grape, and one was on the same chromosome, suggesting that there may be a genetic linkage in the VvCBS22 gene. There were 2 tandem duplication events on chromosomes 3, 5, and 7, and the genes had closer genetic relationships after the tandem duplication events, and these results may provide a reference for functional prediction. In order to gain further insight into the evolutionary relationship of VvCBS genes among different plants, this study employed inter-species covariance analyses of grape, rice, and Arabidopsis thaliana CBS gene families. The results are illustrated in Figure 5B, which shows that 21 covariance gene pairs exist between grape and Arabidopsis thaliana and that the same 7 pairs also exist with rice. The results revealed similarities in the association between grape CBS and monocotyledon rice CBS and dicotyledon Arabidopsis CBS, indicating that grape CBS genes have undergone significant conservation during evolutionary processes.
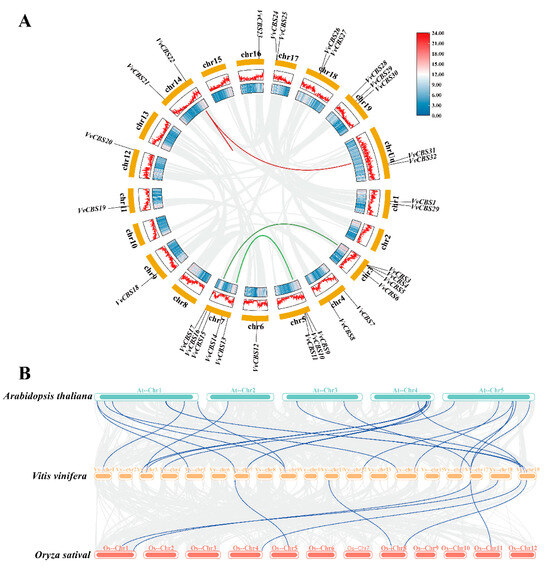
Figure 5.
Chromosomal distribution and collinearity analysis of the CBS genes. (A) Syntenic relationships among synteny blocks carrying the four duplicated VvCBS gene pairs. The annotations on the fragments represent different chromosomes, and the numbers in the outermost circle represent the positions on the corresponding chromosomes. Red and green bars denote synteny blocks that harbored duplicated VvCBS genes. Grey bars in the background indicate all the identified synteny blocks. The colored circles in the outermost circle and the innermost circle represent the gene density. Red indicates a higher density, and blue indicates the lowest density. (B) Collinearity analyses between the CBS genes of Vitis vinifera, Arabidopsis, and rice. The collinear gene pairs with CBS genes between different species were highlighted by the blue lines. The colored boxes indicate the different chromosomes.
The STRING website was used to predict the protein interactions of VvCBS family members (Figure 6). The results show that there may be interactions between 19 VvCBS proteins, and other proteins only have a single function. These 14 VvCBS proteins interact to form two protein interaction networks (PPI networks). It is predicted that the first protein interaction network is mainly related to transport of small molecules, stimuli-sensing channels, and lon channel transport, while VvCBS7, VvCBS14, and VvCBS29 may be related to intracellular membrane-bounded organelles. The second protein interaction network is mainly related to MTOR signaling, signal transduction, and gene expression (transcription).
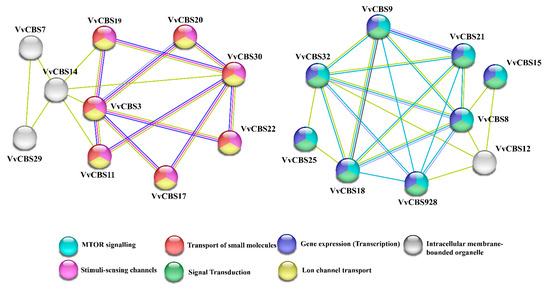
Figure 6.
Analysis of CBS protein interaction in grape. Note: Nodes represent proteins, and different colors represent predicted protein functions. The connections between nodes represent the interactions between proteins, and different colors correspond to different types of interactions.
3.5. Prediction of cis-Acting Elements in the VvCBS Gene Family
To further clarify the potential function of the VvCBS gene family, we analyzed cis-acting elements in the upstream 2000 bp region (Table S4). The results showed that the promoter region of the VvCBS gene was related to hormone response elements, defense and stress response elements, and growth and development related elements. It is worth noting that both VvCBS1 and VvCBS18 promoters enrich hormone response elements, defense and stress response elements, and growth and development-related elements (Figure 7A). The hormone response element includes methyl jasmonate (MeJA) response element, salicylic acid (SA) response element, auxin (IAA) response element, gibberellin (GA), and abscisic acid (ABA) response element. In addition, there are elements related to plant growth and development, including the regulation of zein metabolism regulation, meristem expression, endosperm expression, circadian rhythm, and light response elements, in which light response elements account for a large proportion. We also found many cis-acting elements related to stress conditions, mainly related to plant defense and stress responsiveness, drought-inducibility, low-temperature responsiveness, and anaerobic induction (Figure 7B).
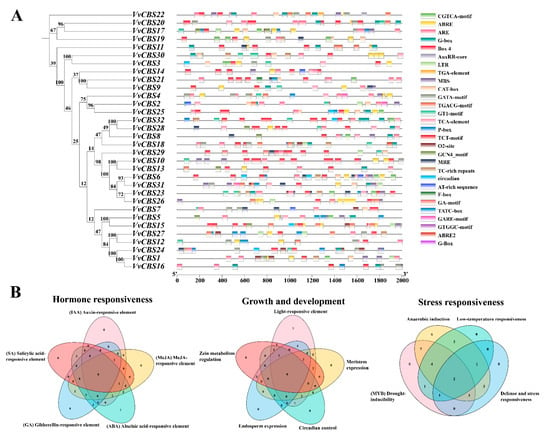
Figure 7.
The first 2000 bp cis-acting element of the VvCBS gene. (A) Analysis of cis-regulatory elements of the VvCBS gene. (B) Venn diagram of the number of cis-acting components of the same. The numbers are the number of cis-acting elements component.
3.6. Analysis of Expression Pattern of CBS Gene Family in Grape
The expression of the VvCBS gene in different grape tissues was analyzed (Table S5). The results showed that 12 VvCBS genes were highly expressed in all grape tissues, and the other 20 VvCBS were expressed at a certain level in different grape tissues. Among them, compared with other grape tissues, the expression level of VvCBS18 in Seed-Veraison and Seed-Mid Ripening was higher, the expression level of VvCBS4 and VvCBS20 in pollen was higher, the expression level of VvCBS23 in root was higher, and the expression level of VvCBS30 in grape tissue was lower. Generally speaking, the VvCBS gene may be closely related to the whole growth and development process of grapes (Figure 8).
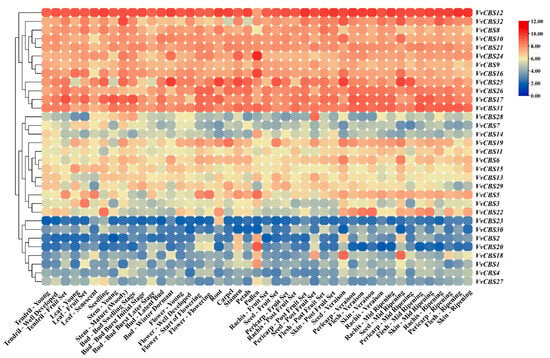
Figure 8.
The tissue differential expression of VvCBS. Red and blue represent up-regulated or down-regulated expression levels, respectively.
3.7. Expression of VvCBS in Grape under Abiotic Stress Treatment
The prediction results of cis-acting elements show that the VvCBS gene may be involved in multiple stress-related cis-acting elements. Therefore, the expression of the VvCBS gene in grapes treated with NaCl, 4 °C, and PEG was studied (Table S6). After 24 h treatment at 4 °C, the expression of all VvCBS members was higher than that of CK, especially VvCBS6, VvCBS13, VvCBS15, and VvCBS23, which were significantly up-regulated under 4 °C treatment. After 24 h treatment at PEG, the expression of VvCBS1, VvCBS5, and VvCBS7 was lower than that of CK, and the expression of other VvCBS members was higher than that of CK, and the expression of VvCBS18, VvCBS21, and VvCBS27 was significantly up-regulated under PEG treatment. After 24 h treatment at NaCl, the expression of all VvCBS members was higher than that of CK, which was positively regulated, and VvCBS1, VvCBS5, VvCBS7, and VvCBS12 were significantly up-regulated under NaCl treatment. The above results showed that the VvCBS gene responded to NaCl, 4 °C, and PEG abiotic stress in grapes (Figure 9).
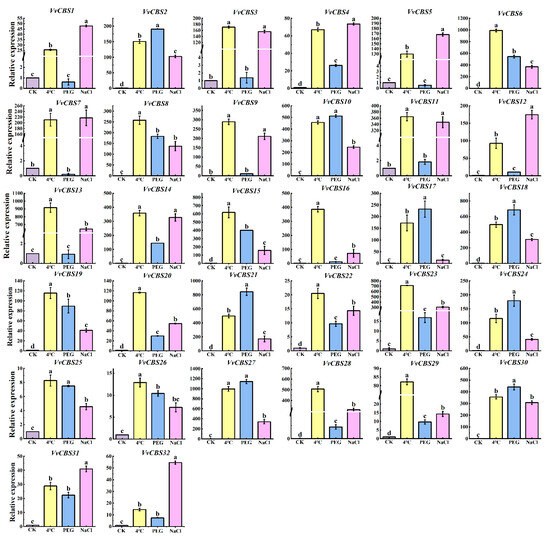
Figure 9.
Expression levels of the VvCBS gene in 4 °C, PEG, and NaCl. Statistical analysis was performed using one-way ANOVA. The expression level of the control group that was not stressed has a value of 1. Black error lines represent the mean ± SE of three biological replicates. Different letters denote significant differences, whereas the same lowercase letters indicate no statistical difference (p < 0.05).
3.8. VvCBS27 Gene Cloning and Subcellular Localization
The VvCBS27 gene was isolated using the cDNA sequence of ‘Pinot Noir’ grape as a template and the VvCBS27-F and VvCBS27-R as primers (Figure S1A). The sequence analysis of VvCBS27 gene showed that the CDS sequence length was 624 bp (Figure S1B).
To verify the localization of VvCBS27 protein in vivo, the instantaneous expression of GFP (green fluorescent protein) labeled fusion protein in tobacco leaves was observed using an Agrobacterium assay (Figure 10). In contrast to the uniform distribution of GFP fluorescence in the control group, the VvCBS27 protein was localized in the cytoplasm, cell membrane, and nucleus.
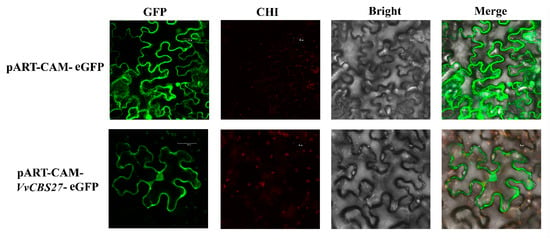
Figure 10.
Subcellular localization of VvCBS27 protein. The GFP fluorescence image, chloroplast autofluorescence (CHI) image, bright field (Bright) image, and the merged GFP, CHI, and Bright images are shown.
4. Discussion
The CBS gene family contains a conserved class of CBS structural domains that are present in proteins of all organisms and regulate plant development and growth by maintaining cellular redox homeostasis, mainly through interaction with the redox regulatory proteins Trxs (thioredoxins) [15,28]. In recent years, the CBS gene family has been identified and analyzed in a few plants, which is of great significance for improving plant resistance and revealing its regulatory mechanism, but the research on the function of CBS proteins in grapevine is basically in a gap. In this study, 32 CBS genes were identified in grape, indicating that the function of CBS genes in grape is relatively conserved. The physicochemical properties show that 62.5% of VvCBS was an acidic amino acid. VvCBS is highly conserved among its members at the protein level. Different members of the VvCBS gene family eventually form mature proteins with similar functions through different transcription and translation processes. There is a pair of tandem repeat genes on chromosome 14, and the collinear relationship further indicates that they are more closely related to each other.
In Arabidopsis, AtCBSX3 was localized to mitochondria and regulates Trx members of the NADP-Trx system within mitochondria [27]. In rice, OsCBSX3 was expressed in the plasma membrane after transient transformation of tobacco leaves [29]. In this study, VvCBS were mainly located in the plasma membrane, and some genes were also located in the mitochondria and nucleus, indicating that the gene family may be involved in material transport, cell recognition, and information transmission.
The expression of some CBS is tissue-specific, and the CBS of some plants will change under external stress. For example, in Arabidopsis, AtCBSX1, AtCBSX2, and AtCBSX3 were highly expressed in pollen and are involved in the development of pollen [26,27]. Based on the data of the maize transcriptome, it was found that the expression of group Ⅱ ZmCBS15 and group Ⅲ ZmCB23 and ZmCB33 in mature pollen was significantly higher than that in other tissues, suggesting that ZmCBS may also be involved in the process of maize pollen development. In addition, some members with only the CBS domain were most highly expressed in embryos (ZmCBS3, ZmCBS9, ZmCBS19, ZmCBS29, ZmCBS30, and ZmCBS32), indicating that they may play a related role in embryonic development [25]. We found that VvCBS18 was highly expressed during seed color conversion and was involved in the seed maturation process. VvCBS4 and VvCBS20 are highly expressed in pollen, which may be closely related to pollen maturation. The expression of VvCBS23 in roots was significantly higher than that in other tissues, indicating that the gene may be involved in the growth and development of grape roots. It is suggested that CBS genes play an important role in the growth and development of grapes.
Low temperature, drought, and saline-alkali are the main environmental stresses that directly affect the growth and development of plants in Northwest China, which are closely related to the yield and quality of grapes [48,49]. During environmental stress, plants must be able to balance endogenous development clues and external signals to coordinate stress responses and development patterns to adapt to the environment throughout the life cycle [50]. The results of promoter element analysis showed that there were many regulatory elements closely related to environmental response in the promoter region of the VvCBS gene, such as methyl jasmonate, abscisic acid, light, hypoxia, drought, and low temperature, indicating that the VvCBS gene plays an important role in the regulation of grape stress.
Previous studies have shown that the expression of AtCBSDUF3, AtCBSCLC7, AtCBSCBSPB4, and AtCBSCBSPB5 in Arabidopsis under cold treatment is up-regulated after 24 h of root stress. Under salt stress, the expression of some CBS genes was up-regulated by oxidative stress at root for 3 h, while almost all differentially expressed CBS genes in bud were up-regulated under oxidative stress. Under drought stress, there was no significant difference in the expression levels of all CBS in roots at different times, while most CBS in stems were up-regulated at 24 h under drought stress [15]. In wheat, the expression of most CBS genes did not change significantly at 5 °C, but the expressions of TaCBS8s, TaCBSCBS1s, and TaCBS10s were significantly up-regulated at −10 °C and −25 °C [22]. In maize, the expression of ZmCBS3 and ZmCBS14 increased significantly under cold stress, and that of ZmCBS22 and ZmCBS14 increased significantly under drought stress, indicating that these genes play a role in the process of drought and low temperature stress [25]. In rice, OsCBSX4 overexpressed in tobacco, a model plant, showed the characteristics of antioxidation, salt resistance, and heavy metal resistance [30]. In addition, phenotypic analysis of GmCBSDUF3 transgenic Arabidopsis was carried out in soybeans under NaCl, PEG, and ABA stress. Overexpression of GmCBSDUF3 can enhance the tolerance of Arabidopsis to drought and salt stress [33].
Similar results were obtained in grape, where VvCBS6, VvCBS13, VvCBS15, and VvCBS23 were significantly up-regulated under 4 °C treatment. The expression of VvCBS18, VvCBS21, and VvCBS27 was significantly up-regulated after PEG treatment for 24 h. The expression of VvCBS1, VvCBS5, VvCBS7, and VvCBS12 was significantly up-regulated after 24 h of NaCl treatment. This indicates that VvCBS genes can respond to abiotic stress. These results revealed the complexity of VvCBS gene regulation and laid the foundation for further studies on the function of grape CBS genes.
In conclusion, in view of the diversity and important functions of CBS genes, it is necessary to conduct systematic and detailed functional studies of the CBS gene family in grape. Through analysis, we found that the CBS gene plays an important role in grape response to abiotic stress and development, which lays a foundation for the functional analysis of the CBS gene in grape. The study of the grape CBS gene family has important theoretical and practical significance for further understanding the characteristics of plant CBS genes and improving grape stress resistance by means of genetic engineering.
5. Conclusions
A total of 32 VvCBS were identified in the grape genome through bioinformatics analysis. These VvCBS genes could be classified into three subfamilies based on their amino acid numbers (ranging from 208 to 811), molecular masses (ranging from 23.13 to 89.20 kDa), and isoelectric points. Of these, 12 were found to be basic proteins, while the remaining 20 were acidic proteins. The results of RT-qPCR analysis indicated that VvCBS family members play a crucial role in regulating the stress response to 4 °C, PEG, and NaCl. The VvCBS27 gene was successfully cloned, and subcellular localization showed that VvCBS27 was located in the plasma membrane and nucleus.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae10091009/s1, Table S1: Design sequence of RT-qPCR primers for VvCBS gene family. Table S2: Physicochemical properties of the VvCBS gene family. Table S3: Subcellular location prediction of VvCBS protein. Table S4: cis-acting elements analysis of the VvCBS promoter region. Table S5: Hierarchical clustering of VvCBS gene expression profiles in different organs of grape. Table S6: Expression levels of VvCBS gene in 4 °C, PEG, and NaCl. Text S1: Sequence files used in evolutionary trees. Figure S1: Cloning of the VvCBS27 gene. (A) Gene clone of VvCBS27. (B) PCR detection of Escherichia coli.
Author Contributions
Conceived and designed the experiments: Y.Y. Conducted experimentation and drafting: X.S. and S.X. Analyzed the data: Y.Y. and X.S. Wrote the paper: X.S. and S.X. Revised the paper: S.X. and Y.L. retrieve and data analysis. X.S., S.X. and Y.L. revised the manuscript and technical input. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by the Huzhou South Taihu Lake Innovation Talent Program (2021RC018).
Data Availability Statement
Data is contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bateman, A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem. Sci. 1997, 22, 12–13. [Google Scholar] [CrossRef] [PubMed]
- Ignoul, S.; Eggermont, J. CBS domains: Structure, function, and pathology in human proteins. Am. J. Physiol. Cell Physiol. 2005, 289, C1369–C1378. [Google Scholar] [CrossRef] [PubMed]
- Ereño-Orbea, J.; Oyenarte, I.; Martínez-Cruz, L.A. CBS domains: Ligand binding sites and conformational variability. Arch. Biochem. Biophys. 2013, 540, 70–81. [Google Scholar] [CrossRef]
- Anashkin, V.A.; Baykov, A.A.; Lahti, R. Enzymes regulated via cystathionine β-synthase domains. Biochemistry 2017, 82, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Kemp, B.E. Bateman domains and adenosine derivatives form a binding contract. J. Clin. Investig. 2004, 113, 182–184. [Google Scholar] [CrossRef]
- Zhang, R.; Evans, G.; Rotella, F.J.; Westbrook, E.M.; Beno, D.; Huberman, E.; Joachimiak, A.; Collart, F.R. Characteristics and crystal structure of bacterial inosine-5 ‘-monophosphate dehydrogenase. Biochemistry 1999, 38, 4691–4700. [Google Scholar] [CrossRef]
- Baykov, A.A.; Tuominen, H.K.; Lahti, R. The CBS domain: A protein module with an emerging prominent role in regulation. ACS Chem. Biol. 2011, 6, 1156–1163. [Google Scholar] [CrossRef]
- Hackenberg, C.; Hakanpää, J.; Cai, F.; Antonyuk, S.; Eigner, C.; Meissner, S.; Laitaoja, M.; Jänis, J.; Kerfeld, C.A.; Dittmann, E.; et al. Structural and functional insights into the unique CBS-CP12 fusion protein family in cyanobacteria. Proc. Natl. Acad. Ences 2018, 115, 7141–7146. [Google Scholar] [CrossRef]
- Xiao, B.; Heath, R.; Saiu, P.; Leiper, F.C.; Leone, P.; Jing, C.; Walker, P.A.; Haire, L.; Eccleston, J.F.; Davis, C.T.; et al. Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature 2007, 449, 496–500. [Google Scholar] [CrossRef]
- Yoo, K.S.; Ok, S.H.; Jeong, B.C.; Jung, K.W.; Cui, M.H.; Hyoung, S.; Lee, M.R.; Song, H.K.; Shin, J.S. Single cystathionine β-synthase domain–containing proteins modulate development by regulating the thioredoxin system in Arabidopsis. Plant Cell 2011, 23, 3577–3594. [Google Scholar] [CrossRef]
- Ke, X.; Xiao, H.; Peng, Y.; Wang, J.; Lv, Q.; Wang, X. Phosphoenolpyruvate reallocation links nitrogen fixation rates to root nodule energy state. Science 2022, 378, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Sintchak, M.D.; Fleming, M.A.; Futer, O.; Raybuck, S.A.; Chambers, S.P.; Caron, P.R.; Murcko, M.A.; Wilson, K.P. Structure and mechanism of inosine monophosphate dehydrogenase in complex with the immunosuppressant mycophenolic acid. Cell 1996, 85, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Buey, R.M.; Fernández-Justel, D.; Marcos-Alcalde, Í.; Winter, G.; Gómez-Puertas, P.; de Pereda, J.M.; Luis Revuelta, J. A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases. Sci. Rep. 2017, 7, 2648. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Rose, T.; Jentsch, T.J. Reconstitution of functional voltage-gated chloride channels from complementary fragments of CLC-1. J. Biol. Chem. 1997, 272, 20515–20521. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, H.R.; Singh, A.K.; Sopory, S.K.; Singla-Pareek, S.L.; Pareek, A. Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genomics 2009, 10, 200. [Google Scholar] [CrossRef]
- Crozet, P.; Margalha, L.; Confraria, A.; Rodrigues, A.; Martinho, C.; Adamo, M.; Elias, C.A.; Baena-González, E. Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Front. Plant Sci. 2014, 5, 190. [Google Scholar] [CrossRef] [PubMed]
- Shahbaaz, M.; Ahmad, F.; Imtaiyaz Hassan, M. Structure-based functional annotation of putative conserved proteins having lyase activity from Haemophilus influenzae. 3 Biotech 2015, 5, 317–336. [Google Scholar] [CrossRef]
- Scott, J.W.; Hawley, S.A.; Green, K.A.; Anis, M.; Stewart, G.; Scullion, G.A.; Norman, D.G.; Hardie, D.G. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J. Clin. Investig. 2004, 113, 274–284. [Google Scholar] [CrossRef]
- Giménez-Mascarell, P.; González-Recio, I.; Fernández-Rodríguez, C.; Oyenarte, I.; Müller, D.; Martínez-Chantar, M.L.; Martínez-Cruz, L.A. Current structural knowledge on the CNNM family of magnesium transport mediators. Int. J. Mol. Sci. 2019, 20, 1135. [Google Scholar] [CrossRef]
- Xiao, B.; Sanders, M.J.; Underwood, E.; Heath, R.; Mayer, F.V.; Carmena, D.; Jing, C.; Walker, P.A.; Eccleston, J.F.; Haire, L.F.; et al. Structure of mammalian AMPK and its regulation by ADP. Nature 2011, 472, 230–233. [Google Scholar] [CrossRef]
- Hardie, D.G.; Hawley, S.A. AMP-activated protein kinase: The energy charge hypothesis revisited. Bioessays 2001, 23, 1112–1119. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.Y.; Cang, J.; Lu, Q.W.; Tian, Y.; Song, C.H.; Ren, Z.P.; Sun, X.Z.; Fan, X.P.; Wang, D.J.; Fu, L.S. Genome-Wide Analysis of CBS gene Family in Hexaploid Wheat. J. Triticeae Crops. 2020, 23, 421–433. (In Chinese) [Google Scholar]
- Liu, H.; Wang, Q.; Xie, L.; Xu, K.; Zhang, F.; Ruan, X.; Li, L.; Tan, G. Genome-wide identification of cystathionine beta synthase genes in wheat and its relationship with anther male sterility under heat stress. Front. Plant Sci. 2022, 13, 1061472. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.R.; Li, J.; Lin, J.X.; Xu, H.M.; Chu, N.; Wang, Q.N.; Gao, S.J. Genome-wide characterization of cys-tathionine-β-synthase domain-containing proteins in sugarcane reveals their role in defense responses under multiple stressors. Front. Plant Sci. 2022, 13, 985653. [Google Scholar] [CrossRef]
- Chen, Z.Q.; Li, G.; Yan, J.W.; Guo, X.R.; Hu, C.Q.; Wang, Y.; Wang, F.; Tian, D.G. Identification and characterisation of CBS gene family in maize. Jiangsu Agric. Sci 2024, 52, 41–49. (In Chinese) [Google Scholar]
- Jung, K.W.; Kim, Y.Y.; Yoo, K.S.; Ok, S.H.; Cui, M.H.; Jeong, B.C.; Yoo, S.D.; Jeung, J.U.; Shin, J.S. A cystathionine-β-synthase domain-containing protein, CBSX2, regulates endothecial secondary cell wall thickening in anther development. Plant Cell Physiol. 2013, 54, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.S.; So, W.M.; Kim, S.Y.; Noh, M.; Hyoung, S.; Yoo, K.S.; Shin, J.S. CBSX3-Trxo-2 regulates ROS generation of mitochondrial complex II (succinate dehydrogenase) in Arabidopsis. Plant Sci. 2020, 294, 110458. [Google Scholar] [CrossRef]
- Wang, X.; Ren, X.; Zhu, L.; He, G.C. OsBi1, a rice gene, encodes a novel protein with a CBS-like domain and its expression is induced in responses to herbivore feeding. Plant Sci. 2004, 166, 1581–1588. [Google Scholar] [CrossRef]
- Mou, S.; Shi, L.; Lin, W.; Liu, Y.; Shen, L.; Guan, D.; He, S. Over-expression of rice CBS domain containing protein, OsCBSX3, confers rice resistance to Magnaporthe oryzae inoculation. Int. J. Mol. Sci. 2015, 16, 15903–15917. [Google Scholar] [CrossRef]
- Singh, A.K.; Kumar, R.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Overexpression of rice CBS domain containing protein improves salinity, oxidative, and heavy metal tolerance in transgenic tobacco. Mol. Biotechnol. 2012, 52, 205–216. [Google Scholar] [CrossRef]
- Tomar, S.; Subba, A.; Bala, M.; Singh, A.K.; Pareek, A.; Singla-Pareek, S.L. Genetic conservation of CBS domain containing protein family in Oryza species and their association with abiotic stress responses. Int. J. Mol. Sci. 2022, 23, 1687. [Google Scholar] [CrossRef] [PubMed]
- Hao, Q.; Shang, W.; Zhang, C.; Chen, H.; Chen, L.; Yuan, S.; Chen, S.; Zhang, X.; Zhou, X. Identification and comparative analysis of CBS domain-containing proteins in soybean (Glycine max) and the primary function of GmCBS21 in enhanced tolerance to low nitrogen stress. Int. J. Mol. Sci. 2016, 17, 620. [Google Scholar] [CrossRef] [PubMed]
- Hao, Q.; Yang, Y.; Shan, Z.; Chen, H.; Zhang, C.; Chen, L.; Yuan, S.; Zhang, X.; Chen, S.; Yang, Z.; et al. Genome-wide investigation and expression profiling under abiotic stresses of a soybean unknown function (DUF21) and cystathionine-β-synthase (CBS) domain-containing protein family. Biochem. Genet. 2021, 59, 83–113. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Li, M.; Harrison, M.T.; Fahad, S.; Wei, M.; Li, X.; Yin, L.; Sha, A.; Zhou, M.; Liu, K.; et al. iTRAQ proteomic analysis of Wheat (Triticum aestivum L.) genotypes differing in waterlogging tolerance. Front. Plant Sci. 2022, 13, 890083. [Google Scholar] [CrossRef]
- Dong, A.; Yang, Y.; Liu, S.; Zenda, T.; Liu, X.; Wang, Y.; Duan, H. Comparative proteomics analysis of two maize hybrids revealed drought-stress tolerance mechanisms. Biotechnol. Biotechnol. Equip. 2020, 34, 763–780. [Google Scholar] [CrossRef]
- Che, L.; Lu, S.; Liang, G.; Gou, H.; Li, M.; Chen, B.; Mao, J. Identification and expression analysis of the grape pentatricopeptide repeat (PPR) gene family in abiotic stress. Physiol. Mol. Biol. Plants. 2022, 28, 1849–1874. [Google Scholar] [CrossRef]
- Airaki, M.; Leterrier, M.; Mateos, R.M.; Valderrama, R.; Chaki, M.; Barroso, J.B.; Del Río, L.A.; Palma, J.M.; Corpas, F.J. Metabolism of reactive oxygen species and reactive nitrogen species in pepper (Capsicum annuum L.) plants under low temperature stress. Plant Cell Environ. 2012, 35, 281–295. [Google Scholar] [CrossRef]
- Basal, O.; Szabó, A.; Veres, S. Physiology of soybean as affected by PEG-induced drought stress. Curr. Plant Biol. 2020, 22, 100135. [Google Scholar] [CrossRef]
- Li, F.; Deng, Y.; Yamamoto, E.; Zhenya, L. Plant omics databases: An online resource guide. In Plant Omics: Advances in Big Data Biology; CABI: Wallingford, UK, 2022; pp. 253–269. [Google Scholar]
- Artimo, P.; Jonnalagedda, M.; Arnold, K. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Ayaz, A.; Huang, H.; Zheng, M.; Zaman, W.; Li, D.; Saqib, S.; Zhao, H.; Lü, S. Molecular cloning and functional analysis of GmLACS2-3 reveals its involvement in cutin and suberin biosynthesis along with abiotic stress tolerance. Int. J. Mol. Sci. 2021, 22, 9175. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.D.; Gao, T.; Chen, J.F.; Yang, J.K.; Huang, H.Y.; Yu, Y.B. The late embryogenesis abundant gene family in tea plant (Camellia sinensis): Genome-wide characterization and expression analysis in response to cold and dehydration stress. Plant Physiol. Bioch. 2018, 135, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.P.; Hou, Y.; Wang, H.; Wang, P.; Mao, J.; Hong, B. VaBAM1 weakens cold tolerance by interacting with the negative regulator VaSR1 to suppress β-amylase expression. Int. J. Biol. Macromol. 2023, 225, 1394–1404. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Che, L.; Lu, S.; Gou, H.; Li, M.; Guo, L.; Yang, J.; Mao, J. VvJAZ13 positively regulates cold tolerance in Arabidopsis and grape. Int. J. Mol. Sci. 2024, 25, 4458. [Google Scholar] [CrossRef] [PubMed]
- Hwarari, D.; Guan, Y.L.; Ahmad, B.; Movahedi, A.; Min, T.; Hao, Z.D.; Lu, Y.; Chen, J.H.; Yang, L.M. ICE-CBF-COR signaling cascade and its regulation in plants responding to cold stress. Int. J. Mol. Sci. 2022, 23, 1549. [Google Scholar] [CrossRef]
- Fraga, H.; Santos, J.A.; Moutinho-Pereira, J.; Carlos, C.; Silvestre, J.; Eiras-Dias, J.; Mota, T.; Malheiro, A.C. Statistical modelling of grapevine phenology in Portuguese wine regions: Observed trends and climate change projections. J. Agric. Sci. 2016, 154, 795–811. [Google Scholar] [CrossRef]
- Chen, L.P.; Zhao, Y.; Xu, S.J.; Zhang, Z.Y.; Xu, Y.Y.; Zhang, J.Y.; Chong, K. OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol. 2018, 218, 219–231. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).