Functional Identification of the Isopentenyl Diphosphate Isomerase Gene from Fritillaria unibracteata
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Strain and Plasmid
2.1.2. Reagents and Plant Materials
2.1.3. Experimental Apparatus
2.2. Experimental Methods
2.2.1. Construction of pTrc-FuIPI Expression Plasmid and Expression Host
2.2.2. Color Complementation Experiment
2.2.3. Determination of β-Carotene Content
2.2.4. Site Mutation at Glu190 Residue
2.2.5. Subcellular Localization of FuIPI Protein
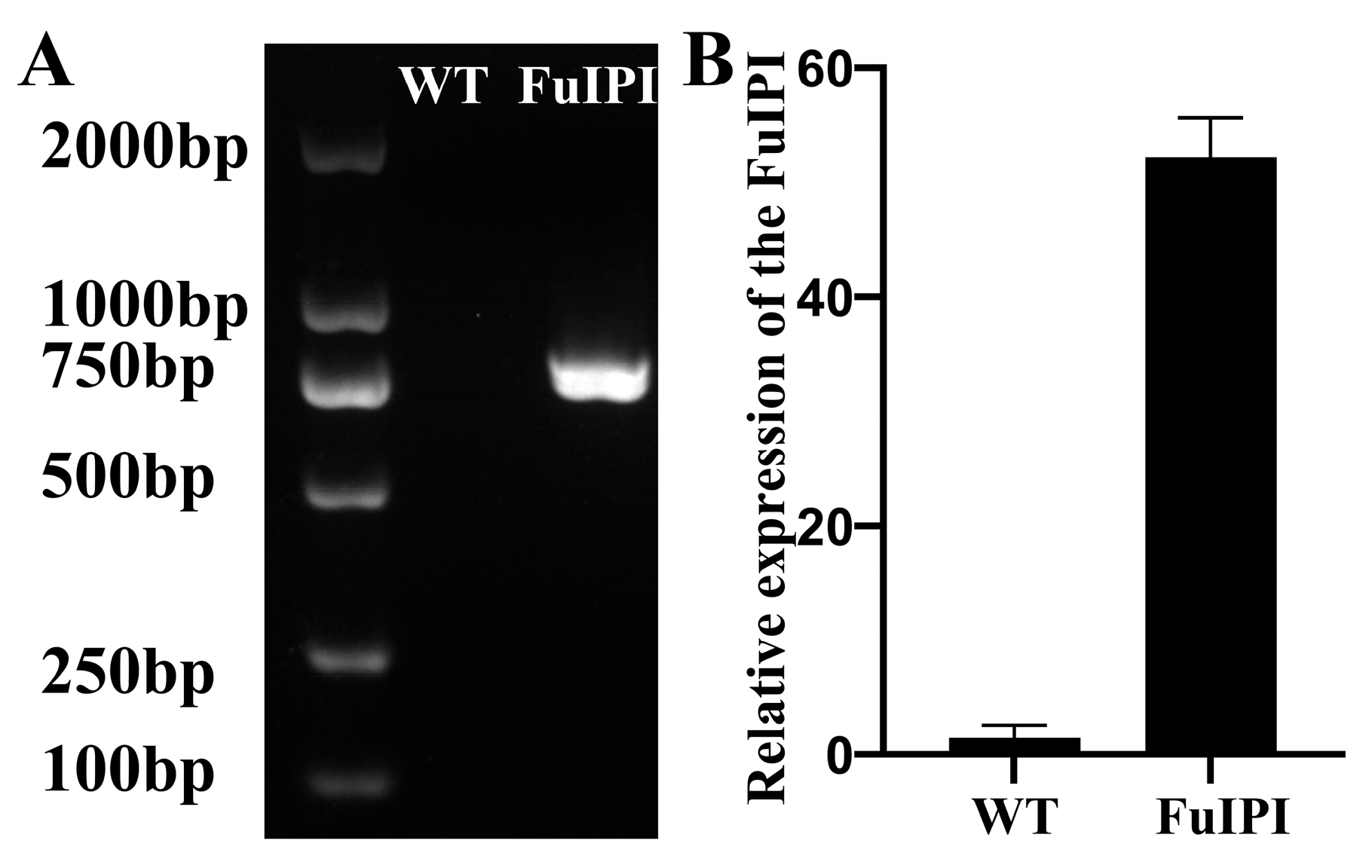
2.2.6. Construction and Identification of FuIPI Transgenic A. thaliana
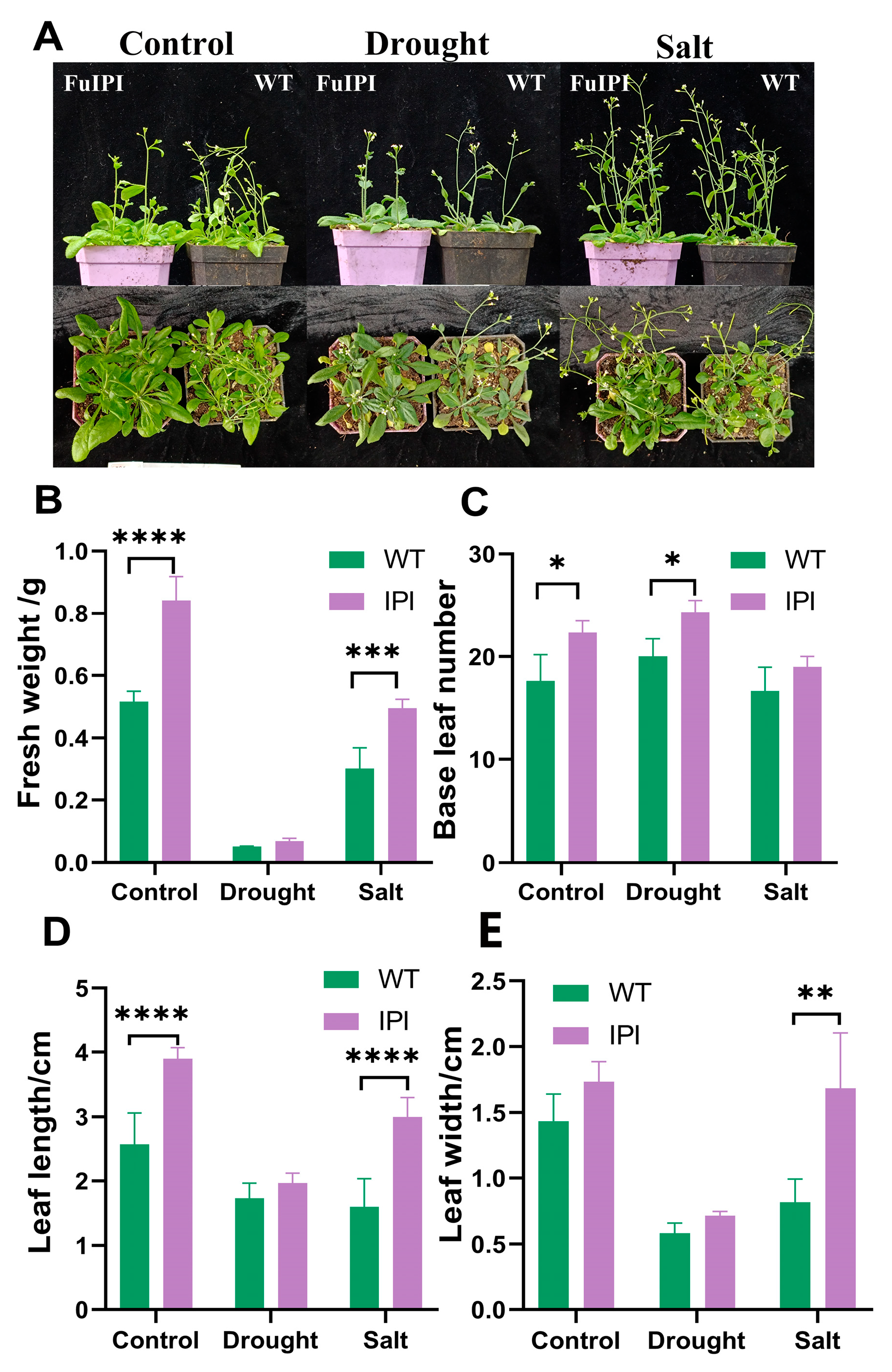
2.2.7. Phenotypic Observation of FuIPI Transgenic Plants under Stress
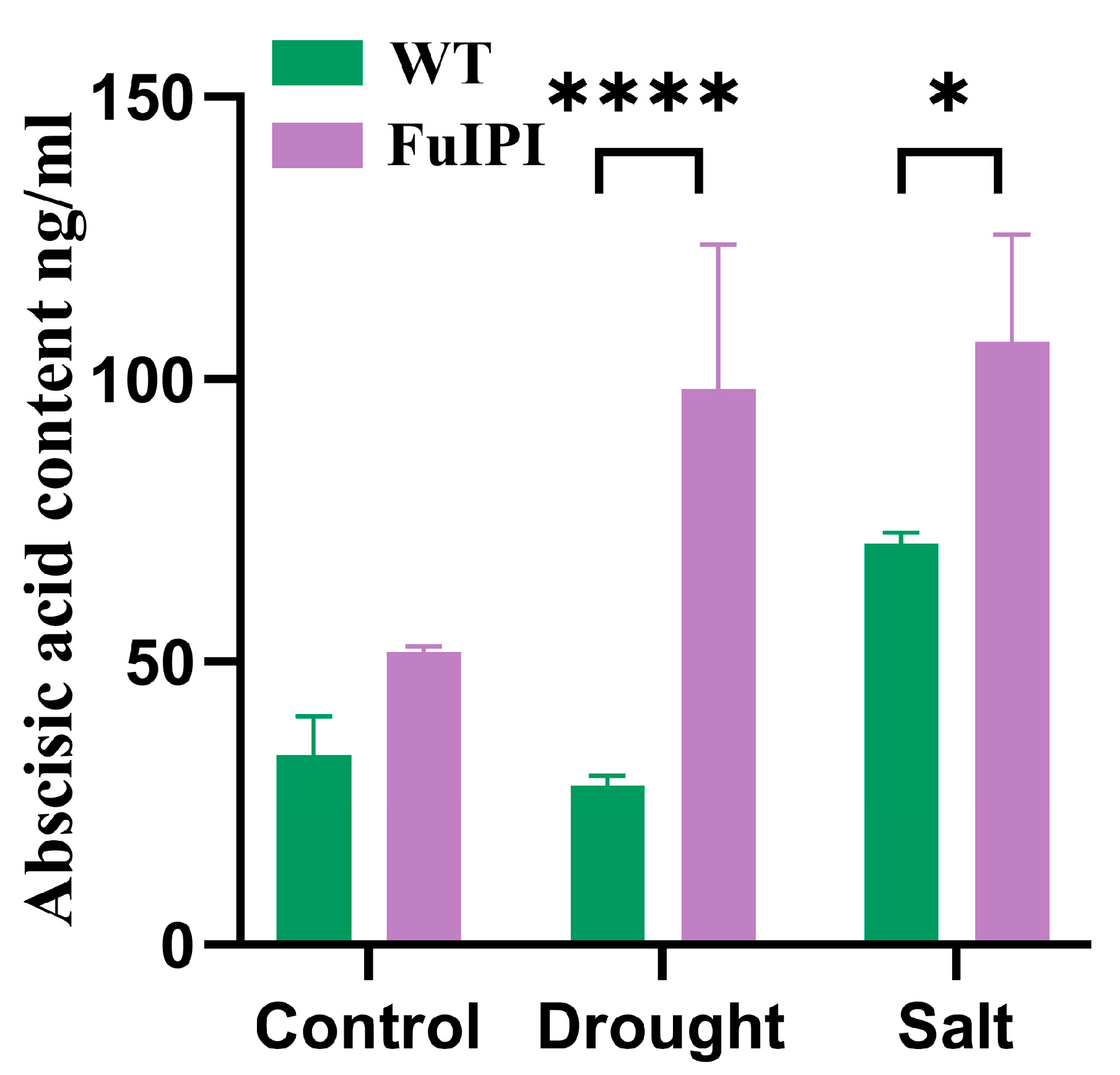
2.2.8. Determination of Abscisic Acid (ABA) Content
3. Result
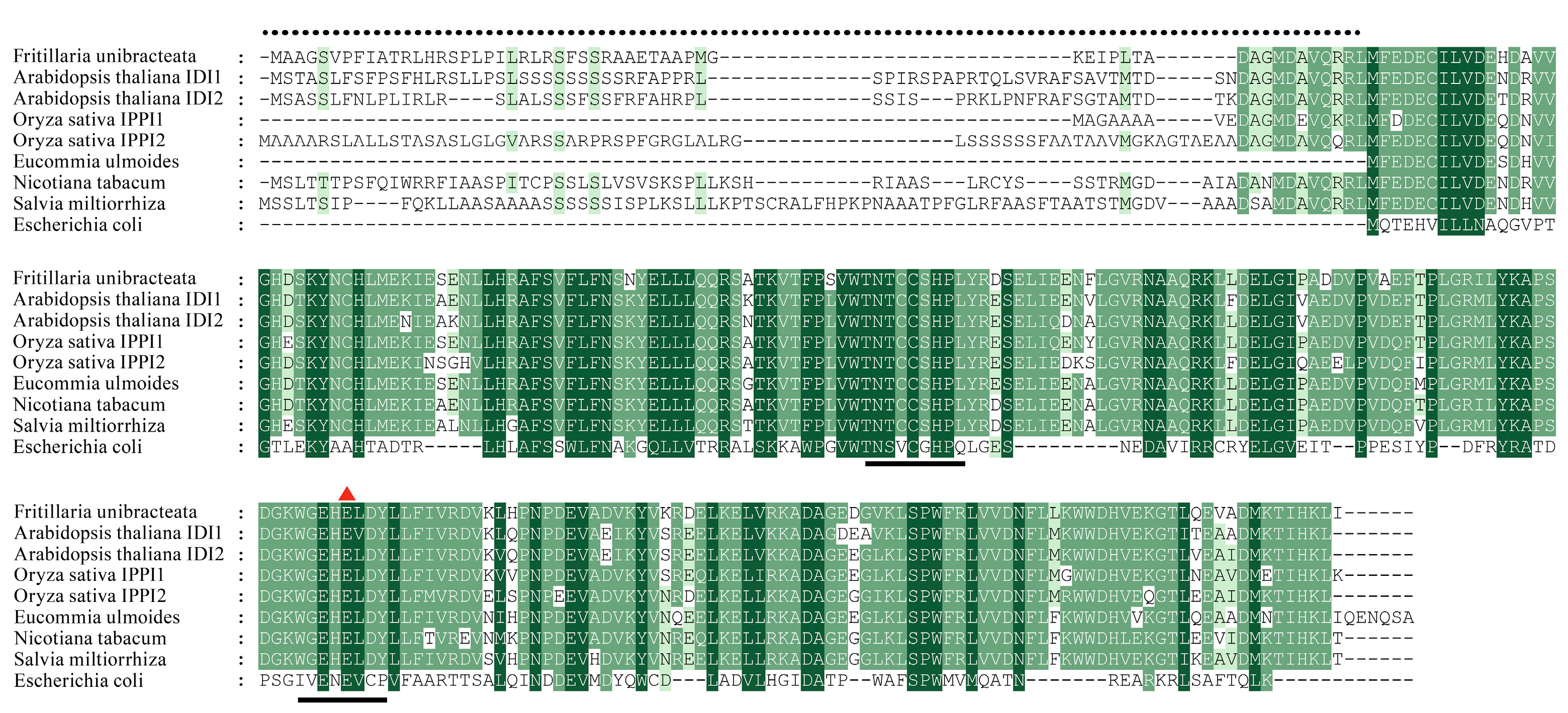
3.1. Functional Identification of FuIPI Gene in E. coli
3.2. Subcellular Localization of FuIPI
3.3. Production of FuIPI Transgenic Plants
3.4. FuIPI Transgenic A. thaliana Strengthened Tolerance to Drought and Salinity Stress
3.5. Overexpression of the FuIPI Gene Enhanced the Accumulation of ABA Content
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, T.; Huang, S.; Song, S.; Zou, M.; Yang, T.; Wang, W.; Zhou, J.; Liao, H. Identification of evolutionary relationships and DNA markers in the medicinally important genus Fritillaria based on chloroplast genomics. PeerJ 2021, 9, e12612. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, M.; Yang, T.; Wai Ming, T.; Wai Gaun, T.K.; Ye, B. LC-MS/MS coupled with chemometric analysis as an approach for the differentiation of bulbus Fritillaria unibracteata and Fritillaria ussuriensis. Phytochem. Anal. 2021, 32, 957–969. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.; Li, P.; Li, S.L.; Chan, S.W. Chromatographic analysis of Fritillaria isosteroidal alkaloids, the active ingredients of Beimu, the antitussive traditional Chinese medicinal herb. J. Chromatogr. A 2001, 935, 321–338. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zong, J.F.; Yili, A.; Yu, M.H.; Aisa, H.A.; Hou, A.J. Isosteroidal alkaloids from the bulbs of Fritillaria tortifolia. Fitoterapia 2018, 131, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hou, H.; Ren, Q.; Hu, H.; Yang, T.; Li, X. Natural drug sources for respiratory diseases from Fritillaria: Chemical and biological analyses. Chin. Med. 2021, 16, 40. [Google Scholar] [CrossRef]
- Zhang, Y.; Han, H.; Li, D.; Fan, Y.; Liu, M.; Ren, H.; Liu, L. Botanical characterization, phytochemistry, biosynthesis, pharmacology clinical application, and breeding techniques of the Chinese herbal medicine Fritillaria unibracteata. Front. Pharmacol. 2024, 15, 1428037. [Google Scholar] [CrossRef]
- Zhao, D.; Wang, J.; Dai, W.; Ye, K.; Chen, J.; Lai, Q.; Li, H.; Zhong, B.; Yu, X. Effects of climate warming and human activities on the distribution patterns of Fritillaria unibracteata in eastern Qinghai-Tibetan Plateau. Sci. Rep. 2023, 13, 15770. [Google Scholar] [CrossRef]
- Jiang, R.; Zou, M.; Qin, Y.; Tan, G.; Huang, S.; Quan, H.; Zhou, J.; Liao, H. Modeling of the Potential Geographical Distribution of Three Fritillaria Species Under Climate Change. Front. Plant Sci. 2022, 12, 749838. [Google Scholar] [CrossRef]
- Liao, H.; Quan, H.; Huang, B.; Ji, H.; Zhang, T.; Chen, J.; Zhou, J. Integrated transcriptomic and metabolomic analysis reveals the molecular basis of tissue-specific accumulation of bioactive steroidal alkaloids in Fritillaria unibracteata. Phytochemistry 2023, 214, 113831. [Google Scholar] [CrossRef]
- Moreno, J.C.; Mi, J.; Agrawal, S.; Kössler, S.; Turečková, V.; Tarkowská, D.; Thiele, W.; Al-Babili, S.; Bock, R.; Schöttler, M.A. Expression of a carotenogenic gene allows faster biomass production by redesigning plant architecture and improving photosynthetic efficiency in tobacco. Plant J. 2020, 103, 1967–1984. [Google Scholar] [CrossRef]
- Tian, C.; Quan, H.; Jiang, R.; Zheng, Q.; Huang, S.; Tan, G.; Yan, C.; Zhou, J.; Liao, H. Differential roles of Cassia tora 1-deoxy-D-xylulose-5-phosphate synthase and 1-deoxy-D-xylulose-5-phosphate reductoisomerase in trade-off between plant growth and drought tolerance. Front. Plant Sci. 2023, 14, 1270396. [Google Scholar] [CrossRef]
- Liao, Z.; Chen, M.; Yang, Y.; Yang, C.; Fu, Y.; Zhang, Q.; Wang, Q. A new isopentenyl diphosphate isomerase gene from sweet potato: Cloning, characterization and color complementation. Biologia 2008, 63, 221–226. [Google Scholar] [CrossRef]
- Zhang, X.; Guan, H.; Dai, Z.; Guo, J.; Shen, Y.; Cui, G.; Gao, W.; Huang, L. Functional Analysis of the Isopentenyl Diphosphate Isomerase of Salvia miltiorrhiza via Color Complementation and RNA Interference. Molecules 2015, 20, 20206–20218. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.W.; Oh, M.K.; Liao, J.C. Engineered isoprenoid pathway enhances astaxanthin production in Escherichia coli. Biotechnol. Bioeng. 1999, 62, 235–241. [Google Scholar] [CrossRef]
- Brüggemann, N.; Schnitzler, J.P. Relationship of isopentenyl diphosphate (IDP) isomerase activity to isoprene emission of oak leaves. Tree Physiol. 2002, 22, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Harada, Y.; Bamba, T.; Nakazawa, Y.; Gyokusen, K. Overexpression of an isopentenyl diphosphate isomerase gene to enhance trans-polyisoprene production in Eucommia ulmoides Oliver. BMC. Biotechnol. 2012, 12, 78. [Google Scholar] [CrossRef]
- Chen, J.; Song, S.; Tang, J.; Xin, J.; Zhang, Q.; Zhang, H.; Chen, X.; Zhou, J.Y.; Liao, H. Cloning and functional analysis of IPI gene from Fritillaria unibracteata Hsiao et K.C.Hsia. Acta Pharm. Sin. 2023, 58, 447–453. [Google Scholar] [CrossRef]
- Song, S.; Chen, A.; Zhu, J.; Yan, Z.; An, Q.; Zhou, J.; Liao, H.; Yu, Y. Structure basis of the caffeic acid O-methyltransferase from Ligusiticum chuanxiong to understand its selective mechanism. Int. J. Biol. Macromol. 2022, 194, 317–330. [Google Scholar] [CrossRef]
- Qin, Y.; Li, Q.; An, Q.; Li, D.; Huang, S.; Zhao, Y.; Chen, W.; Zhou, J.; Liao, H. A phenylalanine ammonia lyase from Fritillaria unibracteata promotes drought tolerance by regulating lignin biosynthesis and SA signaling pathway. Int. J. Biol. Macromol. 2022, 213, 574–588. [Google Scholar] [CrossRef]
- Flowerika; Alok, A.; Kumar, J.; Thakur, N.; Pandey, A.; Pandey, A.K.; Upadhyay, S.K.; Tiwari, S. Characterization and Expression Analysis of Phytoene Synthase from Bread Wheat (Triticum aestivum L.). PLoS ONE 2016, 11, e0162443. [Google Scholar] [CrossRef]
- Yang, J.; Adhikari, M.N.; Liu, H.; Xu, H.; He, G.; Zhan, R.; Wei, J.; Chen, W. Characterization and functional analysis of the genes encoding 1-deoxy-D-xylulose-5-phosphate reductoisomerase and 1-deoxy-D-xylulose-5-phosphate synthase, the two enzymes in the MEP pathway, from Amomum villosum Lour. Mol. Biol. Rep. 2012, 39, 8287–8296. [Google Scholar] [CrossRef]
- Cunningham, F.X., Jr.; Gantt, E. A portfolio of plasmids for identification and analysis of carotenoid pathway enzymes: Adonis aestivalis as a case study. Photosynth. Res. 2007, 92, 245–259. [Google Scholar] [CrossRef]
- Okada, K.; Kasahara, H.; Yamaguchi, S.; Kawaide, H.; Kamiya, Y.; Nojiri, H.; Yamane, H. Genetic evidence for the role of isopentenyl diphosphate isomerases in the mevalonate pathway and plant development in Arabidopsis. Plant Cell. Physiol. 2008, 49, 604–616. [Google Scholar] [CrossRef]
- Llamas, E.; Pulido, P.; Rodriguez-Concepcion, M. Interference with plastome gene expression and Clp protease activity in Arabidopsis triggers a chloroplast unfolded protein response to restore protein homeostasis. PLoS. Genet. 2017, 13, e1007022. [Google Scholar] [CrossRef]
- Yu, T.; Yang, Y.; Wang, H.; Qian, W.; Hu, Y.; Gao, S.; Liao, H. The Variations of C/N/P Stoichiometry, Endogenous Hormones, and Non-Structural Carbohydrate Contents in Micheliamaudiae ‘Rubicunda’ Flower at Five Development Stages. Horticulturae 2023, 9, 1198. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, L.; Xu, H.; Wei, Z.; Wang, Y.; Lin, Y.; Gong, W. Crystal structures of human IPP isomerase: New insights into the catalytic mechanism. J. Mol. Biol. 2007, 366, 1437–1446. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Baysal, C.; Gao, L.; Medina, V.; Drapal, M.; Ni, X.; Sheng, Y.; Shi, L.; Capell, T.; Fraser, P.D.; et al. The subcellular localization of two isopentenyl diphosphate isomerases in rice suggests a role for the endoplasmic reticulum in isoprenoid biosynthesis. Plant Cell Rep. 2020, 39, 119–133. [Google Scholar] [CrossRef]
- Phillips, M.A.; D’Auria, J.C.; Gershenzon, J.; Pichersky, E. The Arabidopsis thaliana type I Isopentenyl Diphosphate Isomerases are targeted to multiple subcellular compartments and have overlapping functions in isoprenoid biosynthesis. Plant Cell 2008, 20, 677–696. [Google Scholar] [CrossRef]
- Nakamura, A.; Shimada, H.; Masuda, T.; Ohta, H.; Takamiya, K. Two distinct isopentenyl diphosphate isomerases in cytosol and plastid are differentially induced by environmental stresses in tobacco. FEBS Lett. 2001, 506, 61–64. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Xu, T.; Wang, J.; Zhang, T.; Yang, J.; Feng, L.; Song, T.; Yang, J.; Wu, Y. Transcriptomic and free monoterpene analyses of aroma reveal that isopentenyl diphosphate isomerase inhibits monoterpene biosynthesis in grape (Vitis vinifera L.). BMC Plant Biol. 2024, 24, 595. [Google Scholar] [CrossRef]
- Pankratov, I.; McQuinn, R.; Schwartz, J.; Bar, E.; Fei, Z.; Lewinsohn, E.; Zamir, D.; Giovannoni, J.J.; Hirschberg, J. Fruit carotenoid-deficient mutants in tomato reveal a function of the plastidial isopentenyl diphosphate isomerase (IDI1) in carotenoid biosynthesis. Plant J. 2016, 88, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant 2013, 147, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Muhammad Aslam, M.; Waseem, M.; Jakada, B.H.; Okal, E.J.; Lei, Z.; Saqib, H.S.A.; Yuan, W.; Xu, W.; Zhang, Q. Mechanisms of Abscisic Acid-Mediated Drought Stress Responses in Plants. Int. J. Mol. Sci. 2022, 23, 1084. [Google Scholar] [CrossRef]
- Marusig, D.; Tombesi, S. Abscisic Acid Mediates Drought and Salt Stress Responses in Vitis vinifera-A Review. Int. J. Mol. Sci. 2020, 21, 8648. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, L.; Huang, L.; Wang, G. The expression of AcIDI1 reveals diterpenoid alkaloids’ allocation strategies in the roots of Aconitum carmichaelii Debx. Gene 2024, 920, 148529. [Google Scholar] [CrossRef]
- Pan, J.; Huang, C.; Yao, W.; Niu, T.; Yang, X.; Wang, R. Full-length transcriptome, proteomics and metabolite analysis reveal candidate genes involved triterpenoid saponin biosynthesis in Dipsacus asperoides. Front. Plant. Sci. 2023, 14, 1134352. [Google Scholar] [CrossRef]
- Deng, Y.A.; Li, L.; Peng, Q.; Feng, L.F.; Yang, J.F.; Zhan, R.T.; Ma, D.M. Isolation and characterization of AaZFP1, a C2H2 zinc finger protein that regulates the AaIPPI1 gene involved in artemisinin biosynthesis in Artemisia annua. Planta 2022, 255, 122. [Google Scholar] [CrossRef]


| Software | Prediction Result |
|---|---|
| iPSORT | mitochondrial targeting or chloroplast transport peptide |
| Plant-mPLoc | chloroplast |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, X.; Chen, J.; Yan, H.; Huang, X.; Chen, J.; Ma, Z.; Zhou, J.; Liao, H. Functional Identification of the Isopentenyl Diphosphate Isomerase Gene from Fritillaria unibracteata. Horticulturae 2024, 10, 887. https://doi.org/10.3390/horticulturae10080887
Yu X, Chen J, Yan H, Huang X, Chen J, Ma Z, Zhou J, Liao H. Functional Identification of the Isopentenyl Diphosphate Isomerase Gene from Fritillaria unibracteata. Horticulturae. 2024; 10(8):887. https://doi.org/10.3390/horticulturae10080887
Chicago/Turabian StyleYu, Xinyi, Jiao Chen, Han Yan, Xue Huang, Jieru Chen, Zichun Ma, Jiayu Zhou, and Hai Liao. 2024. "Functional Identification of the Isopentenyl Diphosphate Isomerase Gene from Fritillaria unibracteata" Horticulturae 10, no. 8: 887. https://doi.org/10.3390/horticulturae10080887
APA StyleYu, X., Chen, J., Yan, H., Huang, X., Chen, J., Ma, Z., Zhou, J., & Liao, H. (2024). Functional Identification of the Isopentenyl Diphosphate Isomerase Gene from Fritillaria unibracteata. Horticulturae, 10(8), 887. https://doi.org/10.3390/horticulturae10080887






