Exiguobacterium acetylicum Strain SI17: A Potential Biocontrol Agent against Peronophythora litchii Causing Post-Harvest Litchi Downy Blight
Abstract
1. Introduction
2. Materials and Methods
2.1. E. acetylicum SI17 Bioinformatics Analysis
2.1.1. Bacterial Culture and DNA Extraction
2.1.2. Genome Sequencing, Assembly, Annotation, and Phylogenomic Analyses
2.2. Biocontrol (Antagonistic) Activity on Litchi Fruit
2.2.1. Inoculation Experiments on Litchi Fruit
2.2.2. Relative Quantification of P. litchii SC18 in Pericarp
2.2.3. Enzyme Activity in the Pericarp
2.3. Data Analysis
3. Results
3.1. Genome and Phylogenomic Analysis of E. acetylicum SI17
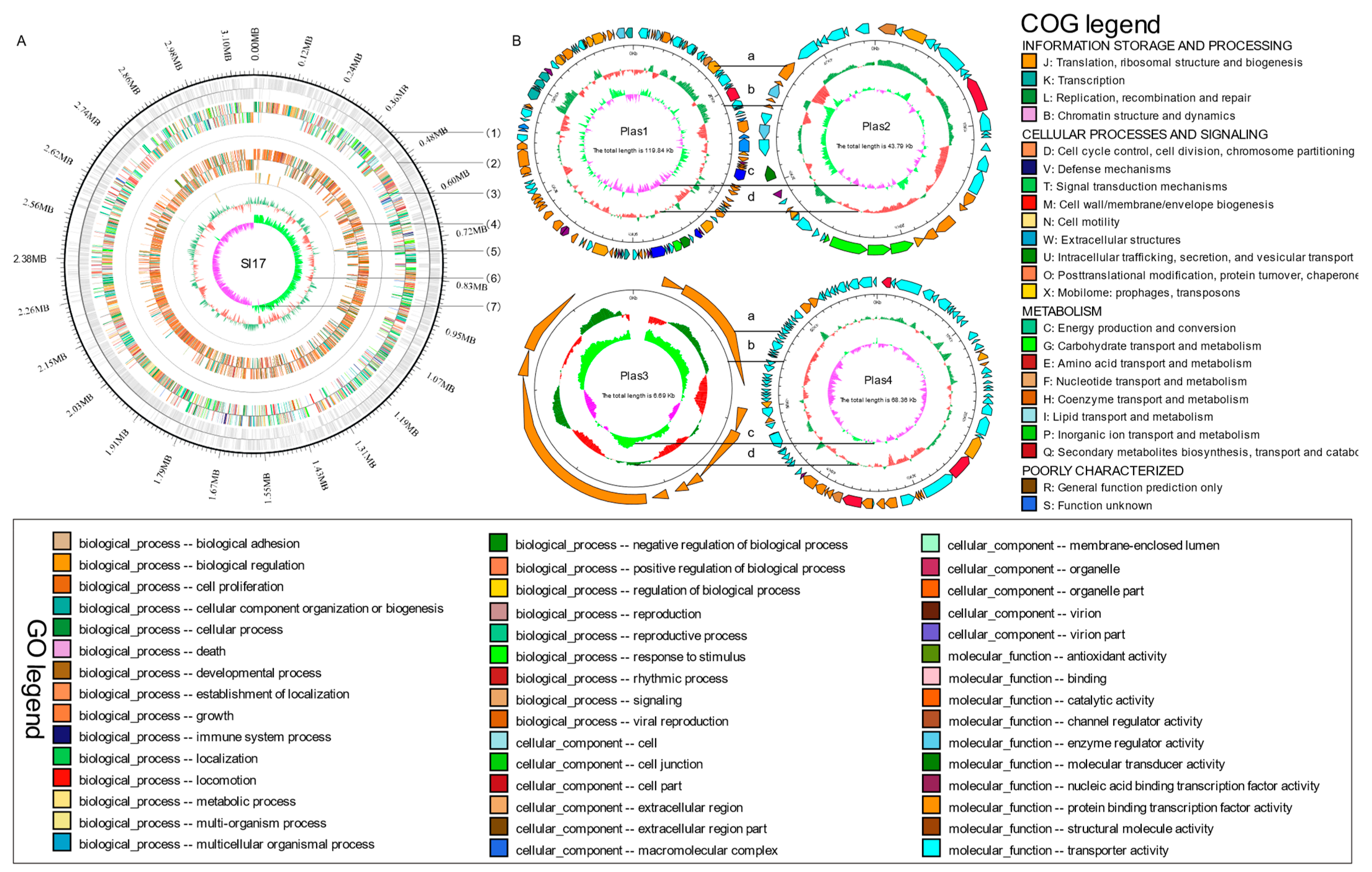
3.1.1. Genome Sequencing, Assembly, and Functional Annotation
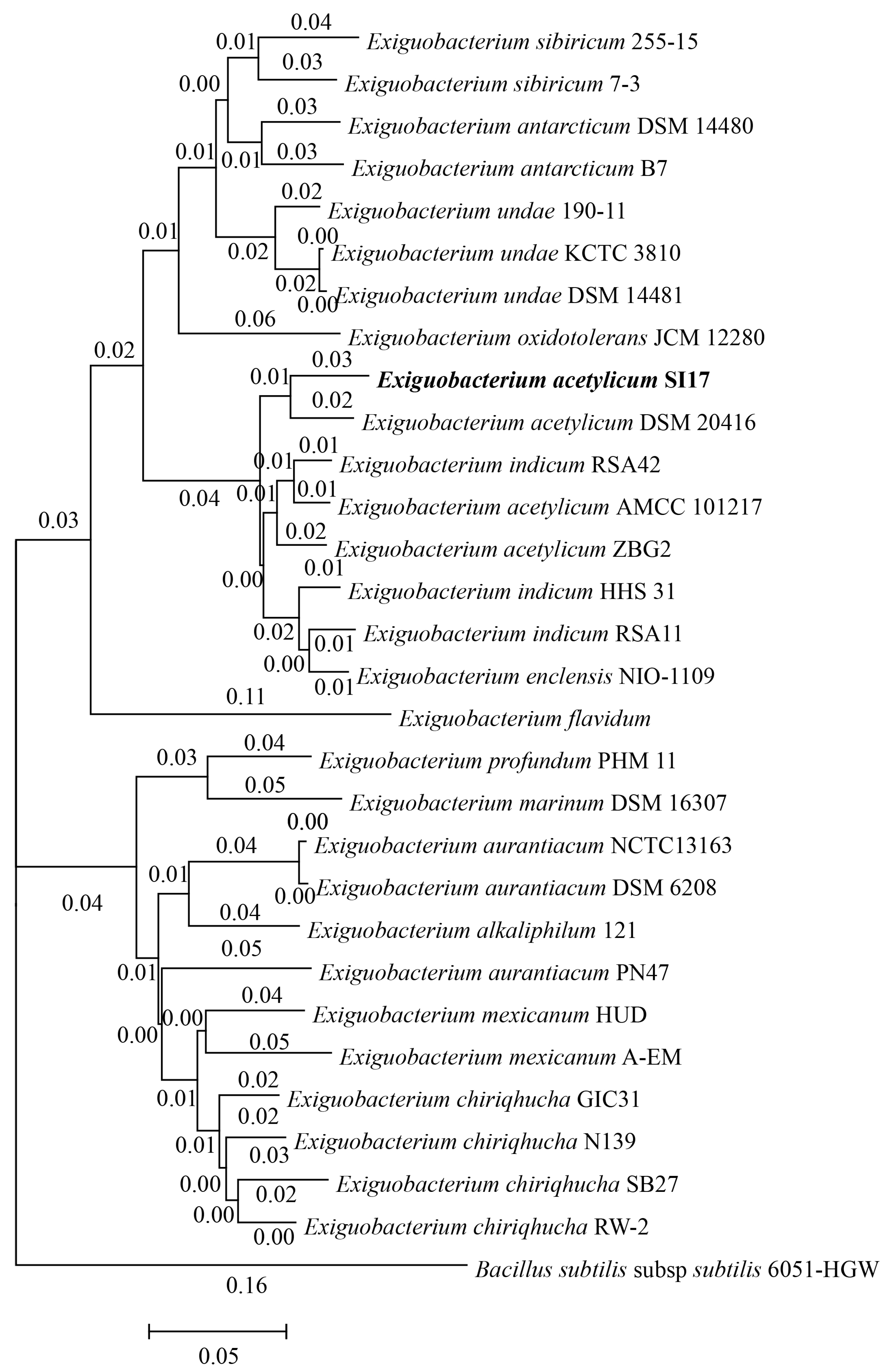
3.1.2. Phylogenomic and Comparative Genomic Analysis
3.2. Biocontrol Activity of E. acetylicum SI17 on LDB
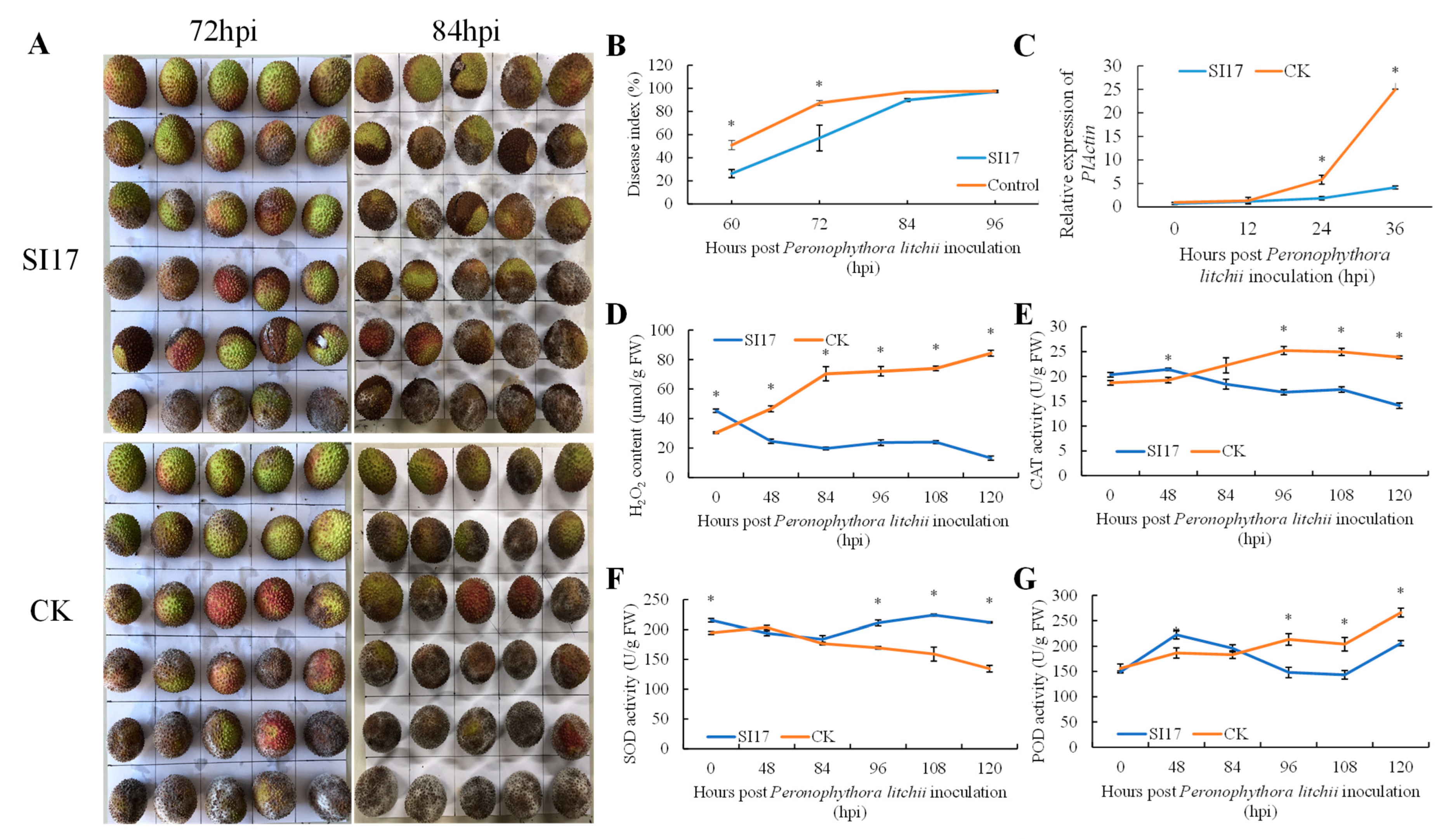
3.2.1. Pre-Harvest E. acetylicum SI17 Treatment Suppressed LDB
3.2.2. Relative Quantification of P. litchii SC18 in Pericarp after E. acetylicum SI17 Treatment
3.2.3. Pre-Harvest E. acetylicum SI17 Treatment Enhanced the Activity of Defense-Related Enzymes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jiang, Y.M.; Zhu, X.R.; Li, Y.B. Postharvest control of litchi fruit rot by Bacillus subtilis. Food Sci. Tech. 2001, 34, 430–436. [Google Scholar] [CrossRef]
- Xu, L.; Xue, J.; Wu, P.; Wang, D.; Lin, L.; Jiang, Y.; Duan, X.; Wei, X. Antifungal activity of hypothemycin against Peronophythora litchii in vitro and in vivo. J. Agric. Food Chem. 2013, 61, 10091–10095. [Google Scholar] [CrossRef] [PubMed]
- Situ, J.; Xi, P.; Lin, L.; Huang, W.; Song, Y.; Jiang, Z.; Kong, G. Signal and regulatory mechanisms involved in spore development of Phytophthora and Peronophythora. Front. Microbiol. 2022, 13, 984672. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, T.; Liu, G.; Hu, M.; Yun, Z.; Duan, X.; Cai, K.; Jiang, G. Inhibition of downy blight and enhancement of resistance in litchi fruit by postharvest application of melatonin. Food Chem. 2021, 347, 129009. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.M.; Wang, Y.; Song, L.; Liu, H.; Lichter, A.; Kerdchoechuen, O.; Joyce, D.C.; Shi, J. Postharvest characteristics and handling of litchi fruit-an overview. Aust. J. Exp. Agric. 2006, 46, 1541. [Google Scholar] [CrossRef]
- Dukare, A.S.; Paul, S.; Nambi, V.E.; Gupta, R.K.; Singh, R.; Sharma, K.; Vishwakarma, R.K. Exploitation of microbial antagonists for the control of postharvest diseases of fruits: A review. Crit. Rev. Food Sci. 2019, 9, 1498–1513. [Google Scholar] [CrossRef]
- Ippolito, A.; Nigro, F. Impact of preharvest application of biological control agents on postharvest diseases of fresh fruits and vegetables. Crop Prot. 2000, 19, 715–723. [Google Scholar] [CrossRef]
- Sivakumar, D.; Zeeman, K.; Korsten, L. Effect of a biocontrol agent (Bacillus subtilis) and modified atmosphere packaging on postharvest decay control and quality retention of litchi during storage. Phytoparasitica 2007, 35, 507–518. [Google Scholar] [CrossRef]
- Sivakumar, D.; Arrebola, E.; Korsten, L. Postharvest decay control and quality retention in litchi (cv. McLean’s Red) by combined application of modified atmosphere packaging and antimicrobial agents. Crop Prot. 2008, 27, 1208–1214. [Google Scholar] [CrossRef]
- Cai, X.Q.; Lin, N.; Chen, W.; Hu, F.P.; DongLiang, Q.; Mitra, S.K.; Diczbalis, Y. Control effects on litchi downy blight disease by endophytic bacterial strain TB2 and its pathogenesis-related proteins. Acta Hortic. 2010, 863, 631–636. [Google Scholar] [CrossRef]
- Wu, Y.; Lin, H.; Lin, Y.; Shi, J.; Xue, S.; Hung, Y.C.; Chen, Y.; Hui, W. Effects of biocontrol bacteria Bacillus amyloliquefaciens LY-1 culture broth on quality attributes and storability of harvested litchi fruit. Postharvest Biol. Tec. 2017, 132, 81–87. [Google Scholar] [CrossRef]
- Martínez-Castellanos, G.; Pelayo-Zaldívar, C.; Pérez-Flores, L.J.; López-Luna, A.; Gimeno, M.; Bárzana, E.; Shirai, K. Postharvest litchi (Litchi chinensis Sonn.) quality preservation by Lactobacillus plantarum. Postharvest Biol. Tec. 2011, 59, 172–178. [Google Scholar] [CrossRef]
- Liao, L.; Zhou, J.; Wang, H.; Fei, H.; Liu, S.; Jiang, Z.; Chen, S.; Zhang, L.H. Control of litchi downy blight by zeamines produced by Dickeya zeae. Sci. Rep. 2015, 5, 15719. [Google Scholar] [CrossRef]
- Xing, M.; Zheng, L.; Deng, Y.; Xu, D.; Xi, P.; Li, M.; Kong, G.; Jiang, Z. Antifungal activity of natural volatile organic compounds against litchi downy blight pathogen Peronophythora litchii. Molecules 2018, 23, 358. [Google Scholar] [CrossRef]
- Stéphane, H.; Schadt, C.W. Towards a holistic understanding of the beneficial interactions across the Populus microbiome. New Phytol. 2015, 205, 1424–1430. [Google Scholar]
- Conrath, U.; Beckers, G.J.; Flors, V.; Garcã-Agustã, P.; Jakab, G.; Mauch, F.; Newman, M.A.; Pieterse, C.M.J.; Poinssot, B.; Pozo, M.J.; et al. Priming: Getting ready for battle. Mol. Plant Microbe Interact. 2006, 19, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Kasana, R.C.; Pandey, C.B. Exiguobacterium: An overview of a versatile genus with potential in industry and agriculture. Crit. Rev. Biotechnol. 2018, 38, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Vijayalaxmi, S.; AnuAppaiah, K.A.; Jayalakshmi, S.K.; Mulimani, V.H.; Sreeramuluet, K. Production of bioethanol from fermented sugars of sugarcane bagasse produced by lignocellulolytic enzymes of Exiguobacterium sp. VSG-1. Appl. Biochem. Biotech. 2013, 2171, 246–260. [Google Scholar] [CrossRef]
- Xie, J.; Xie, W.; Yu, J.; Xin, R.; Shi, Z.; Song, L.; Yang, X. Extraction of chitin from shrimp shell by successive two-step fermentation of Exiguobacterium profundum and Lactobacillus acidophilus. Front. Microbiol. 2021, 12, 677126. [Google Scholar] [CrossRef]
- Ali, C.H.; Zhang, J.J.; Mbadinga, S.M.; Mu, B.Z. Screening, isolation and optimization of an extracellular lipase producing Exiguobacterium sp. BBXS-7 segregated from waste cooking oil contaminated sites. Wulfenia J. 2015, 2, 185–201. [Google Scholar]
- Mojallali, L.; ShahbaniZahiri, H.; Rajaei, S.; Noghabi, K.A.; Haghbeenet, K. A novel ~34-kDa a-amylase from psychrotroph Exiguobacterium sp. SH3: Production, purification, and characterization. Biotechnol. Appl. Bioc. 2014, 61, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Peng, Q.; Yan, J.; Wang, H.; Ding, H.; Shi, B. Gene cloning and enzymatic characterization of alkali-tolerant type I pullulanase from Exiguobacterium acetylicum. Lett. Appl. Microbiol. 2015, 60, 52–59. [Google Scholar] [CrossRef]
- Parthasarathy, A.; Miranda, R.R.; Eddingsaas, N.C.; Chu, J.; Freezman, I.M.; Tyler, A.C.; Hudson, A.O. Polystyrene degradation by Exiguobacterium sp. RIT 594: Preliminary evidence for a pathway containing an atypical oxygenase. Microorganisms 2022, 10, 1619. [Google Scholar] [CrossRef] [PubMed]
- Sakdapetsiri, C.; Kaokhum, N.; Pinyakong, O. Biodegradation of crude oil by immobilized Exiguobacterium sp. AO-11 and shelf life evaluation. Sci. Rep. 2021, 11, 12990. [Google Scholar] [CrossRef]
- Delegan, Y.; Kocharovskaya, Y.; Bogun, A.; Sizova, A.; Solomentsev, V.; Iminova, L.; Lyakhovchenko, N.; Zinovieva, A.; Goyanov, M.; Solyanikova, I. Characterization and genomic analysis of Exiguobacterium alkaliphilum B-3531D, an efficient crude oil degrading strain. Biotechnol. Rep. 2021, 32, e00678. [Google Scholar] [CrossRef]
- Barghoth, M.G.; Desouky, S.E.; Radwan, A.A.; Shah, M.P.; Salem, S.S. Characterizations of highly efficient moderately halophilic toluene degrading Exiguobacterium mexicanum M7 strain isolated from Egyptian saline sediments. Biotechnol. Genet. Eng. 2023, 2, 1–19. [Google Scholar] [CrossRef]
- Alam, M.Z.; Malik, A. Chromate resistance, transport and bioreduction by Exiguobacterium sp. ZM-2 isolated from agricultural soil irrigated with tannery effluent. J. Basic. Microb. 2008, 48, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Okeke, B.C. Bioremoval of hexavalent chromium from water by a salt tolerant bacterium, Exiguobacterium sp. GS1. J. Industrial Microbiol. Biot. 2008, 35, 1571–1579. [Google Scholar] [CrossRef]
- Das, S.; Bikash, C.B.; Ranjan, K.M.; Biswaranjan, P.; Mathummal, S.; Anindita, C.; Hrudayanath, T. Reduction of hexavalent chromium by Exiguobacterium mexicanum isolated from chromite mines soil. Chemosphere 2021, 282, 131135. [Google Scholar] [CrossRef]
- Huang, Y.; Tang, J.; Zhang, B.; Long, Z.E.; Ni, H.; Fu, X.; Zou, L. Influencing factors and mechanism of Cr(VI) reduction by facultative anaerobic Exiguobacterium sp. PY14. Front. Microbiol. 2023, 14, 1242410. [Google Scholar] [CrossRef]
- Dastager, S.G.; Kumaran, D.C.; Pandey, A. Characterization of plant growth-promoting rhizobacterium Exiguobacterium NII-0906 for its growth promotion of cowpea (Vigna unguiculata). Biologia 2010, 65, 197–203. [Google Scholar] [CrossRef]
- Selvakumar, G.; Kundu, S.; Joshi, P.; Nazim, S.; Gupta, A.D.; Gupta, H.S. Growth promotion of wheat seedlings by Exiguobacterium acetylicum 1P (MTCC 8707) a cold tolerant bacterial strain from the Uttarakhand Himalayas. Indian J. Microbiol. 2010, 50, 50–66. [Google Scholar] [CrossRef] [PubMed]
- Bharti, N.; Yadav, D.; Barnawal, D.; Maji, D.; Kalra, A. Exiguobacterium oxidotolerans, a halotolerant plant growth promoting rhizobacteria, improves yield and content of secondary metabolites in Bacopa monnieri (L.) Pennell under primary and secondary salt stress. World J. Microb. Biot. 2013, 29, 379–387. [Google Scholar] [CrossRef]
- Venkadesaperumal, G.; Amaresan, N.; Kumar, K. Plant growth promoting capability and genetic diversity of bacteria isolated from mud volcano and lime cave of Andaman and Nicobar Islands. Braz. J. Microbiol. 2014, 45, 1271–1281. [Google Scholar] [CrossRef]
- Marfetán, J.A.; Gallo, A.L.; Farias, M.E.; Vélez, M.L.; Pescuma, M.; Ordoñez, O.F. Exiguobacterium sp. as a bioinoculant for plant-growth promotion and selenium biofortification strategies in horticultural plants. World J. Microb. Biot. 2023, 39, 134. [Google Scholar] [CrossRef]
- Barnett, S.J.; Anstis, S.T.; Roget, D.K.; Ryder, M.H. Suppression of Rhizoctonia solani AG-8 induced disease on wheat by the interaction between Pantoea, Exiguobacterium, and Microbacteria. Aust. J. Soil. Res. 2006, 44, 331–342. [Google Scholar] [CrossRef]
- Selvakumar, G.; Joshi, P.; Nazim, S.; Mishra, P.K.; Kundu, S.; Gupta, H.S. Exiguobacterium acetylicum strain 1P MTCC 8707, a novel bacterial antagonist from the North Western Indian Himalayas. World J. Microbiol. Biotechnol. 2009, 25, 131–137. [Google Scholar] [CrossRef]
- Vishnivetskaya, T.A.; Kathariou, S.; Tiedje, J.M. The Exiguobacterium genus: Biodiversity and biogeography. Extremophiles 2009, 13, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhu, Z.; Li, Y.; Li, X.; Guan, Z.; Zheng, J. Comparative genomics of Exiguobacterium reveals what makes a cosmopolitan bacterium. mSystems 2021, 6, e0038321. [Google Scholar] [CrossRef]
- Zheng, L.; Situ, J.; Zhu, Q.; Xi, P.; Zheng, Y.; Liu, H.; Zhou, X.; Jiang, Z.D. Identification of volatile organic compounds for the biocontrol of postharvest litchi fruit pathogen Peronophythora litchii. Postharvest Biol. Technol. 2019, 155, 37–46. [Google Scholar] [CrossRef]
- Hoffman, C.S.; Winston, F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 1987, 57, 267–272. [Google Scholar] [CrossRef]
- Zheng, L.; Huang, S.; Hsiang, T.; Yu, G.; Guo, D.; Jiang, Z.; Li, J. Biocontrol using Bacillus amyloliquefaciens PP19 against litchi downy blight caused by Peronophythora litchii. Front. Microbiol. 2021, 11, 619423. [Google Scholar] [CrossRef]
- Zuo, G.; Hao, B. CVTree3 Web server for whole-genome-based and alignment-free prokaryotic phylogeny and taxonomy. Genom. Proteom. Bioinf. 2015, 13, 321–331. [Google Scholar] [CrossRef]
- Sorokulova, I.; Krumnow, A.; Globa, L.; Vodyanoy, V. Efficient decomposition of shrimp shell waste using Bacillus cereus and Exiguobacterium acetylicum. J. Ind. Microbiol. Biot. 2009, 36, 1123–1126. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Srivastava, R.; Bharati, A.P.; Singh, A.K.; Sharma, A.; Das, S.; Tiwari, P.K.; Srivastava, A.K.; Chakdar, H.; Kashyap, P.L.; et al. Analysis of biosynthetic gene clusters, secretory, and antimicrobial peptides reveals environmental suitability of Exiguobacterium profundum PHM11. Front. Microbiol. 2022, 12, 785458. [Google Scholar] [CrossRef] [PubMed]
- Mekuto, L.; Alegbeleye, O.O.; Ntwampe, S.K.; Ngongang, M.M.; Mudumbi, J.B.; Akinpelu, E.A. Co-metabolism of thiocyanate and free cyanide by Exiguobacterium acetylicum and Bacillus marisflavi under alkaline conditions. 3 Biotech. 2016, 6, 173. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, Y.; Hao, X.; Zhang, X.; Ma, Y.; Niu, Z. A novel marine bacterium Exiguobacterium marinum a-1 isolated from in situ plastisphere for degradation of additive-free polypropylene. Environ. Pollut. 2023, 336, 122390. [Google Scholar] [CrossRef] [PubMed]
- Mhamdi, A.; Van Breusegem, F. Reactive oxygen species in plant development. Development 2018, 145, dev164376. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Li, J.; Huang, C.; Zhang, D.; Xu, Y.; Yang, X.; Song, J.; Wang, D.; Qiu, N.; Short, D.P.G.; et al. Cu/Zn superoxide dismutase (VdSOD1) mediates reactive oxygen species detoxification and modulates virulence in Verticillium dahliae. Mol. Plant Pathol. 2021, 22, 1092–1108. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Bioch. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Zhang, X.; Xin, Y.; Wang, J.; Dhanasekaran, S.; Yue, Q.; Feng, F.; Gu, X.; Li, B.; Zhao, L.; Zhang, H. Characterization of a Bacillus velezensis strain as a potential biocontrol agent against soft rot of eggplant fruits. Int. J. Food Microbiol. 2024, 410, 110480. [Google Scholar] [CrossRef] [PubMed]
- Nan, J.; Zhang, S.; Jiang, L. Antibacterial potential of Bacillus amyloliquefaciens GJ1 against citrus Huanglongbing. Plants 2021, 10, 261. [Google Scholar] [CrossRef] [PubMed]
- Chai, C.H.; Hong, C.F.; Huang, J.W. Identification and characterization of a multifunctional biocontrol agent, Streptomyces griseorubiginosus LJS06, against cucumber anthracnose. Front. Microbiol. 2022, 13, 923276. [Google Scholar] [CrossRef]
- Duke, K.A.; Becker, M.G.; Girard, I.J.; Millar, J.L.; Dilantha Fernando, W.G.; Belmonte, M.F.; de Kievit, T.R. The biocontrol agent Pseudomonas chlororaphis PA23 primes Brassica napus defenses through distinct gene networks. BMC Genom. 2017, 18, 467. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, Y.; Qiao, J.; Yu, W.; Pan, X.; Zhang, T.; Liu, Y.; Lu, S.E. Phenazine-1-carboxylic acid produced by Pseudomonas chlororaphis YL-1 is effective against Acidovorax citrulli. Microorganisms 2021, 9, 2012. [Google Scholar] [CrossRef]
- Yang, K.; Chen, C.; Wang, Y.; Li, J.; Dong, X.; Cheng, Y.; Zhang, H.; Zhai, Y.; Ai, G.; Song, Q.; et al. Nep1-like proteins from the biocontrol agent Pythium oligandrum enhance plant disease resistance independent of cell death and reactive oxygen species. Front. Plant Sci. 2022, 13, 830636. [Google Scholar] [CrossRef]
| Features | Value | ||||
|---|---|---|---|---|---|
| Number of reads | 716,365 | ||||
| Number of bases (bp) | 8,189,664,288 | ||||
| Mean read length (bp) | 11,432.3 | ||||
| N50 read length (bp) | 14,606 | ||||
| Mean read quality | 10.1 | ||||
| Gene number | 3541 | ||||
| Gene total length (bp) | 3,022,368 | ||||
| Gene average length (bp) | 846 | ||||
| Gene/genome (%) | 88.98 | ||||
| Features | Chromosome | Plasmid 1 | Plasmid 2 | Plasmid 3 | Plasmid 4 |
| Genome size (bp) | 3,157,929 | 119,838 | 43,790 | 6694 | 68,355 |
| G + C content (%) | 47.28 | 40.05 | 43.16 | 37.50 | 37.92 |
| Non-coding RNA (ncRNA) | ||||
| Type | Number | Average_Length | Total_Length | In Genome (%) |
| tRNA | 70 | 77 | 5394 | 0.1588 |
| 5s rRNA | 9 | 115 | 1035 | 1.2128 |
| 16s rRNA | 9 | 1550 | 13,950 | |
| 23s rRNA | 9 | 2912 | 26,208 | |
| sRNA | 1 | 86 | 86 | 0.0025 |
| Genomics Islands (GIs) | ||||
| GIs_ID | Start | End | Length | GC% |
| GIs001 (chr) | 1,353,417 | 1,359,120 | 5704 | 41.09 |
| GIs002 (chr) | 1,511,852 | 1,532,374 | 20,523 | 39.63 |
| GIs003 (chr) | 1,710,372 | 1,718,135 | 7764 | 42.17 |
| GIs004 (plas 1) | 57,004 | 61,841 | 4838 | 36.59 |
| GIs005 (plas 1) | 96,328 | 103,839 | 7512 | 50.12 |
| GIs006 (plas 2) | 12,956 | 20,897 | 7942 | 36.15 |
| Prophage | ||||
| Prophage_ID | Start | End | Length | GC% |
| Prophage_1 (Chr1) | 804,985 | 828,528 | 23,544 | 47.52 |
| Prophage_2 (Chr1) | 1,237,842 | 1,295,459 | 57,618 | 47.55 |
| Prophage_3 (Chr1) | 1,333,287 | 1,341,796 | 8,510 | 47.56 |
| Prophage_4 (Chr1) | 1,600,271 | 1,629,792 | 29,522 | 49.76 |
| Prophage_5 (Chr1) | 1,845,601 | 1,882,853 | 37,253 | 45.65 |
| Interspersed Repeat | ||||
| Type | Number | Total Length (bp) | Average length (bp) | In Genome (%) |
| LTR | 89 | 7967 | 90 | 0.2346 |
| DNA | 14 | 1005 | 72 | 0.0296 |
| LINE | 35 | 3408 | 103 | 0.1003 |
| SINE | 14 | 1110 | 79 | 0.0327 |
| RC | 2 | 101 | 50 | 0.003 |
| Total | 154 | 13,043 | 0.384 | 90 |
| Tandem Repeat | ||||
| Type | Number | Repeat Size (bp) | Total Length (bp) | In Genome (%) |
| Tandem repeat (TR) | 55 | 8~201 | 5332 | 0.157 |
| Minisatellite DNA | 50 | 11~51 | 4348 | 0.128 |
| Microsatellite DNA | 0 | 0~0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, S.; Lv, X.; Zheng, L.; Guo, D. Exiguobacterium acetylicum Strain SI17: A Potential Biocontrol Agent against Peronophythora litchii Causing Post-Harvest Litchi Downy Blight. Horticulturae 2024, 10, 888. https://doi.org/10.3390/horticulturae10080888
Huang S, Lv X, Zheng L, Guo D. Exiguobacterium acetylicum Strain SI17: A Potential Biocontrol Agent against Peronophythora litchii Causing Post-Harvest Litchi Downy Blight. Horticulturae. 2024; 10(8):888. https://doi.org/10.3390/horticulturae10080888
Chicago/Turabian StyleHuang, Shilian, Xinmin Lv, Li Zheng, and Dongliang Guo. 2024. "Exiguobacterium acetylicum Strain SI17: A Potential Biocontrol Agent against Peronophythora litchii Causing Post-Harvest Litchi Downy Blight" Horticulturae 10, no. 8: 888. https://doi.org/10.3390/horticulturae10080888
APA StyleHuang, S., Lv, X., Zheng, L., & Guo, D. (2024). Exiguobacterium acetylicum Strain SI17: A Potential Biocontrol Agent against Peronophythora litchii Causing Post-Harvest Litchi Downy Blight. Horticulturae, 10(8), 888. https://doi.org/10.3390/horticulturae10080888







