RNA Sequencing Reveals Transcription Factors and Genes in Phenylpropanoid Biosynthesis That Positively Regulate Size and Weight of Oak Tree Seeds
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sample Collection
2.2. Sample Validation
2.3. Measurement of Seed Weight and Size
2.4. Measurement of Soluble Sugar and Starch
2.5. RNA Extraction, cDNA Library Construction, and Sequencing
2.6. Assembly Normalization and Quality Assessment
2.7. Differentially Expressed Genes (DEGs) Analysis
2.8. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes Pathway Analysis
2.9. Gene Expression Analysis by qPCR
2.10. Statistical Analysis
3. Results
3.1. Morphological and Biochemical Analysis of Oak Variety
3.2. Identification of DEGs between Control Oak and Keumsura1ho at Different Developmental Stages
3.3. Validation of RNA-Seq Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bobiec, A.; Reif, A.; Öllerer, K. Seeing the oakscape beyond the forest: A landscape approach to the oak regeneration in Europe. Landsc. Ecol. 2018, 33, 513–528. [Google Scholar] [CrossRef]
- Hill, A.F. Economic Botany. A Textbook of Useful Plants and Plant Products; McGraw-Hill: New York, NY, USA, 1952. [Google Scholar]
- Mason, S.L.R. Acorns in Human Subsistence; University of London: London, UK; University College London (United Kingdom): London, UK, 1992. [Google Scholar]
- Melamed, Y.; Kislev, M.E.; Geffen, E.; Lev-Yadun, S.; Goren-Inbar, N. The plant component of an Acheulian diet at Gesher Benot Ya ‘aqov, Israel. Proc. Natl. Acad. Sci. USA 2016, 113, 14674–14679. [Google Scholar] [CrossRef]
- Xu, J.; Cao, J.; Yue, J.; Zhang, X.; Zhao, Y. New triterpenoids from acorns of Quercus liaotungensis and their inhibitory activity against α-glucosidase, α-amylase and protein-tyrosine phosphatase 1B. J. Funct. Foods 2018, 41, 232–239. [Google Scholar] [CrossRef]
- Taib, M.; Bouyazza, L.; Lyoussi, B. Acorn oil: Chemistry and functionality. J. Food Qual. 2020, 2020, 8898370. [Google Scholar] [CrossRef]
- Monteiro, V.; Soares, C.; Grosso, C.; Delerue-Matos, C.; Ramalhosa, M.J. From Forest to Table: Optimizing the Nutritional Value of Acorns through Effective Tannin Extraction. Biol. Life Sci. Forum 2023, 26, 16. [Google Scholar] [CrossRef]
- Szabłowska, E.; Tańska, M. Acorn flour properties depending on the production method and laboratory baking test results: A review. Compr. Rev. Food Sci. Food Saf. 2021, 20, 980–1008. [Google Scholar] [CrossRef]
- Ceccon, E.; González, E.J.; Martorell, C. Is direct seeding a biologically viable strategy for restoring forest ecosystems? Evidences from a Meta-analysis. Land Degrad. Dev. 2016, 27, 511–520. [Google Scholar] [CrossRef]
- Löf, M.; Madsen, P.; Metslaid, M.; Witzell, J.; Jacobs, D.F. Restoring forests: Regeneration and ecosystem function for the future. New For. 2019, 50, 139–151. [Google Scholar] [CrossRef]
- Hou, X.; Yi, X.; Yang, Y.; Liu, W. Acorn germination and seedling survival of Q. variabilis: Effects of cotyledon excision. Ann. For. Sci. 2010, 67, 711. [Google Scholar] [CrossRef]
- Yi, X.; Zhang, J.; Wang, Z. Large and small acorns contribute equally to early-stage oak seedlings: A multiple species study. Eur. J. For. Res. 2015, 134, 1019–1026. [Google Scholar] [CrossRef]
- Greenberg, C.H. Oak growth and acorn production in southern Appalachian mature forests and shelterwood with reserves regeneration harvests. For. Ecol. Manag. 2021, 481, 118691. [Google Scholar] [CrossRef]
- Valero Galván, J.; Jorrin Novo, J.J.; Cabrera, A.G.; Ariza, D.; García-Olmo, J.; Cerrillo, R.M.N. Population variability based on the morphometry and chemical composition of the acorn in Holm oak (Quercus ilex subsp. ballota [Desf.] Samp.). Eur. J. For. Res. 2012, 131, 893–904. [Google Scholar] [CrossRef]
- National Institute of Forest Science. The characteristics and cultivation of the new cultivar “Keumsuro1ho” of the Quercus Acutissima. For. Sci. News 2017, 17–26. [Google Scholar]
- Tobimatsu, Y.; Schuetz, M. Lignin polymerization: How do plants manage the chemistry so well? Curr. Opin. Biotechnol. 2019, 56, 75–81. [Google Scholar] [CrossRef]
- Ali, M.B.; McNear, D.H. Induced transcriptional profiling of phenylpropanoid pathway genes increased flavonoid and lignin content in Arabidopsis leaves in response to microbial products. BMC Plant Biol. 2014, 14, 84. [Google Scholar] [CrossRef]
- Lu, Y.; Sharkey, T.D. The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta 2004, 218, 466–473. [Google Scholar] [CrossRef]
- Stitt, M.; Lilley, R.M.; Gerhardt, R.; Heldt, H.W. [32] Metabolite levels in specific cells and subcellular compartments of plant leaves. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1989; Volume 174, pp. 518–552. [Google Scholar]
- Walters, R.G.; Ibrahim, D.G.; Horton, P.; Kruger, N.J. A mutant of Arabidopsis lacking the triose-phosphate/phosphate translocator reveals metabolic regulation of starch breakdown in the light. Plant Physiol. 2004, 135, 891–906. [Google Scholar] [CrossRef]
- Bolger, A.; Giorgi, F. Trimmomatic: A flexible read trimming tool for Illumina NGS data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Pant, G.; Bhavnasi, Y.K.; Blanchard Jr, S.G.; Brouwer, C. Pathview Web: User friendly pathway visualization and data integration. Nucleic Acids Res. 2017, 45, W501–W508. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, P.; Ahn, E.; Reddy, U.K.; Perumal, R.; Prom, L.K.; Magill, C. RNA-Sequencing in resistant (QL3) and susceptible (Theis) sorghum cultivars inoculated with Johnsongrass isolates of Colletotrichum sublineola. Front. Genet. 2021, 12, 722519. [Google Scholar] [CrossRef] [PubMed]
- Makela, A.V.; Murrell, D.H.; Parkins, K.M.; Kara, J.; Gaudet, J.M.; Foster, P.J. Cellular imaging with MRI. Top. Magn. Reson. Imaging 2016, 25, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Migueal, P.; Nkongolo, K.; Michael, P.; Djeukam, C. Differential gene transcription in red oak (Quercus rubra) genotypes resistant to copper toxicity. Am. J. Biochem. Biotechnol. 2017, 13, 215–255. [Google Scholar] [CrossRef]
- Simon, P.W.; Jenderek, M.M. Flowering, seed production, and the genesis of garlic breeding. Plant Breed. Rev. 2004, 23, 211–244. [Google Scholar]
- Khush, G.S. Rice breeding: Past, present and future. J. Genet. 1987, 66, 195–216. [Google Scholar] [CrossRef]
- Takeda, K. Inheritance of grain size and its implications for rice breeding. In Rice Genetics II: (In 2 Parts); World Scientific: Singapore, 1991; pp. 181–189. [Google Scholar]
- Sestras, R.E.; Sestras, A.F. Quantitative traits of interest in apple breeding and their implications for selection. Plants 2023, 12, 903. [Google Scholar] [CrossRef] [PubMed]
- Fresnedo-Ramírez, J.; Frett, T.J.; Sandefur, P.J.; Salgado-Rojas, A.; Clark, J.R.; Gasic, K.; Peace, C.P.; Anderson, N.; Hartmann, T.P.; Byrne, D.H. QTL mapping and breeding value estimation through pedigree-based analysis of fruit size and weight in four diverse peach breeding programs. Tree Genet. Genomes 2016, 12, 25. [Google Scholar] [CrossRef]
- Li, Y.-Y.; Tao, H.-J.; Zhao, X.-Q.; Jie, X.; Li, G.-M.; Hu, S.-K.; Dong, G.-J.; Shi, Z.-Y.; Wu, L.-W.; Jiang, H. Molecular improvement of grain weight and yield in rice by using GW6 gene. Rice Sci. 2014, 21, 127–132. [Google Scholar] [CrossRef]
- Wang, P.; Xing, Y.; Li, Z.; Yu, S. Improving rice yield and quality by QTL pyramiding. Mol. Breed. 2012, 29, 903–913. [Google Scholar] [CrossRef]
- Wang, W.; Simmonds, J.; Pan, Q.; Davidson, D.; He, F.; Battal, A.; Akhunova, A.; Trick, H.N.; Uauy, C.; Akhunov, E. Gene editing and mutagenesis reveal inter-cultivar differences and additivity in the contribution of TaGW2 homoeologues to grain size and weight in wheat. Theor. Appl. Genet. 2018, 131, 2463–2475. [Google Scholar] [CrossRef] [PubMed]
- Gautam, T.; Amardeep; Saripalli, G.; Rakhi; Kumar, A.; Gahlaut, V.; Gadekar, D.; Oak, M.; Sharma, P.; Balyan, H. Introgression of a drought insensitive grain yield QTL for improvement of four Indian bread wheat cultivars using marker assisted breeding without background selection. J. Plant Biochem. Biotechnol. 2021, 30, 172–183. [Google Scholar] [CrossRef]
- Li, S.; Zhou, Y.; Liu, M.; Zhang, Y.; Cao, S. Nutrient composition and starch characteristics of Quercus glandulifera Bl. seeds from China. Food Chem. 2015, 185, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.-L.; Gao, C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 12617. [Google Scholar] [CrossRef] [PubMed]
- Vieitez, A.M.; Corredoira, E.; Martínez, M.T.; San-José, M.C.; Sánchez, C.; Valladares, S.; Vidal, N.; Ballester, A. Application of biotechnological tools to Quercus improvement. Eur. J. For. Res. 2012, 131, 519–539. [Google Scholar] [CrossRef]
- Pijut, P.M.; Woeste, K.E.; Vengadesan, G.; Michler, C.H. Technological advances in temperate hardwood tree improvement including breeding and molecular marker applications. In Vitro Cell. Dev. Biol. Plant 2007, 43, 283–303. [Google Scholar] [CrossRef]
- Ramírez-Valiente, J.A.; Valladares, F.; Gil, L.; Aranda, I. Population differences in juvenile survival under increasing drought are mediated by seed size in cork oak (Quercus suber L.). For. Ecol. Manag. 2009, 257, 1676–1683. [Google Scholar] [CrossRef]
- González-Rodríguez, V.; Villar, R.; Navarro-Cerrillo, R.M. Maternal influences on seed mass effect and initial seedling growth in four Quercus species. Acta Oecologica 2011, 37, 1–9. [Google Scholar] [CrossRef]
- Landergott, U.; Gugerli, F.; Hoebee, S.E.; Finkeldey, R.; Holderegger, R. Effects of seed mass on seedling height and competition in European white oaks. Flora-Morphol. Distrib. Funct. Ecol. Plants 2012, 207, 721–725. [Google Scholar] [CrossRef]
- Kormanik, P.P.; Sung, S.; Kormanik, T.; Schlarbaum, S.; Zarnoch, S.J. Effect of acorn size on development of northern red oak 1-0 seedlings. Can. J. For. Res. 1998, 28, 1805–1813. [Google Scholar] [CrossRef]
- Navarro, F.B.; Jiménez, M.N.; Ripoll, M.Á.; Fernández-Ondoño, E.; Gallego, E.; De Simón, E. Direct sowing of holm oak acorns: Effects of acorn size and soil treatment. Ann. For. Sci. 2006, 63, 961–967. [Google Scholar] [CrossRef]
- Tilki, F. Influence of acorn size and storage duration on moisture content, germination and survival of Quercus petraea (Mattuschka). J. Environ. Biol. 2010, 31, 325. [Google Scholar]
- Kriebel, C.H. Team decision models of an inventory supply organization. Nav. Res. Logist. Q. 1965, 12, 139–154. [Google Scholar] [CrossRef]
- Davis, A.; Jacobs, D. Afforestation in the central hardwood forest region of the USA. In Forest Research Information Paper; Ontario Forest Research Institute: Sault Ste. Marie, ON, Canada, 2005. [Google Scholar]
- Son, S.-G.; Kim, H.-J.; Kang, Y.-J.; Oh, C.-J.; Kim, C.-S.; Byun, K.-O. Establishment of breeding population for Quercus glauca and climatic factors. Korean J. Agric. For. Meteorol. 2011, 13, 109–114. [Google Scholar] [CrossRef][Green Version]
- Woeste, K.E.; Pike, C.C.; Warren, J.C.; Coggeshall, M.V. Characterization of stem volume and form tradeoffs in a northern red oak (Quercus rubra) breeding population in early stages of selection. Ann. For. Sci. 2021, 78, 72. [Google Scholar] [CrossRef]
- Li, M.; Cheng, C.; Zhang, X.; Zhou, S.; Li, L.; Yang, S. Overexpression of pear (Pyrus pyrifolia) CAD2 in tomato affects lignin content. Molecules 2019, 24, 2595. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, H.; Zhao, H.; Xia, Y.; Zheng, X.; Fan, R.; Tan, Z.; Duan, C.; Fu, Y.; Li, L. Multi-omics analysis dissects the genetic architecture of seed coat content in Brassica napus. Genome Biol. 2022, 23, 86. [Google Scholar] [CrossRef]
- Noda, S.; Koshiba, T.; Hattori, T.; Yamaguchi, M.; Suzuki, S.; Umezawa, T. The expression of a rice secondary wall-specific cellulose synthase gene, OsCesA7, is directly regulated by a rice transcription factor, OsMYB58/63. Planta 2015, 242, 589–600. [Google Scholar] [CrossRef]
- Zhang, Y.-C.; Yu, Y.; Wang, C.-Y.; Li, Z.-Y.; Liu, Q.; Xu, J.; Liao, J.-Y.; Wang, X.-J.; Qu, L.-H.; Chen, F. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 2013, 31, 848. [Google Scholar] [CrossRef]
- Wang, C.Y.; Zhang, S.; Yu, Y.; Luo, Y.C.; Liu, Q.; Ju, C.; Zhang, Y.C.; Qu, L.H.; Lucas, W.J.; Wang, X. MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol. J. 2014, 12, 1132–1142. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.-Q.; Sun, Y.; Guo, T.; Shi, C.-L.; Zhang, Y.-M.; Kan, Y.; Xiang, Y.-H.; Zhang, H.; Yang, Y.-B.; Li, Y.-C. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 2020, 11, 2629. [Google Scholar] [CrossRef] [PubMed]
- Na, G.; Mu, X.; Grabowski, P.; Schmutz, J.; Lu, C. Enhancing micro RNA 167A expression in seed decreases the α-linolenic acid content and increases seed size in Camelina sativa. Plant J. 2019, 98, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Le, B.H.; Cheng, C.; Bui, A.Q.; Wagmaister, J.A.; Henry, K.F.; Pelletier, J.; Kwong, L.; Belmonte, M.; Kirkbride, R.; Horvath, S. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. USA 2010, 107, 8063–8070. [Google Scholar] [CrossRef] [PubMed]
- Orozco-Arroyo, G.; Paolo, D.; Ezquer, I.; Colombo, L. Networks controlling seed size in Arabidopsis. Plant Reprod. 2015, 28, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.-L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.-L.; Patrick, J.W.; Bouzayen, M.; Osorio, S.; Fernie, A.R. Molecular regulation of seed and fruit set. Trends Plant Sci. 2012, 17, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Guo, D.; Liu, Y.; Zhu, L.; Xie, F.; Xie, L. RNA-Seq combined with population-level analysis reveals important candidate genes related to seed size in flax (Linum usitatissimum L.). Front. Plant Sci. 2022, 13, 1015399. [Google Scholar] [CrossRef]
- Li, M.; Dong, X.; Peng, J.; Xu, W.; Ren, R.; Liu, J.; Cao, F.; Liu, Z. De novo transcriptome sequencing and gene expression analysis reveal potential mechanisms of seed abortion in dove tree (Davidia involucrata Baill.). BMC Plant Biol. 2016, 16, 82. [Google Scholar] [CrossRef]
- Jafari, M.; Shiran, B.; Rabiei, G.; Ravash, R.; Sayed Tabatabaei, B.E.; Martínez-García, P.J.; Martínez-Gómez, P. Identification of genes associated with kernel size in almond [Prunus dulcis (Mill.) DA Webb] using RNA-Seq. Plant Growth Regul. 2022, 97, 357–373. [Google Scholar] [CrossRef]
- Gray, J.; Caparrós-Ruiz, D.; Grotewold, E. Grass phenylpropanoids: Regulate before using! Plant Sci. 2012, 184, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Le Roy, J.; Huss, B.; Creach, A.; Hawkins, S.; Neutelings, G. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 2016, 7, 202190. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q. Lignification: Flexibility, biosynthesis and regulation. Trends Plant Sci. 2016, 21, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Treutter, D. Significance of flavonoids in plant resistance: A review. Environ. Chem. Lett. 2006, 4, 147–157. [Google Scholar] [CrossRef]
- Cao, Y.; Li, K.; Li, Y.; Zhao, X.; Wang, L. MYB transcription factors as regulators of secondary metabolism in plants. Biology 2020, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Xiao, Y.; Lv, Z.; Tan, H.; Chen, R.; Li, Q.; Chen, J.; Wang, Y.; Yin, J.; Zhang, L. AP2/ERF transcription factor, Ii049, positively regulates lignan biosynthesis in Isatis indigotica through activating salicylic acid signaling and lignan/lignin pathway genes. Front. Plant Sci. 2017, 8, 1361. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.k.; Li, X.; Xu, Q.; Chen, J.y.; Yin, X.r.; Ferguson, I.B.; Chen, K.s. Ej AP 2-1, an AP 2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with Ej MYB transcription factors. Plant Biotechnol. J. 2015, 13, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y.; Rochfort, S.; Liu, Z.; Ran, Y.; Griffith, M.; Badenhorst, P.; Louie, G.V.; Bowman, M.E.; Smith, K.F.; Noel, J.P. Functional analyses of caffeic acid O-methyltransferase and cinnamoyl-CoA-reductase genes from perennial ryegrass (Lolium perenne). Plant Cell 2010, 22, 3357–3373. [Google Scholar] [CrossRef]
- Shigeto, J.; Tsutsumi, Y. Diverse functions and reactions of class III peroxidases. New Phytol. 2016, 209, 1395–1402. [Google Scholar] [CrossRef]
- Francoz, E.; Ranocha, P.; Nguyen-Kim, H.; Jamet, E.; Burlat, V.; Dunand, C. Roles of cell wall peroxidases in plant development. Phytochemistry 2015, 112, 15–21. [Google Scholar] [CrossRef]
- Andrews, J.; Malone, M.; Thompson, D.S.; Ho, L.; Burton, K. Peroxidase isozyme patterns in the skin of maturing tomato fruit. Plant Cell Environ. 2000, 23, 415–422. [Google Scholar] [CrossRef]
- Aydin, N.; Kadioglu, A. Changes in the chemical composition, polyphenol oxidase and peroxidase activities during development and ripening of medlar fruits (Mespilus germanica L.). Bulg. J. Plant Physiol 2001, 27, 85–92. [Google Scholar]
- Lee, Y.; Rubio, M.C.; Alassimone, J.; Geldner, N. A mechanism for localized lignin deposition in the endodermis. Cell 2013, 153, 402–412. [Google Scholar] [CrossRef] [PubMed]
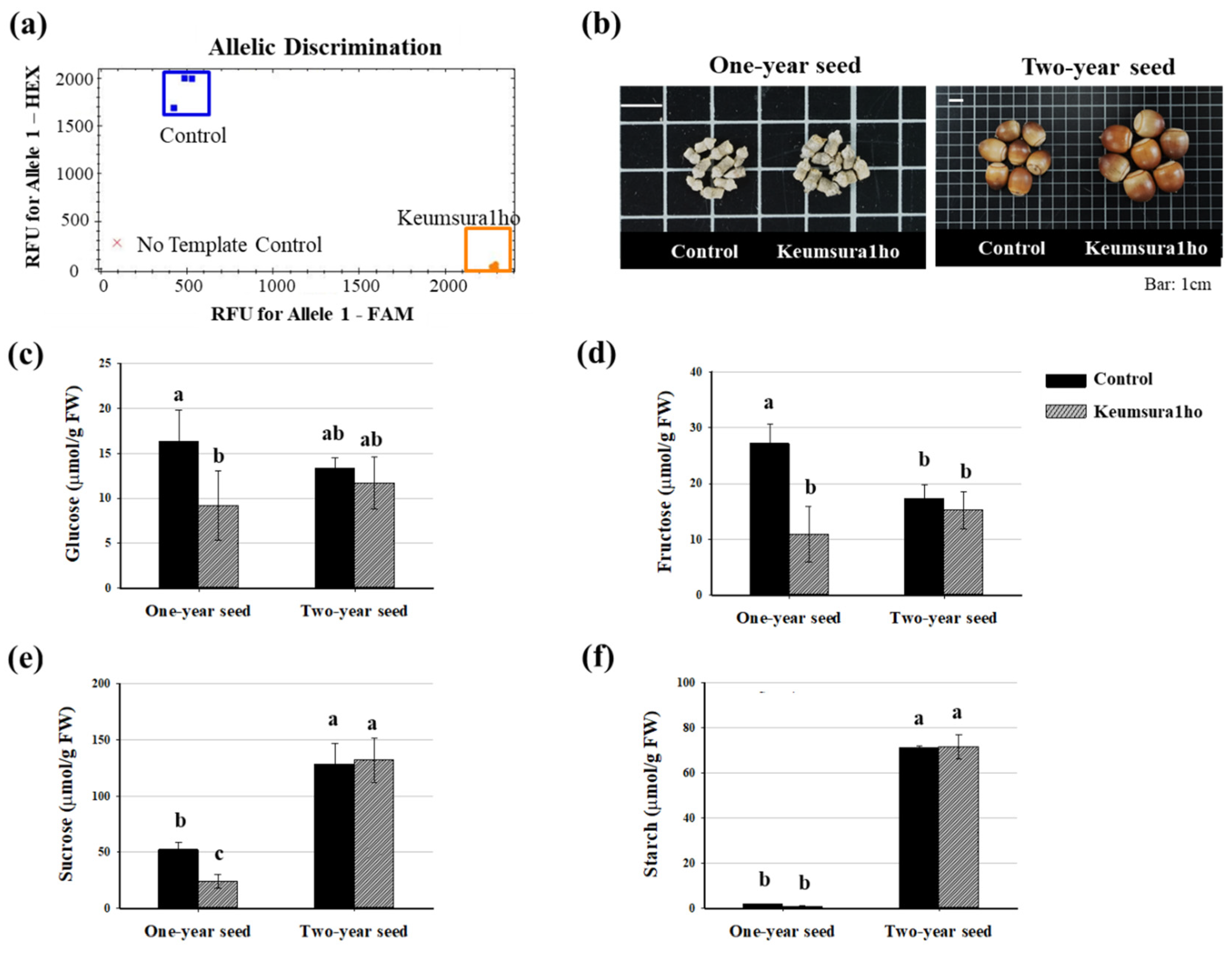
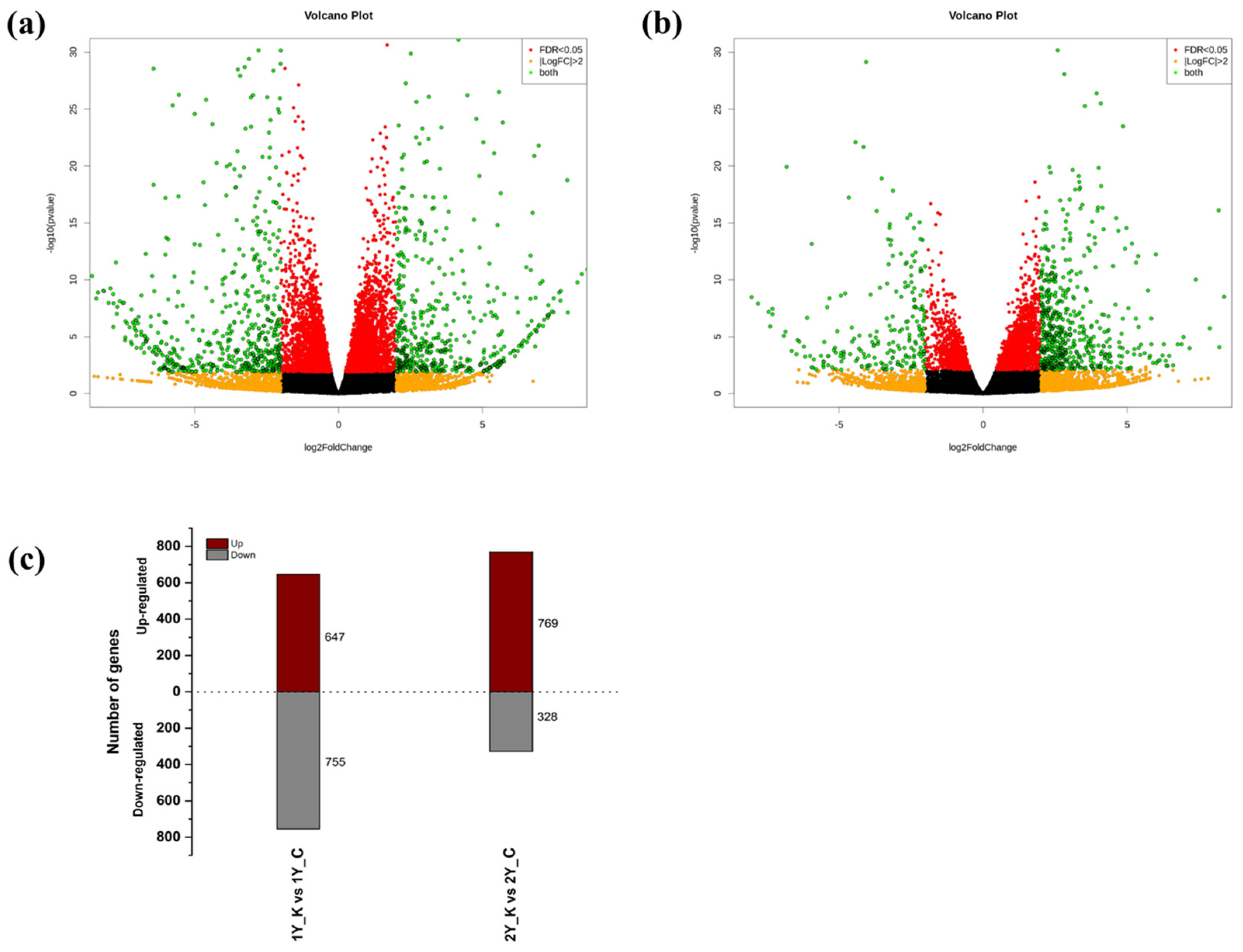
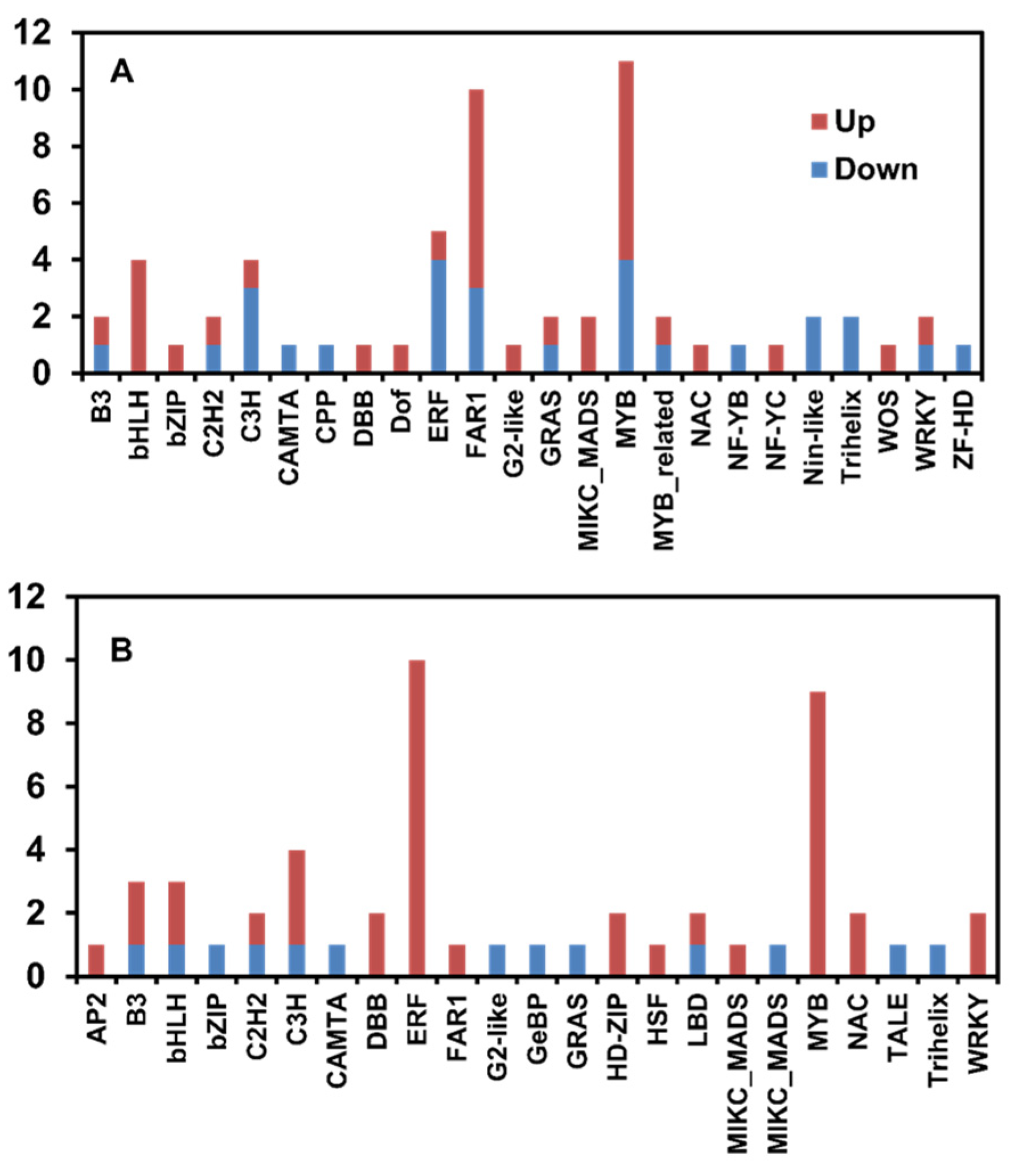
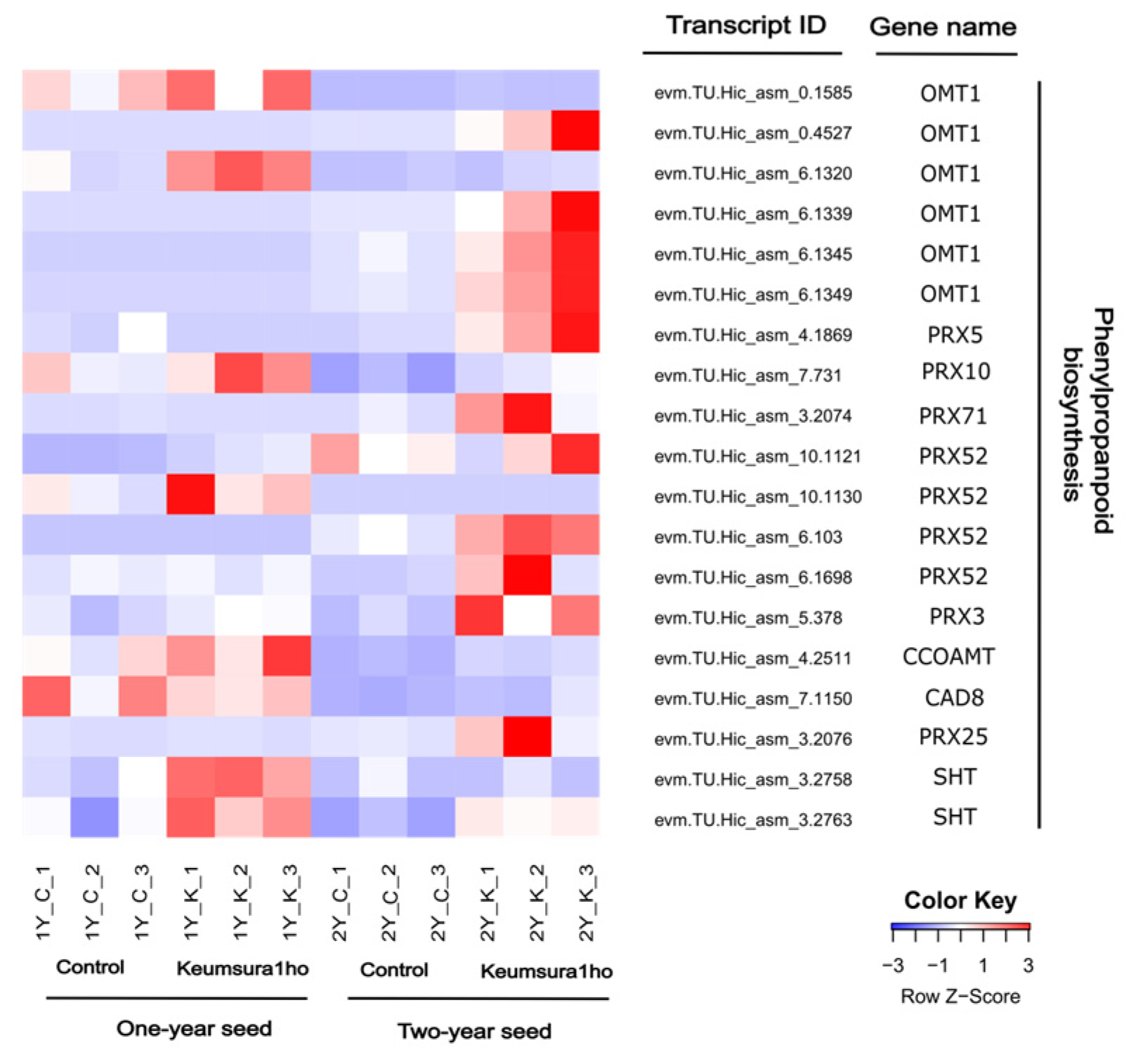

| Sample | Number of Measured Seeds | Average Seed Weight (g) | Seed Height (mm) | Seed Width (mm) | Seed Length (mm) |
|---|---|---|---|---|---|
| Control Oak | 300 | 3.5 ± 0.04 | 20.16 ± 0.09 | 16.53 ± 0.07 | 16.50 ± 0.75 |
| Keumsura1ho | 562 | 7.8 ± 0.04 | 23.93 ± 0.05 | 22.05 ± 0.42 | 22.08 ± 0.04 |
| Comparison | GO-Term | Gene Number | Rich Factor (%) | False Discovery Rate |
|---|---|---|---|---|
| 1Y_K vs 1Y_C | Monoterpenoid biosynthesis | 4 | 0.92 | 0.014 |
| 1Y_K vs 1Y_C | Biosynthesis of secondary metabolites | 38 | 8.78 | 0.022 |
| 1Y_K vs 1Y_C | Biosynthesis of various plant secondary metabolites | 7 | 1.62 | 0.035 |
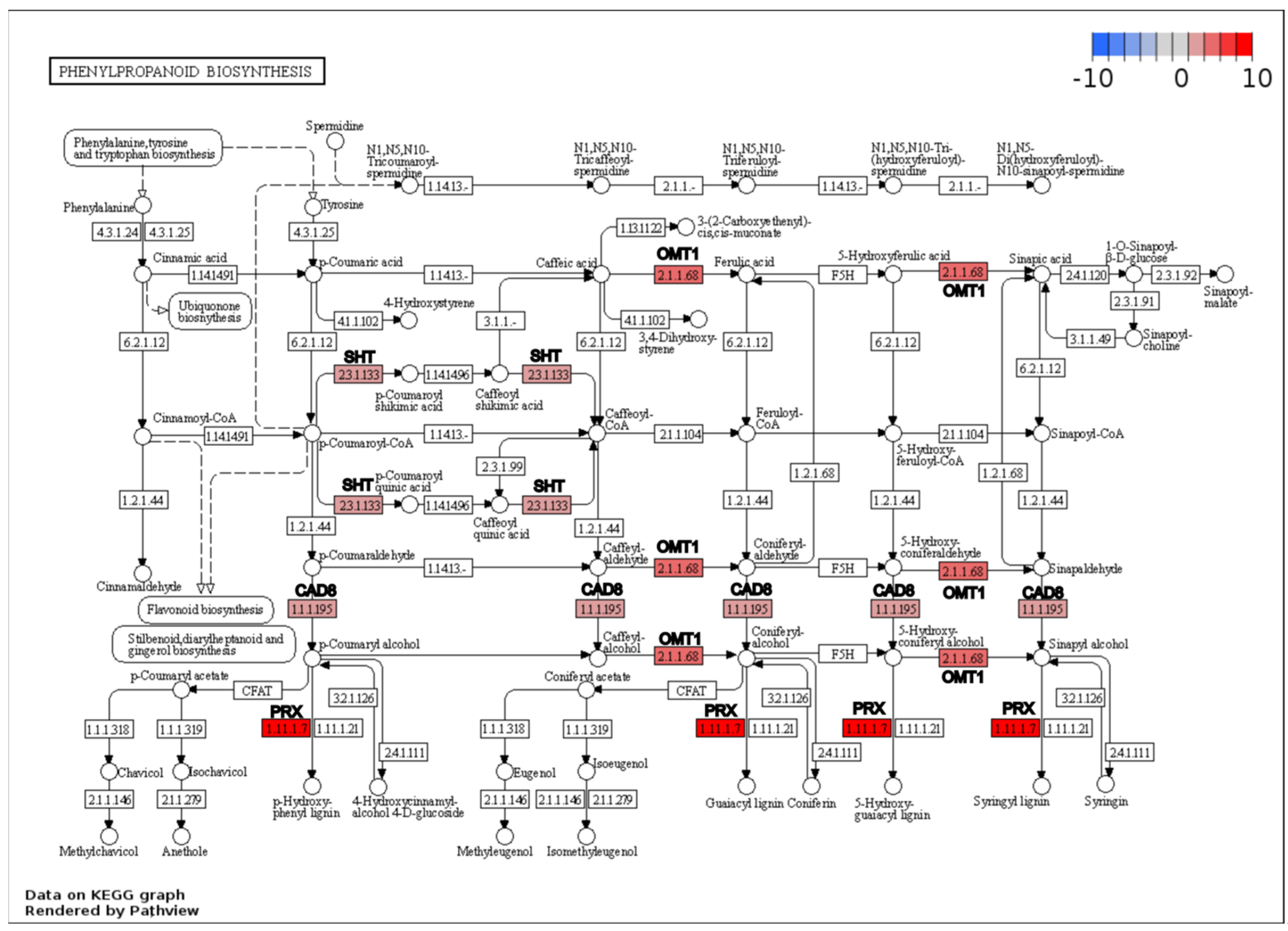
| 2Y_K vs 2Y_C | Phenylpropanoid biosynthesis | 10 | 1.92 | 0.011 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Byeon, S.; Lee, K.; Cheon, K.-S.; Denison, M.I.J.; Kim, T.-L.; Park, D.; Lim, H. RNA Sequencing Reveals Transcription Factors and Genes in Phenylpropanoid Biosynthesis That Positively Regulate Size and Weight of Oak Tree Seeds. Horticulturae 2024, 10, 829. https://doi.org/10.3390/horticulturae10080829
Byeon S, Lee K, Cheon K-S, Denison MIJ, Kim T-L, Park D, Lim H. RNA Sequencing Reveals Transcription Factors and Genes in Phenylpropanoid Biosynthesis That Positively Regulate Size and Weight of Oak Tree Seeds. Horticulturae. 2024; 10(8):829. https://doi.org/10.3390/horticulturae10080829
Chicago/Turabian StyleByeon, Siyeon, Kyungmi Lee, Kyeong-Seong Cheon, Michael Immanuel Jesse Denison, Tae-Lim Kim, Danbe Park, and Hyemin Lim. 2024. "RNA Sequencing Reveals Transcription Factors and Genes in Phenylpropanoid Biosynthesis That Positively Regulate Size and Weight of Oak Tree Seeds" Horticulturae 10, no. 8: 829. https://doi.org/10.3390/horticulturae10080829
APA StyleByeon, S., Lee, K., Cheon, K.-S., Denison, M. I. J., Kim, T.-L., Park, D., & Lim, H. (2024). RNA Sequencing Reveals Transcription Factors and Genes in Phenylpropanoid Biosynthesis That Positively Regulate Size and Weight of Oak Tree Seeds. Horticulturae, 10(8), 829. https://doi.org/10.3390/horticulturae10080829






