Reference Gene Selection and Gene Expression Analysis during Gall Development of Zizania latifolia
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Materials
2.2. RNA Extraction, Quality Testing, and cDNA Reverse Transcription
2.3. Primer Design of Candidate Reference Genes and Pregnancy-Related Genes in Jiaobai
2.4. Stability Analysis of Candidate Reference Genes and Identification of Optimal Reference Genes
3. Results
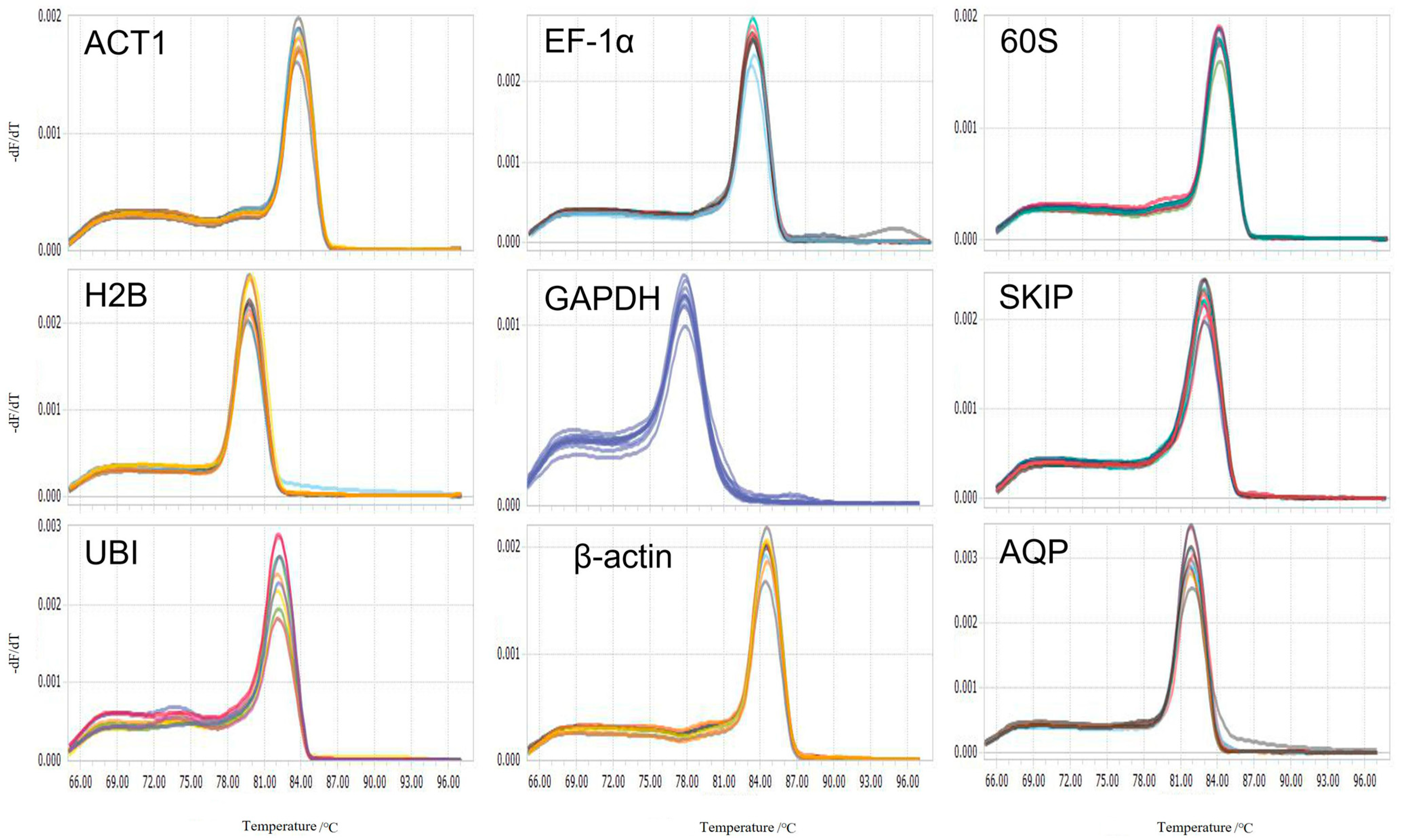
3.1. Total RNA Quality Determination and Primer Specificity Analysis
3.2. Abundance Analysis of Candidate Reference Genes
3.3. Expression Stability of Candidate Reference Genes during Pregnancy of Z. latifolia
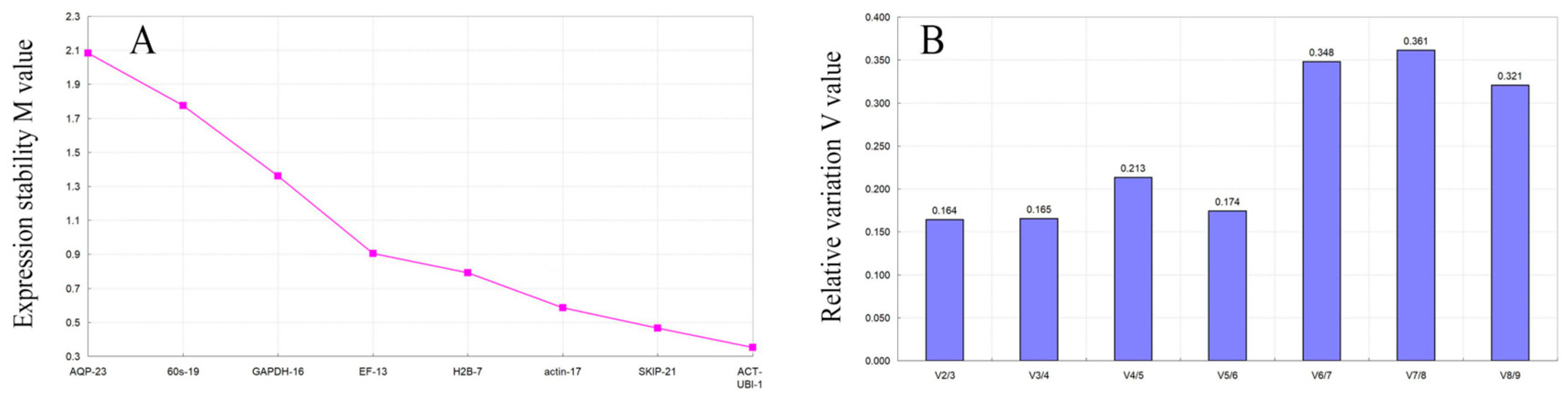
3.3.1. geNorm Analysis
3.3.2. NormFinder Analysis
3.4. Analysis of Key Genes during Gall Development of Jiaobai
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative real-time RT-PCR–a perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Ginzinger, D.G. Gene quantification using real-time quantitative PCR: An emerging technology hits the mainstream. Exp. Hematol. 2002, 30, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Derveaux, S.; Vandesompele, J.; Hellemans, J. How to do successful gene expression analysis using Real-time PCR. Methods 2010, 50, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Meng, C.; Wang, Y.; Zhao, J.; Chen, X.; Shen, S. Reference Genes Selection for Quantitative Real-time PCR in Chinese Cabbage-Cabbage Translocation Lines. Acta Agric. Boreali-Sin. 2018, 33, 60–67. [Google Scholar]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using Real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, G.; Lu, Y.; Yang, Y.; Zheng, X.; Tian, J. Screening of internal reference genes of Chilo suppressalis by real-time fluorescence quantitative PCR and evaluation of expression stability. Chin. Rice Sci. 2019, 33, 75–84. [Google Scholar]
- Wan, H.; Zhao, Z.; Qian, C.; Sui, Y.; Malik, A.A.; Chen, J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 2010, 399, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Wang, Y.; Cao, Y.; He, J.; Jia, M.; Li, Z. Reference genes selection and related genes expression analysis under low and high temperature stress in Taraxacum officinale. Acta Hortic. Sin. 2020, 47, 1153–1164. [Google Scholar]
- Pang, Q.; Li, Z.; Luo, S.; Chen, R.; Jin, Q.; Li, Z. Selection and stability analysis of reference gene for qRT-PCR in eggplant under high temperature stress. Acta Hortic. Sin. 2017, 44, 475–486. [Google Scholar]
- Li, Q.; Sun, S.; Yuan, D.; Yu, H.; Gu, M.; Liu, Q. Validation of candidate reference genes for the accurate normalization of real-time quantitative RT-PCR data in rice during seed development. Plant Mol. Biol. Report. 2010, 28, 49–57. [Google Scholar] [CrossRef]
- Song, X.; Yang, S.; Zhong, Q.; Wang, L.; Zhao, M.; Li, L. Selection of reference genes for quantitative RT-PCR analysis of Helianthus tuberosus. Mol. Plant Breed. 2018, 16, 1190–1196. [Google Scholar]
- Jiang, T.; Gao, Y.; Tong, Z. Selection of reference genes for quantitative real-time PCR in Lycoris. Acta Hortic. Sin. 2015, 42, 1129–1138. [Google Scholar]
- Ke, W.; Zhou, G.; Peng, J.; Huang, X.; Liu, Y.; Li, S. Systematic cluster analysis and comprehensive evaluation of water bamboo resources. Chin. Veg. 2000, 1, 18–21. [Google Scholar]
- Wang, L.; Yang, M.; Li, Y.; Zhang, S.; Zheng, Z. Genetic diversity of Zizania latifolia germplasm resourcesbased on ISSR technology. Zhejiang Agric. Sci. 2019, 60, 732–735. [Google Scholar]
- Guo, L.; Qiu, J.; Han, Z.; Ye, Z.; Chen, C.; Liu, C. A host plant genome (Zizania latifolia) after a century-long endophyte infection. Plant J. 2015, 83, 600–609. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lu, Z.; Yang, Y.; Hou, J.; Yuan, L.; Chen, G. Transcriptome analysis reveals the symbiotic mechanism of Ustilago esculenta-induced gall formation of Zizania latifolia. Mol. Plant-Microbe Interact. 2021, 34, 168–185. [Google Scholar] [CrossRef]
- Wu, J.; He, B.; Du, Y.; Li, W.; Wei, Y. Analysis method of systematically evaluating stability of reference qenes using geNorm, NormFinder and BestKeeper. Mod. Agric. Sci. Technol. 2017, 5, 278–281. [Google Scholar]
- Mashiguchi, K.; Tanaka, K.; Sakai, T. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef] [PubMed]
- Kang, L. Cloning and Functional Analysis of NPR1 Gene in Zizania latifolia. Master’s Thesis, China Jiliang University, Hangzhou, China, 2017. [Google Scholar]
- Yu, L.; Tang, X.; Wu, X.; Yan, B. Segregate cloning and sequence analysis of lea3 gene complete cDNA in Zizania caduciflora. China Veg. 2010, 6, 14–18. [Google Scholar]
- Ma, L.; Duan, Q.; Cui, G.; Du, W.; Jia, W.; Wang, X. Screening of qRT-PCR internal reference genes related to anthocyanidin synthesis in Anemone crassifolia. Acta Hortic. Sin. 2021, 48, 377–388. [Google Scholar]
- Song, X.; Chang, Y.; Liu, H.; Xu, H.; Pei, D. Reference gene selection and genes expression analysis during adventitious root formation in walnut. Acta Hortic. Sin. 2019, 46, 1907–1918. [Google Scholar]
- Zhou, X.; Liu, J.; Zhuang, Y. Selection of appropriate reference genes in Solanum aculeatissimum for quantitative gene expression studies under different experimental conditions. Acta Hortic. Sin. 2014, 41, 1731–1738. [Google Scholar]
- Li, S.; Ye, X.; Wang, B.; Chen, M.; Liu, J.; Zhu, H. Cloning and screening evaluation of real-time fluorescence quantitative PCR reference gene of okra. J. Nucl. Agron. 2021, 35, 60–71. [Google Scholar]
- Ye, X. YUCCA Cloned and Transgenosis Analysis in Rice. Master’s Thesis, Zhejiang Normal University, Jinhua, China, 2012. [Google Scholar]
- Jiang, J.; Cao, B.; Huang, K.; Zhang, Q.; Han, X.; Zhu, Q. Changes of NSC, enzymes and endogenous hormones during Zizania Gall’s Swelling. Acta Hortic. Sin. 2005, 32, 134–137. [Google Scholar]



| Reference Gene | Primer Sequence | Product Length (bp) | Tm (°C) |
|---|---|---|---|
| ACT1 | F: GTCAAGGCAGGTTTTGCTGG R: CACCCACGTAGGCATCCTTT | 118 | 60 |
| H2B | F: AAGAAGGCGAAGAAGAGCGT R: GGCGAGCTTCTCGAAGATGT | 141 | 60 |
| UBI | F: GCACAGATCCTCCCTCTTCG R: CGTCCATCCCAAGCTCAAGT | 143 | 60 |
| EF-1α | F: TTCCGATACCGCCGATCTTG R: GTCTCCGGTAAGACCCTCCT | 115 | 60 |
| GAPDH | F: TGCTGCCTTCTTCTTCCCTG R: GGACATGAAGTCGTCGGAGG | 161 | 60 |
| β-actin | F: GGATTGGGCCTCATCACCAA R: TGGAACCGGAATGGTCAAGG | 145 | 60 |
| 60S | F: AGCTTTTCCCTGGCCTTTGT R: TCTGTTCTGTGCCTGACCAC | 170 | 60 |
| SKIP | F: CAGGTCATATTCCTCCCCGC R: TGAGACACAGTCATGGGCAC | 145 | 60 |
| AQP | F: TGGACCTGGGACTCATTTGC R: CGAGCAGGGTAGGCATGATT | 141 | 60 |
| Pregnancy-Related Gene | Primer Sequence | Product Length (bp) | Tm (°C) |
|---|---|---|---|
| ZlYUCCA | F: CCTAGAAGGTAGCAAGCAGA R: TAAACCCAGTGAAAAAAACG | 179 | 60 |
| ZlNPR1 | 1F: GCTTTGGCAAGGATAATGTTTC 1R: GTTTCCCGAGTTCCACTGTTT | 214 | 56 |
| ZlPR1 | 1F: CACTACACGCAGATCGTGTGG 1R: GTAGTAGTTGCAGGTCATGAAG | 96 | 56 |
| ZlLEA3 | 1F: GCCGGCGAGRCCAAGGSMC 1R:GSCGGWYTTGTCCTTGCCGG | 300 | 58 |
| Sampling Time (Day) | A260/A280 | A260/A230 |
|---|---|---|
| −3 | 2.13 | 2.02 |
| 1 | 2.08 | 1.83 |
| 5 | 2.14 | 1.96 |
| 9 | 2.13 | 1.93 |
| 13 | 2.11 | 1.88 |
| Reference Gene | Stability Value | Ranking |
|---|---|---|
| ACT1 | 0.255 | 1 |
| UBI | 0.333 | 2 |
| H2B | 0.414 | 3 |
| SKIP | 0.442 | 4 |
| β-actin | 0.623 | 5 |
| EF-1α | 0.813 | 6 |
| GAPDH | 1.621 | 7 |
| AQP | 1.970 | 8 |
| 60S | 1.973 | 9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Yi, H.; Gu, Q.; Zheng, Z.; Zhu, M.; Zha, X.; Zhang, S.; Yang, M. Reference Gene Selection and Gene Expression Analysis during Gall Development of Zizania latifolia. Horticulturae 2024, 10, 759. https://doi.org/10.3390/horticulturae10070759
Li Y, Yi H, Gu Q, Zheng Z, Zhu M, Zha X, Zhang S, Yang M. Reference Gene Selection and Gene Expression Analysis during Gall Development of Zizania latifolia. Horticulturae. 2024; 10(7):759. https://doi.org/10.3390/horticulturae10070759
Chicago/Turabian StyleLi, Yipeng, Huan Yi, Qing Gu, Zhaisheng Zheng, Mingxing Zhu, Xiaojun Zha, Shangfa Zhang, and Mengfei Yang. 2024. "Reference Gene Selection and Gene Expression Analysis during Gall Development of Zizania latifolia" Horticulturae 10, no. 7: 759. https://doi.org/10.3390/horticulturae10070759
APA StyleLi, Y., Yi, H., Gu, Q., Zheng, Z., Zhu, M., Zha, X., Zhang, S., & Yang, M. (2024). Reference Gene Selection and Gene Expression Analysis during Gall Development of Zizania latifolia. Horticulturae, 10(7), 759. https://doi.org/10.3390/horticulturae10070759






