Abstract
Nuclear factor Ys (NF-Ys) are heterotrimeric transcription factors that specifically bind to CCAAT boxes present in numerous eukaryotic promoters. In plants, NF-Y proteins consist of the following three subunits: NF-YA, NF-YB, and NF-YC, each encoded by a gene family. Accumulating evidence underscores the crucial roles of NF-Y proteins in various plant development processes and stress responses, such as embryogenesis, flowering time control, drought tolerance, and heat tolerance. Despite this, a comprehensive genome-wide overview of the NF-Y gene family in strawberries is lacking. To bridge this gap, this study was conducted to identify and characterize the NF-Ys in Fragaria vesca. The investigation revealed the presence of six NF-YA, twelve NF-YB, and five NF-YC members in F. vesca. Furthermore, a comprehensive analysis of the FveNF-Ys was performed, including their phylogenetic relationships, gene structures, chromosomal locations, and conserved domains. MiRNA target site prediction found that there were 30 miRNA target sites in 12 (52.2%) FveNF-Y genes. Additionally, the expression profiles of different tissues and developmental stages demonstrated tissue-specific expression patterns among certain members of each NF-Y subfamily. This observation suggests that specific NF-Y subfamily members may play unique roles in different tissues or stages of development. Furthermore, the transient expression assay demonstrated that three selected FveNF-Ys were localized in the nucleus. Our study represents a pioneering effort in the systemic analyses of FveNF-Y genes and will be useful in understanding the functional characterization of NF-Y genes in Fragaria species.
1. Introduction
Nuclear factor Y (NF-Y), also known as CCAAT-box binding factor (CBF) or Heme Activator Protein (HAP), is a heterotrimeric transcription factor complex composed of the following three distinct subunits: NF-YA (CBF-B, HAP2), NF-YB (CBF-A, HAP3), and NF-YC (CBF-C, HAP5). Each of these subunits contains a highly conserved central core region that is essential for subunit interaction and DNA binding. Studies in yeast and mammals have revealed that the assembly of the NF-Y complex follows a strict stepwise pattern. Initially, NF-YB and NF-YC form a heterodimer in the cytoplasm through histone fold motif interaction. Following this initial interaction, this heterodimer transfers to the nucleus. In the nucleus, the mature and functional heterotrimeric NF-Y complex is formed by recruiting the third subunit, NF-YA. The mature NF-Y complex specifically binds to CCAAT-boxes, which are cis-elements found in promoters, thus regulating the transcription of downstream genes. Approximately 25% to 30% of mammalian genome promoters contain CCAAT-boxes [1]. While each NF-Y subunit is encoded by a single gene in yeast, plants encode multiple members for each NF-Y subunit. This difference in encoding leads to a wide range of potential functional heterotrimeric NF-Y combinations.
Plant NF-Y complexes, as well as individual members of each NF-Y subunit family, play important roles in regulating various growth and development processes. For instance, in Arabidopsis, an NF-Y complex composed of NF-YA2, NF-YB2, or NF-YB3, and NF-YC3 or NF-YC9 interacts with CONSTANS in the photoperiod pathway and DELLAs in the gibberellin pathway. These interactions directly regulate the transcription of flowering regulator SOC1 (SUPPERESSOR OF OVEREXPRESSION OF CONSTANS 1) and control flowering time [2,3,4]. Moreover, a distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter, thereby regulating the timing of flowering in Arabidopsis. MiR169 and its target gene AtNF-YA2 have been reported to function in stress-induced flowering [5] and also control root architecture [6]. Interestingly, Arabidopsis NF-YB family members LEC1 (LEAFY COTYLEDON1; AtNF-YB9) and L1L (LEAFY COTYLEDON1-LIKE; AtNF-YB6) play critical roles in embryogenesis, seed maturation, post-embryonic development, and fatty acid biosynthesis [7,8,9,10]. Furthermore, the overexpression of AtNF-YA1, 3, 5, 6, 8, and 9 affects various processes such as male gametogenesis, embryogenesis, seed morphology, and germination [11,12]. In Phaseolus vulgaris, NF-YC1 and NF-YA1 interact with SIN1 and play a central role in nodule organogenesis and lateral root growth [13]. These collective findings demonstrate that plant NF-Y complexes and individual members of each NF-Y subunit family play vital roles in diverse developmental processes and stress responses. Therefore, they make valuable targets for further study in plant biology.
NF-Y proteins have been identified as crucial regulators of stress tolerance. For example, in Arabidopsis, overexpressing AtNF-YA5, a transcriptional factor gene highly expressed in vascular tissues and guard cells, resulted in reduced leaf water loss and increased resistance to drought compared with the wild type. This gene is strongly induced by drought stress in an abscisic acid (ABA)-dependent manner. Conversely, nfya5 knockout plants showed enhanced leaf water loss and increased sensitivity to drought stress compared with wild-type plants [14]. Additionally, the overexpression of DPB3-1 enhanced heat stress tolerance without growth retardation. This was achieved by interacting with NF-YA2 and NF-YB3 to form a transcriptional complex. DREB2A (DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A), a key transcription factor, induces the expression of both dehydration- and heat-stress-inducible genes under the corresponding stress conditions. Interestingly, it selectively enhances heat stress-inducible gene expression during heat stress responses through its interaction with the NF-Y transcriptional complex [15,16]. In Arabidopsis, a transcriptional complex composed of NF-YA4, NF-YB3, and NF-YC2 interacts with the membrane-associated basic domain/leucine zipper (bZIP) transcription factor bZIP28 to regulate stress responses [17]. Similarly, in rice, the expression level of OsHAP2E was induced after inoculation with Magnaporthe oryzae and after treatment with various stressors, such as SA, INA, ABA, and H2O2. Notably, overexpressing OsHAP2E conferred resistance to Magnaporthe oryzae, as well as to salinity and drought [18]. In strawberries, cold treatment induced a more pronounced coloration in Hisone-like transcription factor (CBF/NF-Y) fruits [19].
In this study, a comprehensive analysis of the strawberry NF-Y families was conducted. A total of six FveNF-YAs, 12 FveNF-YBs, and five FveNF-YCs were identified and classified within the Fragaria vesca genome. Subsequently, the phylogenetic relationships, gene structures, chromosomal locations, conserved domains, and protein structure of strawberry NF-Y family genes were examined. Additionally, the expression patterns of FveNF-Ys in different tissues and developmental stages were investigated, and the subcellular location of three selected FveNF-Ys localized in the nucleus was demonstrated. These findings provide a comprehensive analysis of F. vesca NF-Y families and are expected to further characterize their functions and genome-wide analysis in strawberries.
2. Materials and Methods
2.1. Identification and Analysis of Strawberry NF-Y Family Members
In order to identify NF-Y genes in the F. vesca genome, three methods were applied sequentially. Firstly, annotation information, including the GFF3 file, amino acid, and nucleotide sequences of the strawberry, was downloaded from the GDR [20]. Protein sequences of 30 NF-Y genes from Arabidopsis thaliana were downloaded from the TAIR database (https://www.arabidopsis.org/, accessed on 7 January 2024), based on their gene names, to use as query sequences [21]. These 30 sequences were selected based on their known functional relevance and representation of the three NF-Y subfamilies (NF-YA, NF-YB, and NF-YC). The BLASTP program [22] was then utilized with an e-value cutoff of <1 × 10−5 to identify homologous NF-Y genes in F. vesca. Additional filters included a minimum alignment coverage of 50% and a minimum sequence identity of 30%. Secondly, Hidden Markov Model (HMM) profiles of the NF-Y domains (PF00808 and PF02045) were obtained from the Pfam database [23]. The hmmsearch program with an e-value cutoff of <1 × 10−3 was used to identify candidate NF-Y proteins in the F. vesca genome based on these domain profiles. Finally, the presence of conserved domains in the identified candidate NF-Y proteins was confirmed using the Conserved Domains Database (CDD, https://www.ncbi.nlm.nih.gov/Structure/, accessed on 8 January 2024). Additional information about the identified NF-Y proteins, such as sequence length, predicted molecular weight, and predicted isoelectric point, was obtained from the ExPASy website [24].
2.2. Phylogenetic Analysis of FveNF-Ys and AtNF-Ys
To determine the relationship between FveNF-Y proteins and AtNF-Y proteins, a phylogenetic analysis was performed using NF-Y proteins from both the strawberry and Arabidopsis thaliana. Full-length protein alignments were conducted using MUSCLE (Version 5) software [25] with default parameters. The resulting alignments were imported into MEGA11 (Version 11.0.13) software [26] to create phylogenetic trees with the following parameters: Maximum Likelihood (ML) method, Jones–Taylors–Thornton (JTT) model, Gamma Distributed (G), partial deletion, and 1000 bootstrap replicates. No additional adjustments were made to the default settings in MEGA11. The resulting tree was viewed using Evolview (V3) software [27].
2.3. Gene Structures, Conserved Motif, and Domain Analysis
The exon/intron structure of each FveNF-Y gene was analyzed using TBtools V1.098769 [28]. NF-Y conserved motifs were identified using the MEME-Suite online program [29]. The analysis was performed using the following parameters: a maximum of 10 motifs to be identified, with a motif length set between 6 and 50 amino acids, and an E-value threshold of 0.05 to accept a motif as conserved. The results were visualized using TBtools V1.098769 software [28]. Additionally, the conserved domains of the identified NF-Y proteins were identified using Batch Web CD-search tools [30] with default parameters. The alignments of domains were then analyzed and visualized using Jalview [31].
2.4. Chromosomal Distribution and Gene Duplication of FveNF-Y Genes
The physical position information of each strawberry NF-Y gene was obtained from the strawberry GFF3 file and visualized using TBtools [28]. Gene duplication events were analyzed using multiple collinear scanning toolkits (MCScanX) with the default settings, which include a match score of 50, gap penalty of −1, E-value of 1 × 10−5, and minimum block size of 5 genes [32]. The syntenic relationship among FveNF-Ys, OsNF-Ys, and AtNF-Ys was determined using MCScanX with the default settings. Syntenic regions were defined based on these parameters, ensuring that regions with significant collinearity (E-value < 1 × 10−5) were considered. The results were visualized using multiple synteny plot tools in TBtools [28].
2.5. miRNA Target Site Prediction
The miRNA sequences of the strawberry were obtained from our previous works [33], while the FveNF-Y transcript sequences were used for miRNA target site prediction using the psRNATarget website (https://www.zhaolab.org/psRNATarget/analysis?function=3, accessed on 1 February 2024) with the default settings, which include a maximum expectation value of 5, length for complementarity scoring (hspsize) of 19, and flanking region length around target site of 17 bp upstream and 13 bp downstream. Target sites were considered valid based on these default parameters, and additional filtering was applied to eliminate false positive predictions by considering only targets with expectation values less than 5.0 and high complementarity scores.
2.6. Cis-Acting Element Analysis for FveNF-Ys Promoters
The upstream 2 kb region of the start codon of each FveNF-Y gene was defined as the promoter. Promoter sequences were retrieved from the F. vesca genome, and putative cis-acting elements for each FveNF-Y gene were predicted using PlantCARE [34]. The entire 2 kb upstream region was considered in the analysis without additional criteria. The identified cis-regulatory elements were categorized based on their involvement in plant growth and development, abiotic stress, and phytohormone responsiveness. Elements with higher functional relevance were selected based on their frequency of occurrence and known functional annotations in the literature.
2.7. Protein Structure Analysis
Firstly, the protein secondary structure of FveNF-Y proteins was predicted using the GOR IV algorithm available at https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_gor4.html (accessed on 21 February 2024) with default parameters. Subsequently, the 3D protein structure was predicted using ColabFold, an accelerated structure prediction tool based on AlphaFold2. ColabFold structures were generated utilizing Google ColabFold (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb (accessed on 21 February 2024)) with the following configurations: MSA mode set to MMseqs2 (UniRef + Environmental), pair mode set to unpaired_paired, and other parameters set to their default values.
2.8. Gene Expression Analysis
Transcripts Per Million (TPM) values for each FveNF-Y gene in various tissues and stages were extracted from the transcriptome datasets in our previous study [33]. The corresponding TPM values for each FveNF-Y gene in different tissues and stages can be found in Table S1. A heatmap was generated based on the normalized expression values of each FveNF-Y gene.
2.9. Subcellular Localization
The coding sequences of FveNF-YA3, FveNF-YA4, and FveNF-YC5 were amplified from F. vesca and cloned into the GFP::pCAMBIA 1302 vector. The primers used for this vector construction are listed in Table S5. Following the successful construction of the plasmids, each was separately transformed into the Agrobacterium strain GV3101. For the purpose of transient expression, the Agrobacterium culture was resuspended in an infiltration medium and then introduced into the leaves of Nicotiana benthamiana. The transformed 35S::GFP served as a negative control. The nucleus localization was determined using 4′,6-diamidino-2phenylindole (DAPI). Results were assessed 3–5 days post-infection using fluorescence microscopy (Leica CTR6000, Leica Microsystems, Wetzlar, Germany).
3. Results
3.1. Identification and Phylogenetic Analysis of NF-Y Genes in F. vesca
To identify the NF-Y gene families in the strawberry, a combination of BLASTP and the HMM algorithm was used to analysis of the F. vesca genome. A total of 23 NF-Y family genes were identified, comprising six FveNF-YAs, 12 FveNF-YBs, and five FveNF-YCs. The 23 FveNF-Y genes were subsequently named as follows: FveNF-YA1 to FveNF-YA6, FveNF-YB1 to FveNF-YB12, and FveNF-YC1 to FveNF-YC5, respectively (Table 1). The lengths of the 26 FveNF-Ys proteins varied between 126 AA and 340 AA, indicating a wide range of FveNF-Y lengths. Among the three subfamilies of FveNF-Y, FveNF-YAs generally exhibited the longest lengths, ranging from 242 AA to 340 AA with an average length of 305.7 AA. FveNF-YBs displayed shorter lengths, ranging from 155 AA to 259 AA with an average length of 214.3 AA. On the other hand, FveNF-YCs showed the shortest lengths, ranging from 126 AA to 284 AA with an average length of 239.6 AA. The predicted molecular weights (Mws) of the 26 FveNF-Ys ranged from 13.84 (FveNF-YC3) to 37.23 (FveNF-YA2), while the predicted theoretical isoelectric points (pIs) ranged from 4.99 (FveNF-YB4) to 9.55 (FveNF-YA4). The coding sequences and protein sequences of the identified FveNF-Y gene members are listed in Table S3.

Table 1.
Basic information of the nuclear factor Y (NF-Y) gene family in F. vesca.
To examine the phylogenetic relationship among strawberry NF-Y proteins, an unrooted phylogenetic tree was constructed using MEGA11 software [26] based on 30 AtNF-Y and 23 FveNF-Y protein sequences (Figure 1). The phylogenetic analysis revealed that these 63 NF-Y proteins were categorized into three subunits as follows: NF-YA (depicted in pink), NF-YB (depicted in yellow), and NF-YC (depicted in orange). Among them, 10 of the AtNF-YAs and six FveNF-YAs were classified under the NF-YA subunit, while 10 AtNF-YBs and 12 FveNF-YBs were assigned to the NF-YB subunit. Similarly, 10 AtNF-YCs and five FveNF-YCs were allocated to the NF-YC subunit.
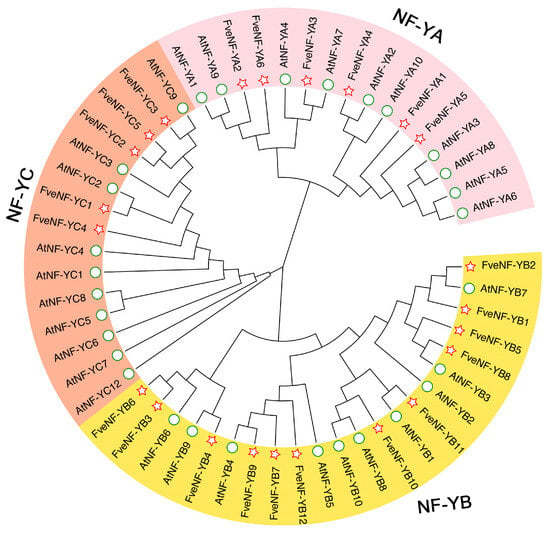
Figure 1.
Phylogenetic tree of NF-Y proteins from F. vesca and Arabidopsis thaliana. The tree was constructed based on full-length protein sequences using MEGA11 software. The three NF-Y subunits are marked with different colors. The green circles represent AtNF-Ys, and the red stars represent FveNF-Ys. The numbers for the branches of the tree are bootstrap confidence out of 1000 replications.
3.2. Gene Structure and Conserved Motif Analysis
Gene structure analysis provides valuable insights into the evolution aspects of a gene family. In this context, the gene structures of FveNF-Ys were compared. Out of the 23 FveNF-Ys examined, the majority, accounting for 73.9%, exhibited multiple exons, while only six members were found to be intronless (Figure 2A). The NF-YA family members displayed a complex exon/intron organization, with four to six introns per gene within their coding regions. In contrast, the exon/intron structure of NF-YB and NF-YC exhibited lower complexity or were less complex (Figure 2A). Most NF-YB genes had only one or two exons exclusively within the coding regions.
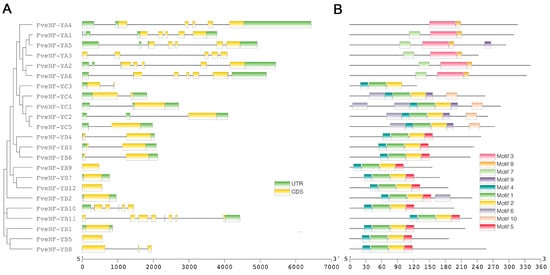
Figure 2.
Conserved motifs and gene structure of FveNF-Y family members. (A) Gene structures of FveNF-Y according to their evolutionary relationship. Green boxes represent UTRs, yellow boxes represent CDS, and black lines represent introns. The size of exons and intros can be estimated using the scale at the bottom. (B) Conserved motifs of FveNF-Y proteins. The conserved motifs of the FveNF-Y proteins are indicated by colored boxes.
Through motif analysis, ten distinct conserved motifs (designated as motifs 1–10) were identified, each demonstrating subfamily-specific patterns. For instance, motif 3 was exclusively present in FveNF-YAs, while motif 10 and motif 5 were specific to FveNF-YCs and FveNF-YBs, respectively. Furthermore, three motifs (motifs 4, 1, and 2) were absent in FveNF-YAs. Overall, different subfamilies exhibited different structural compositions, implying potential functional diversity among them.
3.3. Prediction of the miRNA Target Site of FveNF-Y Genes
The miRNA target site prediction revealed the presence of a total of 30 miRNA target sites in 12 FveNF-Y genes (Table 2), accounting for 52.2% of the total. Among all the FveNF-Y genes, notably, FveNF-YB4/5/6 exhibited the highest number of miRNA target sites, with 5, 5, and 4 miRNAs targeting them, respectively. Conversely, FveNF-YA1, FveNF-YA3, FveNF-YB9, and FveNF-YC1 contained only one miRNA target site each. Of particular interest, within the same subfamily, certain members were targeted by the same miRNA. For instance, both FveNF-YB3 and FveNF-YB6, which belong to the NF-YB subfamily, were simultaneously targeted by miR395a and miR395b. Moreover, FveNF-YB11 was found to be targeted by the following three novel miRNAs simultaneously: miRN21, miRN22, and miRN23. Additionally, the NF-YC subfamily genes were not targeted by known miRNAs but instead by novel miRNAs. For instance, FveNF-YC1 was targeted by miRN34, while FveNF-YC4 was targeted by miRN44 and miRN37.

Table 2.
The potential miRNA target sites of NF-Y genes.
3.4. Conserved Regions of FveNF-Ys
To explore the conserved regions within the three subfamilies of strawberry NF-Y proteins, multiple protein sequence alignments of FveNF-YAs, FveNF-YBs, and FveNF-YCs were conducted. All three subfamilies exhibited conserved regions. The FveNF-YA proteins displayed a conserved core region of approximately 50 amino acids, consisting of two putative alpha helix domains flanked by a relatively conserved linked sequence (Figure 3A). Previous studies in yeast and mammals have demonstrated that the first domain, composed of 20 amino acids, is essential for subunit interaction with NF-YB/NF-YC heterodimers (A1, subunit interaction domain). Similarly, the second domain, comprising 21 amino acids, plays a crucial role in sequence specificity when binding to DNA at CCAAT boxes (A2, DNA binding domain) [35,36,37,38]. Furthermore, FveNF-YC proteins contain a highly conserved domain closely related to the HFM domain of core histone H2A (Figure 3C).
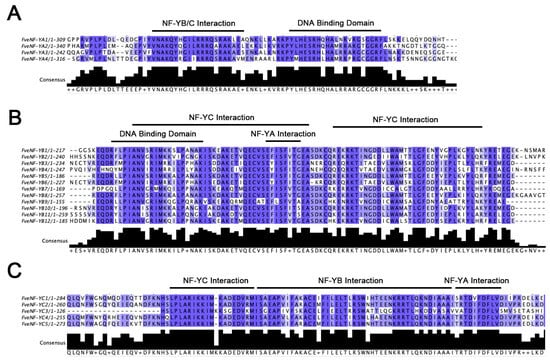
Figure 3.
Multiple alignments of strawberry NF-Y conserved domains. The details of conserved domains in the three subfamilies of FveNF-Ys subunit interaction and DNA binding regions of conserved domains are marked. The completely conserved amino acids are colored by blue boxes. Below each figure is the consensus sequence, which represents the position of the last amino acid in its whole protein. (A) FveNF-YA subfamily; (B) FveNF-YB subfamily; and (C) FveNF-YC subfamily.
3.5. Chromosomal Distribution and Synteny Analysis of FveNF-Y Genes
The FveNF-Y genes were unevenly distributed across the seven chromosomes of the strawberry (Figure 4). Fvb4, Fvb5, and Fvb7 contained two FveNF-Y genes each, followed by three in Fvb1, six in Fvb6, seven in Fvb3, and a single FveNF-Y locus in Contig_6. However, Fvb2 did not have any FveNF-Y genes. The amplification of the FveNF-Y family was attributed to gene duplication events, as indicated by the identification of four pairs of segmental duplication genes (highlighted by red lines) between chromosomes as follows: Fvb1/Fvb6, Fvb41/Fvb6 (two pairs), and Fvb6/Fvb7 (Figure 4A).
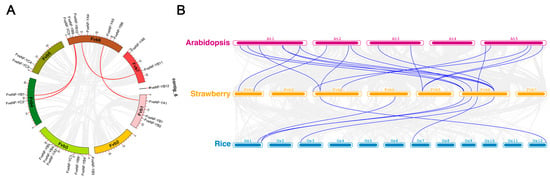
Figure 4.
Genomic positions, duplication events, and syntenic relationships of FveNF-Y genes. (A) Distribution of NF-Y family genes on each chromosome in the strawberry. FveNF-Y genes likely resulting from segmental duplication events are connected by orange lines. Gray lines represent syntenic blocks in the strawberry genome. (B) At1-At5, Fve1-Fve7, and Os1-Os12 represent five chromosomes of Arabidopsis, 7 chromosomes of the strawberry, and 12 chromosomes of rice, respectively. Gray lines in the background indicate the colinear blocks between three plant genomes (strawberry, Arabidopsis, and rice), while blue lines highlight the syntenic NF-Y gene pairs in three species (strawberry, Arabidopsis, and rice).
To further investigate the relationship among the FveNF-Y genes, their synteny with NF-Y genes from a monocotyledonous plant (rice) and one monocotyledonous plant (Arabidopsis) was examined (Figure 4B). Among the FveNF-Y genes, a total of 11 exhibited syntenic relationships with AtNF-Y genes, with 6 of them having syntenic loci in rice as well. Specifically, FveNF-YB6, FveNF-YB9, FveNF-YB10, and FveNF-YA5 had syntenic loci in both Arabidopsis and rice (Figure 4B).
3.6. Cis-Acting Elements Analysis in the Promoter Regions of FveNF-Ys
Cis-acting regulatory sequences located in gene promoters play a crucial role in regulating transcriptional activity in response to external and internal signals [39]. Within the 2000 bp upstream sequences of the start codon for all FveNF-Y genes, 92 putative cis-acting elements were identified (Table S4), which can be categorized into three categories as follows: plant growth and development, abiotic stress, and phytohormone-responsive (Figure 5A,B). Among these elements, a significant proportion, accounting for 91.4%, were related to light responsiveness, including motifs like GG1-motif, G-box, and TCT-motif, which are associated with plant growth and development (Figure 5C). In the category of abiotic stress, elements such as MYC (responds to abiotic stress signals), MYB (responds to ABA and dehydration), ARE (AU-rich element), STRE (stress response element), MBS (MYB binding site), and DRE core (dehydration-responsive element) were identified, which respond to stress signals like ABA and dehydration. It is noteworthy that MYC was present in all FveNF-Ys, indicating a potential role for FveNF-Ys in responding to abiotic stresses. In the phytohormone-responsive category, the ABA-responsive ABRE/ABRE3a element was the most frequently occurring cis-acting element, found 92 times in 21 FveNF-Ys and accounting for 37% of the hormone-responsive cis-acting elements (Figure 5C). Additionally, the TGACG-motif and CGTCA-motif were identified for MeJA-responsiveness, as well as the P-box for gibberellin-responsive elements, suggesting that FveNF-Ys may be regulated by various hormones (Figure 5C).
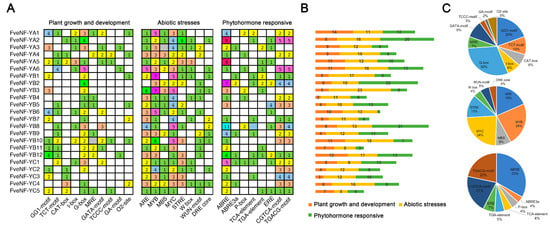
Figure 5.
Investigation of all cis-acting elements in FveNF-Ys. (A) Three categories of cis-acting elements in the FveNF-Ys. Different colors and numbers of the gird represent the numbers of different elements in these FveNF-Ys. (B) Histogram of the cis-acting elements in all genes. The different colored histograms represent the total number of the cis-acting elements in each category. (C) Proportion of each promoter element in each category.
3.7. Secondary and Three-Dimensional Structures of FveNF-Y Proteins
The secondary structures analysis revealed that FveNF-Y proteins exhibit a combination of α-helix, extended chain, and random coil structure. Among these, random coiled amino acids constituted the largest proportion (>38%), followed by α-helix (16.46~56.77%) and extended chain (5.81~28.48%) (Table S5). Three-dimensional structure predictions indicated that the different subfamilies of FveNF-Y share structural similarities, implying potential functional similarities (Figure 6). Specifically, the NF-YA family displayed the simplest structure, while the structures of NF-YB and NF-YC were comparatively more complex.
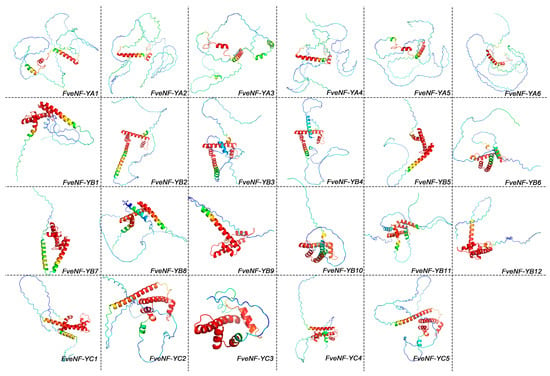
Figure 6.
Prediction of the three-dimensional domain of FveNF-Y proteins (the color from the blue, green, and yellow to red shows the sequence order from the N-terminus to C-terminus).
3.8. Expression Profiles of NF-Y Genes in Different Tissues
In this study, the expression profiles of FveNF-Y genes in different tissues and developmental stages of the woodland strawberry were investigated by analyzing the expression data from our previous study [33]. These tissues included various stages of floral organs, early-stage fruit, green-stage fruit, turning-stage fruit, shoot apical meristem, floral meristem, receptacle meristem, and roots of the woodland strawberry variety “Hawaii 4” (Figure 7, Table S4). Upon examining the NF-YA subfamily, the relative expression levels of FveNF-YB2 were significantly higher in anther12 and pollen compared with the other tissues. Additionally, FveNF-YB3 exhibited high expression in various tissues of achenes, including the embryo, ghost, and wall, indicating its potential involvement in seed development. In addition, FveNF-YC5 exhibited a specific expression pattern in the style. Furthermore, we observed that some FveNF-Y genes displayed ubiquitous expression across multiple tissues. For instance, the FveNF-YB10, FveNF-YA2, and FveNF-YA3 genes generally showed higher expression levels in all tissues, suggesting their broad role in various physiological processes.
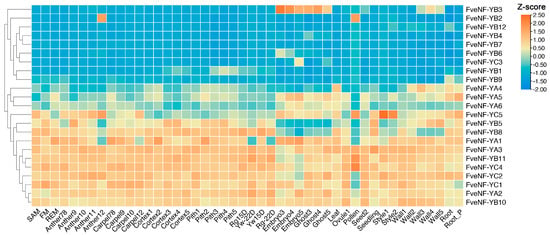
Figure 7.
Expression profile analysis of FveNF-Y genes across a total of 46 different tissues in the strawberry based on the transcriptome data. The color in the heat map represents the Transcripts Per Million (TPM) value based on variance-stabilized trans-formed values on a log2 scale.
3.9. Subcellular Localization of FveNF-YA3, FveNF-YA4, and FveNF-YC5
Transcription factors, which play a critical role in regulatory functions, are predominantly located in the nucleus. To further explore this phenomenon in the strawberry, the FveNF-YA3, FveNF-YA4, and FveNF-YC5 proteins were transiently expressed as GFP fusion constructs. The control group, transformed with a non-target GFP, showed diffuse GFP signals, indicating non-specific localization. However, when FveNF-YA3, FveNF-YA4, and FveNF-YC5 were expressed, cells displayed a strong nuclear GFP signal, which is a clear contrast to the control group (Figure 8). This pronounced nuclear localization suggests that these proteins, like other transcription factors, mainly localize in the nucleus, further reinforcing their potential roles in nuclear processes.
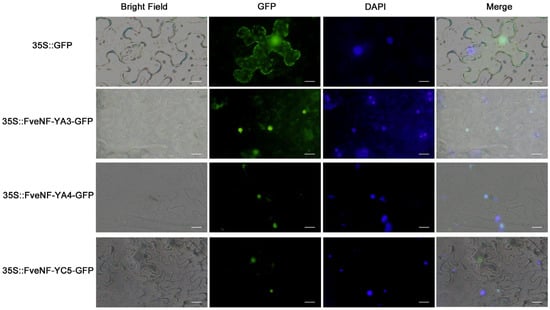
Figure 8.
Subcellular localization of FveNF-Y-GFP fusion proteins. Tobacco leaves were transformed with the fusion construct (FveNF-YA3-GFP, FveNF-YA4-GFP, and FveNF-YC5-GFP) and a control (GFP). GFP fluorescence, bright-field, nucleoid localization (DAPI), and merged images are shown. Bars = 20 μm.
4. Discussion
4.1. Identification of FveNF-Ys
NF-Y is a crucial heterodimeric transcription factor composed of NF-YA, NF-YB, and NF-YC subunits. Over the past decade, increasing evidence has highlighted the significance of NF-Y family genes in various plant growth and development processes, as well as their involvement in stress responses. While the functions of some NF-Y family members in Arabidopsis have been reported [40,41,42], the identification and characterization of NF-Y family genes in strawberries have remained elusive. In this study, 37 NF-YC gene family members were identified in F. vesca, compared to 30 members in Arabidopsis, 34 members in rice, 33 members in canola [43], and 68 members in soybeans [43,44]. This variation in the number of NF-Y members could potentially be linked to the size of the genome in each species. Our analysis of the 37 FveNF-Y members revealed a wide range of diversity in terms of molecular weight, gene structures, conserved motifs, theoretical isoelectric points, and expression patterns. These characteristics are similar to findings in other plant species [43,44,45], indicating the relative accuracy of our FveNF-Y identification and the conservation of NF-Y characteristics across different plant species.
When comparing the gene structures of FveNF-Ys, the majority of them (73.9%) possessed multiple exons, indicating the presence of introns. However, six members were intronless. NF-YA family members exhibited a complex exon/intron organization, with four to six introns per gene in their coding regions, while NF-YB and NF-YC subfamilies displayed a less complex exon/intron structure, with NF-YB genes containing only one or two exons within the coding regions. This finding is consistent with observations made in NF-Y genes of other plant species, such as the potato and soybean [44,46]. The presence of introns and the complexity of exon/intron organization can contribute to gene regulation and alternative splicing, which play significant roles in modulating gene expression and protein diversity.
It is worth noting that while the DNA binding domain shares some similarity with the CCT domain of flowering time regulator CONSTANS, NF-YA proteins do not exhibit obvious homology to other transcription factor families [47]. FveNF-YB proteins are characterized by a highly conserved central domain, which bears similarity to the histone fold motif (HFM) found in the core histone H2B in terms of amino acid sequence and structure [48]. This central domain, consisting of four alpha helices, functions in DNA binding to CCAAT boxes and interacting with NF-YA and NF-YC subunits [49]. This conserved domain plays a critical role in both DNA binding and subunit interaction [50]. NF-YAs contained the CBFB_NFYA domain, motif 3, motif 8, and motif 7. NF-YBs contained the CBFD_ NFYB_HMF domain, motif 1, motif 2, motif 4, motif 6, motif 9, and motif 10. NF-YCs contained the CBFD_NFYB_HMF and HAP5 domains, motif 1, motif 2, motif 4, and motif 5 (Figure 2). Despite the absence of certain motifs in individual members, each subfamily demonstrated a unique composition of domains and motifs. This suggests preserved function across the subfamily and supports the reliability of the classification system used. Despite the absence of certain motifs in individual members, each subfamily demonstrated a unique composition of domains and motifs, and the composition of motifs is consistent with other species, such as the potato and soybean [44,46]. This implies a maintained functional consistency within each subfamily and affirms the dependability of the classification methodology. The secondary structure analysis revealed the presence of α-helices, extended chains, and random coils in FveNF-Y proteins (Figure 6). The high proportion of random coils suggests the existence of flexible regions that may facilitate protein–protein interactions or undergo conformational changes upon binding to specific targets [51]. The α-helices and extended chains likely contribute to the stability and structural integrity of FveNF-Y proteins [52]. The tertiary structures exhibited greater consistency within FveNF-YAs compared with FveNF-YBs and FveNF-YCs, indicating a more preserved function in FveNF-YAs. These tertiary structure models of FveNF-Y proteins are consistent with NF-Y structure in tobacco [53], which suggests the conservation of the structure of NF-Y among species. Predicted protein interactions revealed intricate interplays among the three FveNF-Y subfamilies.
MiRNA is considered a post-transcriptional regulator and plays a critical role in regulating biological processes associated with various molecular mechanisms. NF-Y transcription factors are no exception, as they interact with different families of miRNAs to influence plant biological processes. Several miRNAs targeting NF-YA members have been identified. Putative target sites of miR169b, miR169c, and miR169d have been recognized in the 3′-UTRs of almost all NF-YA members of Phaseolus vulgaris, which are presumably involved in DNA binding, repair, and subunit interactions [54]. In our study, 12 (52.2%) FveNF-Y genes were found to have 30 miRNA target sites, suggesting that post-transcriptional regulation of FveNF-Y genes by miRNA might play a role in the strawberry.
4.2. Roles of FveNF-Ys in Flower Development
NF-Y proteins were reported to regulate different developmental stages, such as flowering, gametogenesis, and seed development [12,55]. For example, SlNF-YA3b binds to the SINGLE FLOWER TRUSS (SFT) promoter to regulate flowering time in tomatoes [55]. In our study, expression profiles were unitized to examine the functions of FveNF-Ys in strawberry flower and fruit development (Figure 7). The transcriptome data derived from various tissues revealed that FveNF-Ys display unique organ-specific expression patterns, underscoring their crucial roles in flower and fruit development. Further scrutiny revealed that two FveNF-Ys (FveNF-YB2 and FveNF-YC5) demonstrated high expression levels in anther12 and pollen, indicating their significant contribution to the pollen mature process. Interestingly, the promoter of FveNF-YB2 contains a noticeably higher number of G-boxes compared with other genes (Figure 5A). This abundance of G-boxes in the promoter region could potentially be a key factor contributing to the specific expression of the FveNF-YB2 gene in pollen. This finding aligns with previous research on the ZmSTK2 gene in maize, which is also specifically expressed in pollen and contains G-boxes in its promoter [56]. This parallel suggests a possible common regulatory mechanism governing pollen-specific gene expression in different plant species, potentially mediated by the G-box element. Our findings underscore the importance of further investigating the role and mechanism of G-box elements in regulating gene expression, particularly in relation to plant reproductive processes. Additionally, the FveNF-YC4 and FveNF-YB11 genes, which possess the G-box motif in their promoters, were found to exhibit high levels of expression in pollen.
4.3. Roles of FveNF-Ys in Fruit Development
Members of the NF-Y family exhibit unique expression patterns that include temporal, spatial, and organ-specific expression patterns in Arabidopsis [57], rice [58], poplar [59], canola [43], wheat [60], soybeans [44], and tomatoes [45]. Analyzing the expression levels of FveNF-Y transcripts in different tissues and stages of fruits revealed that most of them exhibit similar expression patterns, especially in the receptacle (including cortex and pitch), indicating wide expression throughout the strawberry fruit (Figure 7). An interesting finding is the tissue-specific expression of FveNF-YB3, which shows high expression in various achenes tissues, including the embryo, ghost, and wall. Achenes play a vital role in seed development, and the distinct expression of FveNF-YB3 in different achenes tissues suggests its contribution to specific processes during seed maturation. FveNF-YB3 is homologous to AtNF-YB6 in Arabidopsis, which encodes LEC1-Like (L1L) and functions as a regulator of embryo development [61]. In addition, FveNF-YB3 was targeted by the miR395 family (Table 2), which was abundant in ovules of Gui 99 and fsv1 during ovule development in rice [62]. These results suggest that miR396 may play an important in strawberry ovule development.
Additionally, FveNF-YC5 demonstrates a specific expression pattern in the style, aligning with the known role of NF-YC genes in regulating female reproductive organ development in maize [63]. The style plays a crucial role in pollen tube guidance and fertilization, hence indicating the potential involvement of FveNF-YC5 in these processes. Overall, the tissue-specific expression patterns of FveNF-Y genes in the woodland strawberry provide important insights into their functional roles in specific developmental processes. These observations underscore the complexity and specificity of gene regulation throughout various developmental stages in the woodland strawberry. The expression profiles suggest that different NF-Y subfamilies may have distinct roles in specific tissues and developmental processes. Further research is necessary to elucidate fully the precise functions of these genes and their regulatory networks in the woodland strawberry.
4.4. Roles of FveNF-Ys in Response to Different Abiotic Stresses and Hormonal Signaling
Environmental factors, including abiotic and biotic stresses, can adversely affect plant growth and development [64]. Prior research has indicated that the majority of BBX genes are susceptible to a variety of stress conditions [17,41,65]. In the present study, cis-acting element analysis was used to delve into the roles of FveNF-Ys in plant growth and development, abiotic stresses, and hormone treatments (Figure 5). Until now, the functions of NF-Ys responding to plant biotic stresses had rarely been studied. In Arabidopsis, the expression of AtNF-YB3 has been reported to be elevated under NaCl, mannitol, and cold treatments, suggesting its functional role in stress responses and the interplay among these three abiotic stresses [66]. Furthermore, the overexpression of NF-YB3 from Picea wilsonii imparts tolerance to salinity and drought stress in transformed Arabidopsis [67]. In our study, cis-acting elements responsive to abiotic stresses were found in the promoter regions of FveNF-YB1 and FveNF-YB3, suggesting its potential role in resistance to abiotic stresses.
Previous studies have verified that the NF-Y gene family plays critical roles in response to stresses and can be regulated by exogenous hormones [65]. In our study, various cis-acting elements were identified in the promoter regions of FveNF-Ys, including ABRE (ABA-responsive element), ERE, CGTCA-motif, TGACG-motif, and P-box. Further analysis revealed that all FveNF-Ys contain at least one hormone-responsive and abiotic stress-related cis-element, suggesting their potential involvement in responses to abiotic stresses and hormones. In rice, global genome expression analysis in response to drought and high-salinity stresses showed that the promoter regions of genes responsive to both stresses contain more ABRE elements compared with those responsive to a single stress [68]. Overall, the characterization of FveNF-Ys in response to various stresses in our study enhances the understanding of NF-Y functions. However, further research is needed to determine the functions of FveNF-Ys precisely.
5. Conclusions
In this study, 23 NF-Y members were identified in F. vesca and appropriately divided into corresponding NF-Y subfamilies. The conservation and evaluation of gene and protein sequences, including gene structure, synteny, phylogeny, and cis-elements, provide valuable insights into the evolutionary relationships and functional diversity of the FveNF-Y gene family in strawberries. miRNA target site prediction revealed potential regulatory mechanisms, suggesting post-transcriptional regulation of NF-Y genes by miRNAs. Furthermore, the expression profiles across various tissues and developmental stages highlight the potential roles of NF-Y genes in specific developmental processes in the woodland strawberry. These findings contribute to our understanding of gene family evolution and the molecular mechanisms underlying gene regulation in plants. Further research focusing on the functional characterization of these genes will contribute to a better understanding of the regulatory mechanisms underlying tissue-specific gene expression in plants.
6. Future Perspectives
To strengthen our conclusions about the roles of NF-Y genes in stress response and development, future studies should include functional validation using advanced techniques such as CRISPR/Cas9 and RNA interference (RNAi). These approaches will provide more definitive insights into the genetic and molecular mechanisms underlying NF-Y gene functions.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae10070755/s1, Table S1. Expression profiles of all NF-Y genes across different tissues. Table S2. Primers of subcellular localization. Table S3. Coding sequences and protein sequences of the identified strawberry NF-Y gene members. Table S4. Promoter analysis of the F. vesca NF-Y gene family. Table S5. Two-dimensional structures of FveNF-Y proteins.
Author Contributions
Conceptualization, T.L. and M.W.; data curation, Y.Z. and F.G.; funding acquisition, M.W.; investigation, Y.Z.; methodology, T.L.; software, W.Z.; supervision, T.L.; writing—original draft, T.L.; writing—review and editing, M.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Hainan Provincial Natural Science Foundation of China (RZ2400002197), the Scientific Research Start-up Fund Project of Hainan University (RZ2300002729 and RZ2200001399), and the Collaborative Innovation Center of Nanfan and High-Efficiency Tropical Agriculture, Hainan University (XTCX2022NYC21).
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors on request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Mantovani, R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998, 26, 1135–1143. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Zhou, J.; Liu, C.; Liu, L.; Shen, L.; Yu, H. Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 2014, 5, 4601. [Google Scholar] [CrossRef] [PubMed]
- Kumimoto, R.W.; Adam, L.; Hymus, G.J.; Repetti, P.P.; Reuber, T.L.; Marion, C.M.; Hempel, F.D.; Ratcliffe, O.J. The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 2008, 228, 709–723. [Google Scholar] [CrossRef] [PubMed]
- Kumimoto, R.W.; Zhang, Y.; Siefers, N.; Holt, B.F., 3rd. NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2010, 63, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.Y.; Zhang, L.; Li, W.W.; Hu, X.L.; Wang, M.B.; Fan, Y.L.; Zhang, C.Y.; Wang, L. Stress-induced early flowering is mediated by miR169 in Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Sorin, C.; Declerck, M.; Christ, A.; Blein, T.; Ma, L.; Lelandais-Briere, C.; Njo, M.F.; Beeckman, T.; Crespi, M.; Hartmann, C. A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol. 2014, 202, 1197–1211. [Google Scholar] [CrossRef]
- Huang, M.; Hu, Y.; Liu, X.; Li, Y.; Hou, X. Arabidopsis LEAFY COTYLEDON1 controls cell fate determination during post-embryonic development. Front. Plant Sci. 2015, 6, 955. [Google Scholar] [CrossRef]
- Kwong, R.W.; Bui, A.Q.; Lee, H.; Kwong, L.W.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 2003, 15, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Tan, H.; Zheng, Q.; Fu, F.; Liang, Y.; Zhang, J.; Yang, X.; Wang, T.; Chong, K.; Wang, X.J.; et al. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 1042–1054. [Google Scholar] [CrossRef]
- West, M.; Yee, K.M.; Danao, J.; Zimmerman, J.L.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON1 Is an Essential Regulator of Late Embryogenesis and Cotyledon Identity in Arabidopsis. Plant Cell 1994, 6, 1731–1745. [Google Scholar] [CrossRef]
- Fornari, M.; Calvenzani, V.; Masiero, S.; Tonelli, C.; Petroni, K. The Arabidopsis NF-YA3 and NF-YA8 genes are functionally redundant and are required in early embryogenesis. PLoS ONE 2013, 8, e82043. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Tan, H.; Hong, S.; Liang, Y.; Zuo, J. Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol. Plant 2013, 6, 188–201. [Google Scholar] [CrossRef]
- Battaglia, M.; Ripodas, C.; Clua, J.; Baudin, M.; Aguilar, O.M.; Niebel, A.; Zanetti, M.E.; Blanco, F.A. A nuclear factor Y interacting protein of the GRAS family is required for nodule organogenesis, infection thread progression, and lateral root growth. Plant Physiol. 2014, 164, 1430–1442. [Google Scholar] [CrossRef] [PubMed]
- Li, W.X.; Oono, Y.; Zhu, J.; He, X.J.; Wu, J.M.; Iida, K.; Lu, X.Y.; Cui, X.; Jin, H.; Zhu, J.K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 2008, 20, 2238–2251. [Google Scholar] [CrossRef]
- Sato, H.; Mizoi, J.; Tanaka, H.; Maruyama, K.; Qin, F.; Osakabe, Y.; Morimoto, K.; Ohori, T.; Kusakabe, K.; Nagata, M.; et al. Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. Plant Cell 2014, 26, 4954–4973. [Google Scholar] [CrossRef]
- Sato, H.; Todaka, D.; Kudo, M.; Mizoi, J.; Kidokoro, S.; Zhao, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. The Arabidopsis transcriptional regulator DPB3-1 enhances heat stress tolerance without growth retardation in rice. Plant Biotechnol. J. 2016, 14, 1756–1767. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.X.; Howell, S.H. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 2010, 22, 782–796. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.M.; Tanaka, T.; Nakamura, H.; Ichikawa, H.; Kobayashi, K.; Yaeno, T.; Yamaoka, N.; Shimomoto, K.; Takayama, K.; Nishina, H.; et al. Overexpression of a rice heme activator protein gene (OsHAP2E) confers resistance to pathogens, salinity and drought, and increases photosynthesis and tiller number. Plant Biotechnol. J. 2015, 13, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Lv, J.; Sadeghnezhad, E.; Cheng, J.; Jia, H. Transcriptomic and metabolomic profiling of strawberry during postharvest cooling and heat storage. Front. Plant Sci. 2022, 13, 1009747. [Google Scholar] [CrossRef]
- Jung, S.; Lee, T.; Cheng, C.-H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef]
- Petroni, K.; Kumimoto, R.W.; Gnesutta, N.; Calvenzani, V.; Fornari, M.; Tonelli, C.; Holt III, B.F.; Mantovani, R. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 2012, 24, 4777–4792. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Waterhouse, A.; Procter, J.; Martin, D.A.; Barton, G.J. Jalview: Visualization and analysis of molecular sequences, alignments, and structures. BMC Bioinform. 2005, 6, P28. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pi, M.; Gao, Q.; Liu, Z.; Kang, C. Updated annotation of the wild strawberry Fragaria vesca V4 genome. Hortic. Res. 2019, 6, 61. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, R.; Li, X.Y.; Pessara, U.; Hooft van Huisjduijnen, R.; Benoist, C.; Mathis, D. Dominant negative analogs of NF-YA. J. Biol. Chem. 1994, 269, 20340–20346. [Google Scholar] [CrossRef]
- Xing, Y.; Fikes, J.D.; Guarente, L. Mutations in yeast HAP2/HAP3 define a hybrid CCAAT box binding domain. EMBO J. 1993, 12, 4647–4655. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Zhang, S.; Olesen, J.T.; Rich, A.; Guarente, L. Subunit interaction in the CCAAT-binding heteromeric complex is mediated by a very short alpha-helix in HAP2. Proc. Natl. Acad. Sci. USA 1994, 91, 3009–3013. [Google Scholar] [CrossRef] [PubMed]
- Hackenberg, D.; Wu, Y.; Voigt, A.; Adams, R.; Schramm, P.; Grimm, B. Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Mol. Plant 2012, 5, 876–888. [Google Scholar] [CrossRef]
- Singh, K.B.; Foley, R.C.; Oñate-Sánchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wu, D.; Kong, F.; Lin, K.; Zhang, H.; Li, G. The Arabidopsis thaliana nuclear factor Y transcription factors. Front. Plant Sci. 2017, 7, 2045. [Google Scholar] [CrossRef]
- Swain, S.; Myers, Z.A.; Siriwardana, C.L.; Holt, B.F., III. The multifaceted roles of NUCLEAR FACTOR-Y in Arabidopsis thaliana development and stress responses. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2017, 1860, 636–644. [Google Scholar] [CrossRef]
- Lv, X.; Zeng, X.; Hu, H.; Chen, L.; Zhang, F.; Liu, R.; Liu, Y.; Zhou, X.; Wang, C.; Wu, Z. Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO–NF–Y master transcription factor complex. Plant Cell 2021, 33, 1182–1195. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Yin, X.; Lin, Z.; Zheng, Q.; Liu, G.; Zhao, G. Identification and characterization of NF-Y transcription factor families in Canola (Brassica napus L.). Planta 2014, 239, 107–126. [Google Scholar] [CrossRef]
- Quach, T.N.; Nguyen, H.T.; Valliyodan, B.; Joshi, T.; Xu, D.; Nguyen, H.T. Genome-wide expression analysis of soybean NF-Y genes reveals potential function in development and drought response. Mol. Genet. Genom. 2015, 290, 1095–1115. [Google Scholar] [CrossRef]
- Li, S.; Li, K.; Ju, Z.; Cao, D.; Fu, D.; Zhu, H.; Zhu, B.; Luo, Y. Genome-wide analysis of tomato NF-Y factors and their role in fruit ripening. BMC Genom. 2016, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, Y.; Zhu, J.; Ma, W.; Li, Z.; Bi, Z.; Sun, C.; Bai, J.; Zhang, J.; Liu, Y. Genome-wide identification and analysis of the NF-Y gene family in potato (Solanum tuberosum L.). Front. Genet. 2021, 12, 739989. [Google Scholar] [CrossRef] [PubMed]
- Wenkel, S.; Turck, F.; Singer, K.; Gissot, L.; Le Gourrierec, J.; Samach, A.; Coupland, G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 2006, 18, 2971–2984. [Google Scholar] [CrossRef] [PubMed]
- Dolfini, D.; Gatta, R.; Mantovani, R. NF-Y and the transcriptional activation of CCAAT promoters. Crit. Rev. Biochem. Mol. Biol. 2012, 47, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Kim, I.S.; Sohn, K.Y.; de Crombrugghe, B.; Maity, S.N. Three classes of mutations in the A subunit of the CCAAT-binding factor CBF delineate functional domains involved in the three-step assembly of the CBF-DNA complex. Mol. Cell. Biol. 1996, 16, 328–337. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Romier, C.; Cocchiarella, F.; Mantovani, R.; Moras, D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 2003, 278, 1336–1345. [Google Scholar] [CrossRef]
- Miller, W.G.; Goebel, C.V. Dimensions of protein random coils. Biochemistry 1968, 7, 3925–3935. [Google Scholar] [CrossRef]
- Pagel, K.; Wagner, S.C.; Samedov, K.; von Berlepsch, H.; Böttcher, C.; Koksch, B. Random coils, β-sheet ribbons, and α-helical fibers: One peptide adopting three different secondary structures at will. J. Am. Chem. Soc. 2006, 128, 2196–2197. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Song, K.; Li, B.; Song, Y.; Zhang, X.; Li, H.; Yang, L. Genome-wide identification and expression analysis of NF-Y gene family in tobacco (Nicotiana tabacum L.). Sci. Rep. 2024, 14, 5257. [Google Scholar] [CrossRef] [PubMed]
- Rípodas, C.; Castaingts, M.; Clúa, J.; Blanco, F.; Zanetti, M.E. Annotation, phylogeny and expression analysis of the nuclear factor Y gene families in common bean (Phaseolus vulgaris). Front. Plant Sci. 2015, 5, 761. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Ji, K.; Wang, J.; Liu, X.; Zhou, Z.; Huang, R.; Ai, G.; Li, Y.; Wang, X.; Wang, T. Nuclear factor Y-A3b binds to the SINGLE FLOWER TRUSS promoter and regulates flowering time in tomato. Hortic. Res. 2024, 11, uhae088. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Fan, M.; Wang, G.; Zhang, C.; Shi, L.; Wei, Z.; Ma, W.; Chang, J.; Huang, S.; Lin, F. Isolation and characterization of a novel pollen-specific promoter in maize (Zea mays L.). Genome 2017, 60, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Siefers, N.; Dang, K.K.; Kumimoto, R.W.; Bynum, W.E.t.; Tayrose, G.; Holt, B.F., 3rd. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009, 149, 625–641. [Google Scholar] [CrossRef]
- Thirumurugan, T.; Ito, Y.; Kubo, T.; Serizawa, A.; Kurata, N. Identification, characterization and interaction of HAP family genes in rice. Mol. Genet. Genom. 2008, 279, 279–289. [Google Scholar] [CrossRef]
- Yan, D.-H.; Xia, X.; Yin, W. NF-YB Family Genes Identified in a Poplar Genome-wide Analysis and Expressed in Populus euphratica Are Responsive to Drought Stress. Plant Mol. Biol. Report. 2012, 31, 363–370. [Google Scholar] [CrossRef]
- Stephenson, T.J.; McIntyre, C.L.; Collet, C.; Xue, G.P. Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol. Biol. 2007, 65, 77–92. [Google Scholar] [CrossRef]
- Calvenzani, V.; Testoni, B.; Gusmaroli, G.; Lorenzo, M.; Gnesutta, N.; Petroni, K.; Mantovani, R.; Tonelli, C. Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits. PLoS ONE 2012, 7, e42902. [Google Scholar] [CrossRef]
- Yang, L.; Wu, Y.; Wang, W.; Mao, B.; Zhao, B.; Wang, J. Genetic subtraction profiling identifies candidate miRNAs involved in rice female gametophyte abortion. G3 Genes Genomes Genet. 2017, 7, 2281–2293. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Liu, C.; Yu, T.; Liu, X.; Xu, D.; Wang, J.; Wang, G.; Cai, Y. Identification and characterization of paternal-preferentially expressed gene NF-YC8 in maize endosperm. Mol. Genet. Genom. 2015, 290, 1819–1831. [Google Scholar] [CrossRef]
- Sardhara, K.; Mehta, K. Effects of abiotic and biotic stress on the plant. Acad J. Bot. Sci. 2018, 1, 5–9. [Google Scholar]
- Kavi Kishor, P.B.; Ganie, S.A.; Wani, S.H.; Guddimalli, R.; Karumanchi, A.R.; Edupuganti, S.; Naravula, J.; Kumar, V.; Polavarapu, R.; Suravajhala, P. Nuclear factor-y (NF-y): Developmental and stress-responsive roles in the plant lineage. J. Plant Growth Regul. 2023, 42, 2711–2735. [Google Scholar] [CrossRef]
- Sato, H.; Suzuki, T.; Takahashi, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NF-YB2 and NF-YB3 have functionally diverged and differentially induce drought and heat stress-specific genes. Plant Physiol. 2019, 180, 1677–1690. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, D.; Liu, Y.; Luo, C.; Zhou, Y.; Zhang, L. Overexpression of a NF-YB3 transcription factor from Picea wilsonii confers tolerance to salinity and drought stress in transformed Arabidopsis thaliana. Plant Physiol. Biochem. 2015, 94, 153–164. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, X.; Jiao, Y.; Qin, Y.; Liu, X.; He, K.; Chen, C.; Ma, L.; Wang, J.; Xiong, L. Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol. Biol. 2007, 63, 591–608. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).