Genome-Wide Identification of the WUSCHEL-Related Homeobox (WOX) Gene Family in Three Cymbidium Species and Expression Patterns in C. goeringii
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of the WOX Genes
2.2. Phylogenetic Analysis and Physicochemical Properties
2.3. Conserved Motif, Gene Structure Analysis, and Cis-Acting Element Prediction
2.4. Chromosome Location and Collinearity Analysis
2.5. RNA Extraction and RT-qPCR Analysis
3. Results
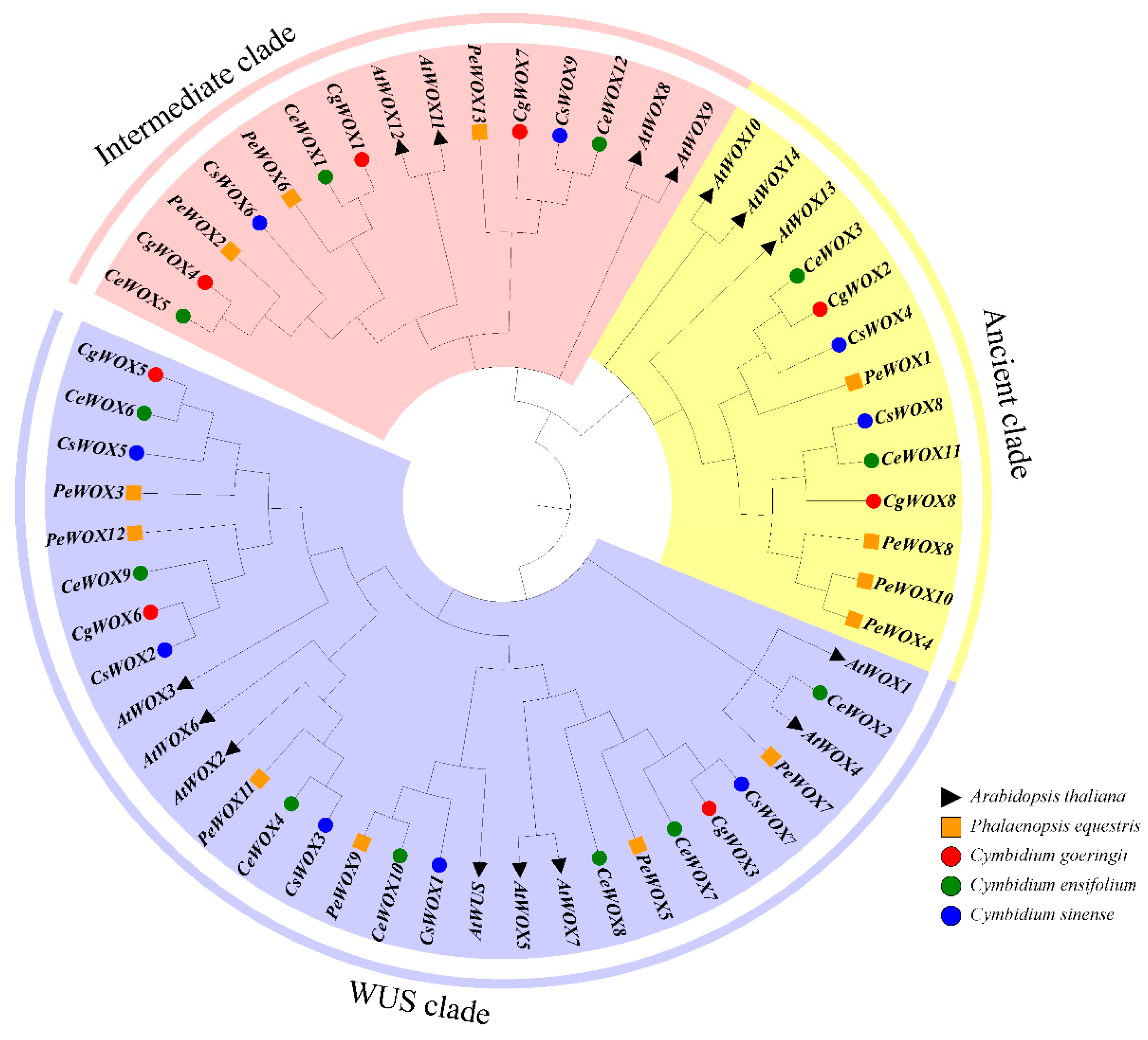
3.1. Phylogeny Analysis of the WOX Genes
3.2. Identification and Characterization of the WOX Genes in Three Cymbidium Species
3.3. Sequence Alignment, Gene Structure, and Conserved Motif Analysis of WOX Genes
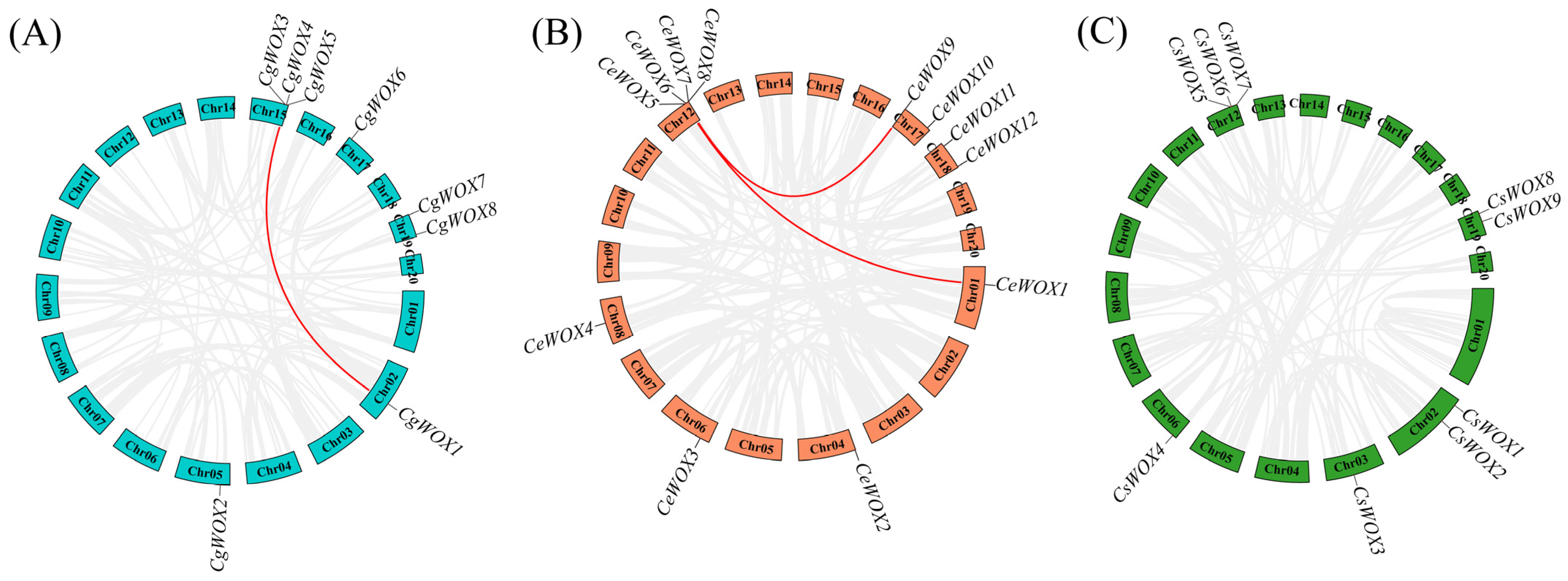
3.4. Chromosomal Localization and Collinearity Analysis of WOX Genes
3.5. Cis-Acting Element Analysis of Three Cymbidium Species
3.6. Expression Patterns and qRT-PCR Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kong, D.Y.; Hao, Y.L.; Cui, H.C. The WUSCHEL related homeobox protein WOX7 regulates the sugar response of lateral root development in Arabidopsis thaliana. Mol. Plant 2016, 9, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Laux, T.; Mayer, K.F.X.; Berger, J.; Jürgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Graaff, E.V.D.; Laux, T.; Rensing, S.A. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009, 10, 248. [Google Scholar] [CrossRef] [PubMed]
- Haecker, A.; Groß-Hardt, R.; Geiges, B.; Sarkar, A.; Breuninger, H.; Herrmann, M.; Laux, T. Expression dynamics of wox genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004, 131, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.K.; Luijten, M.; Miyashima, S.; Lenhard, M.; Hashimoto, T.; Nakajima, K.; Scheres, B.; Heidstra, R.; Laux, T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 2007, 446, 811–814. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Zheng, Q.; He, X.; Zhao, X.; Zhang, M.; Huang, Y.; Cai, B.; Liu, Z. The evolution of the WUSCHEL-related homeobox gene family in Dendrobium species and its role in sex organ development in D. Chrysotoxum. Int. J. Mol. Sci. 2024, 25, 5352. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zong, J.; Liu, J.H.; Yin, J.Y.; Zhang, D.B. Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. J. Integr. Plant Biol. 2010, 52, 1016–1026. [Google Scholar] [CrossRef]
- Minh-Thu, P.T.; Kim, J.S.; Chae, S.; Jun, K.M.; Lee, G.S.; Kim, D.E.; Cheong, J.J.; Song, S.I.; Nahm, B.H.; Kim, Y.K. A WUSCHEL homeobox transcription factor, OSWOX13, enhances drought tolerance and triggers early flowering in rice. Mol. Cells 2018, 41, 781–798. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hu, Y.; Dai, M.; Huang, L.; Zhou, D.X. The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 2009, 21, 736–748. [Google Scholar] [CrossRef]
- Dai, M.Q.; Hu, Y.F.; Zhao, Y.; Liu, H.F.; Zhou, D.X. A WUSCHEL-like homeobox gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 2007, 144, 380–390. [Google Scholar] [CrossRef]
- Hinsley, A.; De Boer, H.J.; Fay, M.F.; Gale, S.W.; Gardiner, L.M.; Gunasekara, R.S.; Kumar, P.; Masters, S.; Metusala, D.; Rob-erts, D.L.; et al. A review of the trade in orchids and its implications for conservation. Bot. J. Linn. Soc. 2018, 186, 435–455. [Google Scholar] [CrossRef]
- Li, H.F.; Li, C.; Wang, Y.H.; Qin, X.S.; Meng, L.H.; Sun, X.D. Genome-wide analysis of the WOX transcription factor genes in Dendrobium catenatum lindl. Genes 2022, 13, 1481. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, T.R.; Madhvi, K.; Kumar, U.S.; Sembi, J.K. Identification and characterization of WUSCHEL-related homeobox (WOX) gene family in economically important orchid species Phalaenopsis equestris and Dendrobium catenatum. Plant Gene 2018, 14, 37–45. [Google Scholar] [CrossRef]
- Ai, Y.; Li, Z.; Sun, W.H.; Chen, J.; Zhang, D.Y.; Ma, L.; Zhang, Q.H.; Chen, M.K.; Zheng, Q.D.; Liu, J.F.; et al. The Cymbidium genome reveals the evolution of unique morphological traits. Hortic. Res. 2021, 8, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Chen, G.Z.; Huang, J.; Liu, D.K.; Xue, F.; Chen, X.L.; Chen, S.Q.; Liu, C.G.; Liu, H.; Ma, H.; et al. The Cymbidium goeringii genome provides insight into organ development and adaptive evolution in orchids. Ornam. Plant Res. 2021, 1, 10–22. [Google Scholar] [CrossRef]
- Yang, F.X.; Gao, J.; Wei, Y.L.; Ren, R.; Zhang, G.Q.; Lu, C.Q.; Jin, J.P.; Ai, Y.; Wang, Y.Q.; Chen, L.J.; et al. The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 2021, 19, 2501–2516. [Google Scholar] [CrossRef]
- Edgar, R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. In the cipres science gateway: A community resource for phylogenetic analyses. In Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery, Salt Lake City, UT, USA, 18–21 July 2011; pp. 1–8. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Clamp, M.; Cuff, J.; Searle, S.M.; Barton, G.J. The Jalview Java alignment editor. Bioinformatics 2004, 20, 426–427. [Google Scholar] [CrossRef]
- Bailey, T.L.; Mikael, B.; Buske, F.A.; Martin, F.; Grant, C.E.; Luca, C.; Jingyuan, R.; Li, W.W.; Noble, W.S. Meme suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. Tbtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Rychlik, W. Oligo 7 primer analysis software. PCR Primer Des. 2007, 402, 35–59. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.Z.; Huang, J.; Zhou, X.Q.; Hao, Y.; Chen, J.L.; Zhou, Y.Z.; Ahmad, S.; Lan, S.R.; Liu, Z.J.; Peng, D.H. Comprehensive analysis for GRF transcription factors in sacred lotus (Nelumbo nucifera). Int. J. Mol. Sci. 2022, 23, 6673. [Google Scholar] [CrossRef] [PubMed]
- Khuraijam, J.S.; Sharma, S.C.; Roy, R.K. Orchids: Potential ornamental crop in north India. Int. J. Hortic. Crop Sci. Res. 2017, 7, 1–8. [Google Scholar]
- Balilashaki, K.; Vahedi, M.; Ho, T.T.; Niu, S.C.; Cardoso, J.C.; Zotz, G.; Khodamzadeh, A.A. Biochemical, cellular and molecular aspects of Cymbidium orchids: An ecological and economic overview. Acta Physiol. Plant. 2022, 44, 24–30. [Google Scholar] [CrossRef]
- Sevilleno, S.S.; Cabahug-Braza, R.A.; An, H.R.; Lim, K.B.; Hwang, Y.J. The role of cytogenetic tools in orchid breeding. Korean J. Agric. Sci. 2023, 50, 193–206. [Google Scholar]
- Shefferson, R.P.; Jacquemyn, H.; Kull, T.; Hutchings, M.J. The demography of terrestrial orchids: Life history, population dynamics and conservation. Bot. J. Linn. Soc. 2020, 192, 315–332. [Google Scholar] [CrossRef]
- Cheng, S.F.; Zhou, D.X.; Zhao, Y. WUSCHEL-related homeobox gene WOX11 increases rice drought resistance by controlling root hair formation and root system development. Plant Signal. Behav. 2016, 11, e1130198. [Google Scholar] [CrossRef]
- Deveaux, Y.; Toffano-Nioche, C.; Claisse, G.; Thareau, V.; Morin, H.; Laufs, P.; Moreau, H.; Kreis, M.; Lecharny, A. Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol. Biol. 2008, 8, 291. [Google Scholar] [CrossRef]
- Hu, X.M.; Xu, L. Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol. 2016, 172, 2363–2373. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, M.; Iwase, A.; Ito, T.; Tanaka, H.; Favero, D.S.; Kawamura, A.; Sakamoto, S.; Wakazaki, M.; Tameshige, T.; Fujii, H.; et al. Wound-inducible WUSCHEL-related homeobox 13 is required for callus growth and organ reconnection. Plant Physiol. 2022, 188, 425–441. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Shi, L.; Liang, X.; Zhao, P.; Wang, W.; Liu, J.; Chang, Y.; Hiei, Y.; Yanagihara, C.; Du, L.; et al. The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat. Plants 2022, 8, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.L.; Dong, L.Q.; Wei, C.; Wani, M.A.; Yang, C.M.; Li, S.C.; Li, F. Creating novel ornamentals via new strategies in the era of genome editing. Front. Plant Sci. 2023, 14, 1142866. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Wang, K.; Du, L.P.; Song, Y.X.; Li, H.H.; Ye, X.G. Genome-wide identification and expression profiling analysis of WOX family protein-encoded genes in Triticeae species. Int. J. Mol. Sci. 2021, 22, 9325–9349. [Google Scholar] [CrossRef] [PubMed]
- Carles, C.C.; Fletcher, J.C. Shoot apical meristem maintenance: The art of a dynamic balance. Trends Plant Sci. 2003, 8, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 2009, 21, 3493–3505. [Google Scholar] [CrossRef]
- Chen, G.Z.; Huang, J.; Lin, Z.C.; Wang, F.; Yang, S.M.; Jiang, X.; Ahmad, S.; Zhou, Y.Z.; Lan, S.; Liu, Z.J.; et al. Genome-wide analysis of WUSCHEL-related homeobox gene family in sacred lotus (Nelumbo nucifera). Int. J. Mol. Sci. 2023, 24, 14216. [Google Scholar] [CrossRef]





| Gene Name | Gene ID | Number of Amino Acids (AA) | Theoretical PI | Molecular Weight (Mw) | Instability Index (II) | Aliphatic Index (AI) | Grand Average of Hydropathicity (GRAVY) | Clade | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| CgWOX1 | GL10772 | 267 | 5.17 | 28,478.85 | 64.82 | 73.52 | −0.194 | Intermediate | Nucleus |
| CgWOX2 | GL12480 | 246 | 6.46 | 28,129.68 | 66.80 | 65.45 | −0.725 | Ancient | Nucleus |
| CgWOX3 | GL01022 | 185 | 8.5 | 21,245.19 | 56.37 | 64.81 | −0.685 | WUS | Nucleus |
| CgWOX4 | GL17509 | 270 | 6.16 | 29,728.17 | 68.62 | 77.59 | −0.366 | Intermediate | Nucleus |
| CgWOX5 | GL12104 | 265 | 5.69 | 30,696.42 | 72.51 | 59.66 | −0.908 | WUS | Nucleus |
| CgWOX6 | GL17950 | 193 | 9.04 | 21,862.46 | 69.93 | 50.57 | −0.798 | WUS | Nucleus |
| CgWOX7 | GL20683 | 334 | 7.76 | 36,147.56 | 52.44 | 78.02 | −0.295 | Intermediate | Nucleus |
| CgWOX8 | GL21289 | 199 | 5.78 | 22,446.3 | 74.02 | 69.20 | −0.716 | Ancient | Nucleus |
| CeWOX1 | JL010469 | 259 | 5.13 | 27,488.5 | 66.44 | 70.15 | −0.283 | Intermediate | Nucleus |
| CeWOX2 | JL008369 | 164 | 9.73 | 19,245.09 | 48.15 | 86.89 | −0.416 | WUS | Nucleus |
| CeWOX3 | JL002473 | 246 | 6.26 | 28,201.74 | 67.93 | 65.45 | −0.737 | Ancient | Nucleus |
| CeWOX4 | JL000437 | 249 | 8.29 | 28,049.87 | 71.75 | 70.88 | −0.576 | WUS | Nucleus |
| CeWOX5 | JL017101 | 249 | 5.95 | 27,247.28 | 68.05 | 70.84 | −0.418 | Intermediate | Nucleus |
| CeWOX6 | JL014141 | 212 | 6.05 | 24,084.04 | 79.61 | 57.08 | −0.753 | WUS | Nucleus |
| CeWOX7 | JL011816 | 143 | 10.13 | 16,583.28 | 60.02 | 72.94 | −0.691 | WUS | Nucleus |
| CeWOX8 | JL011815 | 149 | 7.06 | 17,178.42 | 45.65 | 62.15 | −0.692 | WUS | Nucleus |
| CeWOX9 | JL017647 | 108 | 10.28 | 12,931.8 | 75.10 | 56.85 | −0.780 | WUS | Nucleus |
| CeWOX10 | JL000993 | 198 | 4.89 | 22,612.75 | 68.78 | 54.80 | −0.956 | WUS | Nucleus |
| CeWOX11 | JL000483 | 255 | 5.26 | 28,775.24 | 69.52 | 65.45 | −0.709 | Ancient | Nucleus |
| CeWOX12 | JL015470 | 335 | 7.76 | 36,218.63 | 52.31 | 78.09 | −0.289 | Intermediate | Nucleus |
| CsWOX1 | cymsin_Mol009245 | 102 | 9.88 | 12,008.36 | 62.39 | 60.29 | −1.154 | WUS | Nucleus |
| CsWOX2 | cymsin_Mol021168 | 177 | 9.14 | 20,024.33 | 60.67 | 51.30 | −0.801 | WUS | Nucleus |
| CsWOX3 | cymsin_Mol007512 | 249 | 8.76 | 27,992.9 | 70.28 | 69.32 | −0.573 | WUS | Nucleus |
| CsWOX4 | cymsin_Mol017948 | 246 | 6.09 | 28,160.71 | 68.71 | 64.27 | −0.735 | Ancient | Nucleus |
| CsWOX5 | cymsin_Mol019643 | 212 | 6.23 | 24,098.11 | 79.39 | 57.08 | −0.767 | WUS | Nucleus |
| CsWOX6 | cymsin_Mol006735 | 128 | 11.63 | 14,521.36 | 77.48 | 72.42 | −0.766 | Intermediate | Nucleus |
| CsWOX7 | cymsin_Mol012795 | 194 | 9.6 | 21,959.27 | 40.74 | 76.86 | −0.375 | WUS | Nucleus |
| CsWOX8 | cymsin_Mol012174 | 228 | 5.9 | 25,686.97 | 66.95 | 68.46 | −0.606 | Ancient | Nucleus |
| CsWOX9 | cymsin_Mol005406 | 334 | 7.77 | 36,197.62 | 51.70 | 78.02 | −0.302 | Intermediate | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Chen, X.; Cheng, M.; Zhou, C.; Zheng, R.; Wu, X.; Duan, Y.; Ahmad, S.; Liu, Z.; Chen, J.; et al. Genome-Wide Identification of the WUSCHEL-Related Homeobox (WOX) Gene Family in Three Cymbidium Species and Expression Patterns in C. goeringii. Horticulturae 2024, 10, 645. https://doi.org/10.3390/horticulturae10060645
Wang F, Chen X, Cheng M, Zhou C, Zheng R, Wu X, Duan Y, Ahmad S, Liu Z, Chen J, et al. Genome-Wide Identification of the WUSCHEL-Related Homeobox (WOX) Gene Family in Three Cymbidium Species and Expression Patterns in C. goeringii. Horticulturae. 2024; 10(6):645. https://doi.org/10.3390/horticulturae10060645
Chicago/Turabian StyleWang, Fei, Xiuming Chen, Mengya Cheng, Chengcheng Zhou, Ruiyue Zheng, Xiaopei Wu, Yanru Duan, Sagheer Ahmad, Zhongjian Liu, Jinliao Chen, and et al. 2024. "Genome-Wide Identification of the WUSCHEL-Related Homeobox (WOX) Gene Family in Three Cymbidium Species and Expression Patterns in C. goeringii" Horticulturae 10, no. 6: 645. https://doi.org/10.3390/horticulturae10060645
APA StyleWang, F., Chen, X., Cheng, M., Zhou, C., Zheng, R., Wu, X., Duan, Y., Ahmad, S., Liu, Z., Chen, J., & Peng, D. (2024). Genome-Wide Identification of the WUSCHEL-Related Homeobox (WOX) Gene Family in Three Cymbidium Species and Expression Patterns in C. goeringii. Horticulturae, 10(6), 645. https://doi.org/10.3390/horticulturae10060645







