Abstract
Reactive oxygen species (ROS) are pivotal in signal transduction processes in plant–pathogen interactions. The ROS signaling pathways involved in Candidatus Liberibacter asiaticus (CLas) and Xanthomonas citri subspecies citri (Xcc) infections in Citrus sinensis (sweet orange) are unclear. In this study, we comprehensively identified ROS metabolism-associated genes, including 9 NADPH oxidase (RBOH), 14 superoxide dismutase (SOD), 1 catalase (CAT), 9 peroxiredoxin (PrxR), 5 ascorbate peroxidase (APX), 4 glutathione peroxidase (GPX), 3 monodehydroascorbate reductase (MDAR), 2 dehydroascorbate reductase (DHAR), 2 glutathione reductase (GR), 24 thioredoxin (Trx), and 18 glutaredoxin (GLR) genes in C. sinensis. An analysis revealed variable gene structures but conserved motifs and domains in ROS subfamilies. A comparative synteny analysis with Arabidopsis thaliana and Vitis vinifera indicated evolutionary conservation of most ROS metabolism-associated genes, with some originating from gene duplication events post-species divergence in C. sinensis. Expression profiling revealed five up-regulated genes and four down-regulated genes during both CLas and Xcc infections. Promoter analysis revealed numerous stress-responsive elements in the promoter of ROS metabolism-associated genes. Protein–protein interaction network analysis highlighted the involvement of ROS metabolism in various biological processes. A comparison of ROS metabolism-associated genes between C. sinensis and Poncirus trifoliata indicated multiple gene gain and loss events within ROS subfamilies of C. sinensis. This study enhances our understanding of ROS metabolism in C. sinensis and sheds light on citrus–pathogen interactions.
1. Introduction
Citrus huanglongbing (HLB) and citrus bacterial canker are two major bacterial diseases in citrus production that cause severe losses. HLB is mainly caused by the phloem-limited bacterium Candidatus Liberibacter asiaticus (CLas). CLas infection strongly affects various biological processes and metabolic pathways in the leaves and fruits of citrus plants [1,2,3,4,5]. RNA-seq analysis of fruit peel tissues from Citrus sinensis cv. Valencia revealed an up-regulation of genes associated with the light reactions of photosynthesis, ATP synthesis, various WRKY transcription factor genes, and genes involved in the signaling pathways of ethylene, salicylic acid, and jasmonic acid. Conversely, genes associated with sucrose and starch metabolism, in addition to those involved in cytokinin and gibberellin pathways, were inhibited [4]. However, in HLB-tolerant C. hystrix (kaffir lime), the genes related to starch synthesis and the photosynthetic process remained undisturbed, while an induction of peroxidases, Cu/Zn-SOD, and POD4 genes was observed [2]. The HLB-tolerant ‘LB8-9’ mandarin (C. reticulata) demonstrated superior reactive oxygen species (ROS) scavenging activity compared with the HLB-susceptible ‘Hamlin’ C. sinensis [6]. In HLB-tolerant citrus varieties, the stimulation of ROS-scavenging genes, such as catalases and ascorbate peroxidases, may contribute to the mitigation of ROS in plants, especially in phloem tissues [7]. Prior research has revealed that HLB is an immunity-triggered disease prompted by pathogens, characterized by significant ROS accumulation in plants infected with CLas [8]. ROS accumulation initiates programmed cell death, a process that can be curtailed through the use of antioxidants [8]. However, the identification of genes related to ROS metabolism, including NADPH oxidase (RBOH), superoxide dismutase (SOD), catalase (CAT), peroxiredoxin (PrxR), thioredoxin (Trx), ascorbate peroxidase (APX), glutathione peroxidase (GPX), monodehydroascorbate reductase (MDAR), dehydroascorbate reductase (DHAR), glutaredoxin (GLR), and glutathione reductase (GR), within the genome of C. sinensis and their roles in the process of citrus–CLas interactions remain unclear. Citrus bacterial canker is another bacterial disease in citrus production caused by Xanthomonas citri subspecies citri (Xcc) [9]. Since Xcc can be cultured in vitro (unlike CLas), its pathogenesis is fairly clear. However, the correlation between ROS metabolism-associated genes and Xcc infection remains inadequately elucidated.
Comprehending the mechanisms of transcriptional regulation of plants in response to pathogen attack is integral to elucidating the interactions between plants and pathogens [10]. ROS serve as primary signaling entities in plant physiology, orchestrating a plethora of processes encompassing growth, development, responses to abiotic stress, and particularly, resistance against pathogens [11]. The regulation of ROS levels is critical in plants, as an imbalance can lead to oxidative stress, causing damage to cellular structures [12]. The RBOHs are primary producers of ROS in plants. They generate superoxide by transferring an electron from NADPH to molecular oxygen [13]. SOD, CAT, and PrxR are key enzymes in ROS detoxification. SOD catalyzes the dismutation of superoxide into oxygen and hydrogen peroxide, which is then detoxified by CAT and PrxR [14]. Furthermore, APX and GPX, two key enzymes in the ascorbate-glutathione cycle, transform hydrogen peroxide into water, playing a critical role in maintaining ROS homeostasis [12,15]. Moreover, MDAR, DHAR, and GR are crucial for the regeneration of antioxidants in the ascorbate-glutathione cycle, thereby maintaining the balance of ROS [16]. Additionally, Trx and GLR, two small redox-active proteins, modulate the redox state of target proteins and scavenge ROS, contributing to the redox homeostasis in plant cells [17]. ROS emerges as a vital element in the preliminary signal transduction pathways incited by pathogen detection [18,19]. The genes integral to ROS metabolism govern important roles in ROS signaling. ROS engendered by RBOH proteins not only function as a direct defense against invading pathogens but also perform as secondary messengers, instigating consequent defense reactions [20]. Enzymes such as SOD, CAT, PrxR, APX, GPX, MDAR, DHAR, GR, Trx and GLR, via their ROS detoxification activities, facilitate positive ROS signaling regulation, harmonizing the balance between ROS production and scavenging, thereby assuring optimal ROS signaling [11,14,21].
In this study, we aimed to identify and elucidate the crucial genes involved in ROS metabolism, which are responsive to pathogenic stress in C. sinensis. Furthermore, we sought to elucidate the functional roles of ROS metabolism-related genes during the infection stages of CLas and Xcc. We systematically identified and analyzed the expression profiles of the ROS metabolism-associated genes including RBOH, SOD, CAT, PrxR, APX, GPX, MDAR, DHAR, GR, Trx, and GLR in C. sinensis. Especially, differentially expressed ROS metabolism-associated genes’ responses to HLB and Xcc were achieved. Moreover, we analyzed the synteny events between C. sinensis and Arabidopsis thaliana and Vitis vinifera. We also characterized the cis-acting elements in the promoter regions of all identified ROS metabolism-associated genes. Moreover, the protein–protein interaction (PPI) network analysis of the core differential expressed ROS metabolism-associated genes was constructed. Additionally, the potential gain and loss events of ROS metabolism-associated genes between HLB-susceptible C. sinensis and HLB-tolerant P. trifoliata were analyzed. Our study lays the foundation for further in-depth research into the functions of ROS metabolism-associated genes and provides new insights into the immune response mechanisms of citrus to bacterial diseases.
2. Materials and Methods
2.1. Identification of Genes Related to Metabolism of ROS
The gene family members of RBOH, SOD, CAT, PrxR, APX, GPX, MDAR, DHAR, GR, Trx, and GLR in the genome of C. sinensis (Citrus Pan-enome to Breeding Database, CPBD, version 3.0) were identified through blastp utilizing the sequences of their A. thaliana homologs, applying an E-value threshold of less than 1 × 10−5 [22,23]. A subsequent confirmation of the identified gene candidates was accomplished through sequence analysis utilizing SMART and blastp in the National Center for Biotechnology Information (NCBI) database [24,25]. In addition, the potential candidates underwent further validation via InterPro, which facilitated the identification of conserved domains within the protein sequences [26]. Eventually, the authenticated sequences were classified into different gene families. The chromosomal locations of RBOH, SOD, CAT, PrxR, APX, GPX, MDAR, DHAR, GR, Trx, and GLR genes were illustrated utilizing the ‘Gene Location Visualize from GTF/GFF’ plugin within the TBtools software suite version 2.096 [27].
2.2. Gene Structure Analysis and Motif and Conserved Domain Identification
The exon–intron gene structure of RBOH, SOD, CAT, PrxR, APX, GPX, MDAR, DHAR, GR, Trx, and GLR genes in C. sinensis was delineated using the annotation files from the CPBD [23]. Additionally, the motifs present in the protein sequences of these genes were identified employing the MEME suite [28]. Furthermore, the conserved domains within these proteins were discerned utilizing the Batch CD-Search tool available on NCBI [29]. The visualization for the phylogenetic tree, gene structures, motifs, and conserved domains was conducted using the ‘Gene Structure View (Advanced)’ plugin within the TBtools software suite [27].
2.3. Synteny Analysis
Genomic sequences and Generic Feature Format Version 3 (GFF3) files of A. thaliana, C. sinensis, and V. vinifera were obtained for synteny analysis. Specifically, the TAIR10 chromosome files and GFF3 file of A. thaliana were acquired from the TAIR database [30], while the genome sequences and GFF3 file of C. sinensis (version 3.0) were sourced from CPBD [23]. Moreover, the genomic sequences and GFF3 file of V. vinifera (release 52) were retrieved from EnsemblPlants for analysis [31]. Synteny analysis was executed with MCScanX software version 1.1 utilizing the downloaded genomic sequences and GFF3 files [32]. Visualization of the identified synteny events containing genes related to the metabolism of ROS within C. sinensis was accomplished using the ‘Dual Synteny Plot’ plugin available in the TBtools software suite [27].
2.4. Expression Profile Analysis
The expression profiles of RBOH, SOD, CAT, PrxR, APX, GPX, MDAR, DHAR, GR, Trx, and GLR genes in C. sinensis during CLas and Xcc infection were analyzed. In the investigation of CLas infection, the RNA-seq analysis involved the use of leaves from both CLas-free and CLas-infected C. sinensis cv. Newhall, each with three biological replicates [5]. Similarly, in the study of Xcc infection, RNA-seq analysis was conducted using leaves from Xcc-free and Xcc-infected C. sinensis cv. Newhall at 24 and 48 h post-infection, with three biological replicates for each condition. The RNA-seq data have been deposited in the NCBI Sequence Read Archive (SRA) database under the accession numbers PRJNA953196 [5] and PRJNA1088720. The raw RNA-seq data were first trimmed using Trimmomatic version 0.40 with default parameters [33]. Next, the gene expression profiles in C. sinensis were determined by utilizing Kallisto with a k-mer length of 31, as described by Bray et al. [34]. The expression values for each gene in different samples were normalized using the Z score transformation method [35]. Heatmaps were generated using the ‘HeatMap’ plugin within the TBtools software suite [27].
2.5. RT-qPCR Validation
The extraction of total RNA from samples for RT-qPCR was carried out using the EasyPure® Plant RNA Kit (Transgen Biotech, Beijing, China). The RNA was reverse-transcribed to cDNA using EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (Transgen Biotech). Then, the RT-qPCR procedure was conducted on a StepOnePlus real-time fluorescence quantitative PCR instrument (ABI, South San Francisco, CA, USA) with TransStart® Top Green qPCR SuperMix (Transgen Biotech). The specific primers used for RT-qPCR analysis are listed in Table S1 with the GAPDH gene of C. sinensis as the internal control.
2.6. Hydrogen Peroxide Content, CAT, and SOD Enzyme Activities
The supernatant from the ground leaves was extracted using a cold 50 mM phosphate buffer (pH 7.8) for subsequent measurements of hydrogen peroxide levels as well as CAT and SOD enzyme activities. The hydrogen peroxide content was determined using the potassium iodide method [8]. For CAT enzyme activity measurement, the supernatant was preheated in a constant temperature water bath at 25 °C, and then 50 μL was taken and mixed with 1.95 mL of 0.01 mol/L hydrogen peroxide. After adding hydrogen peroxide, the mixture was immediately placed into a UV spectrophotometer and the absorbance changes were measured at a wavelength of 240 nm, with absorbance readings taken every 1 min for 4 min. An enzyme activity unit (U) was defined as a decrease of 0.1 in absorbance per min. For SOD enzyme activity measurement, a reaction mixture (2 mL 14.5 mmol/L methionine, 0.8 mL 5 mmol/L nitro blue tetrazolium solution, 0.8 mL 5 mmol/L riboflavin solution, 0.8 mL EDTA) was added to the test tubes as a blank control. In the experimental group, 50 μL of supernatant extract was added to the control group. The absorbance was measured at 560 nm using a spectrophotometer, and an enzyme unit (U) was defined as the amount of enzyme needed to inhibit the reaction by 50%.
2.7. Identification of Cis-Acting Elements in the Promoters
The 2000 bp sequences upstream the coding sequence of RBOH, SOD, CAT, PrxR, APX, GPX, MDAR, DHAR, GR, Trx, and GLR genes in C. sinensis were obtained from the genome and subsequently queried against the PlantCARE database [36]. The abiotic and biotic cis-acting elements were displayed utilizing the ‘Simple BioSequence Viewer’ plugin in the TBtools software suite [27].
2.8. PPI Network Analysis In Silico
The PPI networks analysis of the ROS metabolism genes with similar expression patterns under CLas and Xcc infection was conducted using the STRING online database [37]. The ‘Single/Multiple Proteins by Sequence’ search module was utilized to construct PPI networks by querying against C. sinensis.
2.9. Gene Gain and Loss Events
The Orthologous Gene Search feature of the CPBD database was employed to identify gene gain and loss events in genes associated with ROS metabolism in C. sinensis, in comparison with Poncirus trifoliata (trifoliate orange) [23]. Orthologous genes corresponding to each ROS metabolism-associated gene in C. sinensis from P. trifoliata were gathered. Subsequently, an analysis of gene gain and loss was performed utilizing the ‘Gene Lost & Gain Analysis (based on Notung)’ plugin within the TBtools software suite [27]. Protein sequences of RBOH, SOD, CAT, PrxR, APX, GPX, MDAR, DHAR, GR, Trx, and GLR from C. sinensis were retrieved from CPBD [23]. Multiple sequence alignment was performed utilizing MAFFT [38], followed by alignment refinement with Gblocks [39], model selection using PartitionFinder 2 [40], and phylogenetic tree reconstruction via IQ-tree [41].
2.10. Statistical Analysis
The statistical analyses in this study were completed using SPSS software (version 25), which included t-tests and one-way ANOVA tests. Pairwise comparisons for the one-way ANOVA tests were conducted using the Student–Newman–Keuls method.
3. Results
3.1. Identification of ROS Metabolism-Associated Genes in C. sinensis
We utilized methods such as BLAST and conserved domain analysis to identify RBOH, SOD, CAT, PrxR, APX, GPX, MDAR, DHAR, GR, Trx, and GLR genes in C. sinensis. Altogether, 91 ROS metabolism-associated genes, including 9 RBOH, 14 SOD, 1 CAT, 9 PrxR, 5 APX, 4 GPX, 3 MDAR, 2 DHAR, 2 GR, 24 Trx and 18 GLR genes, were identified in C. sinensis (Figure 1A, Table S2). The distribution of these genes was non-uniform, with clusters observed on the chromosomes of C. sinensis, suggesting an intriguing pattern (Figure 1B). For example, the 7 SOD genes, from CsSOD1 to CsSOD7, were closely positioned within a confined segment on chromosome 3 (Figure 1B).
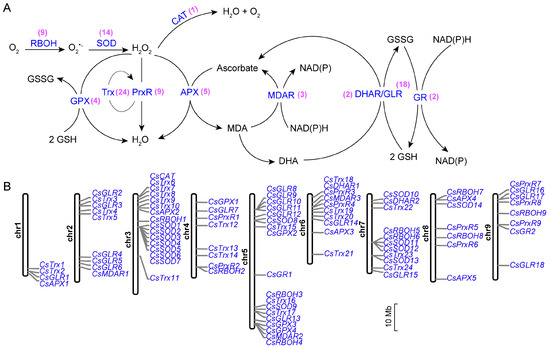
Figure 1.
Identification of reactive oxygen species (ROS) metabolism-associated genes in Citrus sinensis. (A) The ROS metabolism pathway. Enzymes catalyzing corresponding metabolic reactions are highlighted in blue, while the number of genes encoding these enzymes in C. sinensis is represented in purple. (B) Chromosomal localization of ROS metabolism-associated genes. The scale represents 10 million base pairs (Mb). RBOH: NADPH oxidase. SOD: superoxide dismutase. CAT: catalase. PrxR: peroxiredoxin. Trx: thioredoxin. APX: ascorbate peroxidase. GPX: glutathione peroxidase. MDAR: monodehydroascorbate reductase. DHAR: dehydroascorbate reductase. GLR: glutaredoxin. GR: glutathione reductase.
3.2. Characterization of ROS Metabolism-Associated Genes in C. sinensis
To elucidate the characteristics of ROS metabolism-associated genes, we analyzed the gene structures, conserved motifs, and conserved domains of these genes in C. sinensis. The exon counts of ROS metabolism-associated genes varied from 1 to 16 (CsMDAR2), while 11 out of 18 GLR genes comprised a single exon devoid of introns (Figure 2A). An analysis of the conserved motifs of ROS metabolism-associated proteins indicated that members of the same gene family have similar or identical conserved motifs (Figure 2B). For example, in some proteins such as CsMDAR1, no conserved motifs were detected, while multiple conserved motifs were identified in members of the RBOH family (Figure 2B). There are also differences in the presence of conserved motifs within the same gene family. For instance, CsRBOH8 contains 1 conserved motif, while CsRBOH1 contains 10 conserved motifs (Figure 2B). Most of the ROS metabolism-associated proteins contained a conserved domain (Figure 2C). However, CsCAT contained two conserved Catalase domains, and CsSOD1 contains two conserved Sod_Cu domains. Conversely, no conserved domains were predicted in CsDHAR1, CsRBOH9, and CsTrx17 (Figure 2C).
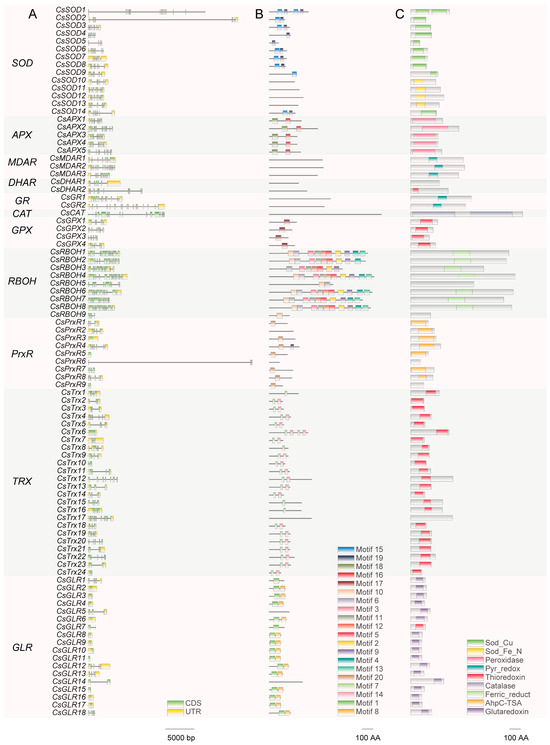
Figure 2.
Characterization of ROS metabolism-associated genes in C. sinensis. (A) Gene structure. Line represents introns, green rectangles represent coding sequences (CDs), and yellow rectangles represent untranslated regions (UTRs). Scale represents 5000 base pairs (bp). (B) Conserved motifs. Rectangles of different colors represent 20 different motifs. Scale represents 100 amino acids (AA). (C) Conserved domains. Rectangles of different colors represent nine different domains. Scale represents 100 (AA).
3.3. Identification of Synteny Events between C. sinensis and Other Species
Synteny analysis is instrumental in elucidating gene evolution [32]. To comprehend the functionality of ROS metabolism-associated genes through the lens of their evolutionary relationships, synteny analysis between C. sinensis and A. thaliana and V. vinifera was performed. A total of 73 Synteny events were detected between C. sinensis and A. thaliana, and 70 Synteny events were identified between C. sinensis and V. vinifera (Figure 3). These findings indicate that a significant proportion of genes associated with ROS metabolism in C. sinensis have evolutionary origins shared with A. thaliana and V. vinifera (Figure 3). This information is valuable for elucidating the timing of gene occurrence relative to the divergence from a common ancestor.
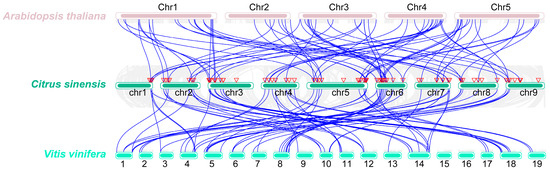
Figure 3.
Identification of synteny events for ROS metabolism-associated genes between C. sinensis and Arabidopsis thaliana and Vitis vinifera. Thick rosy-brown lines represent chromosomes of A. thaliana, thick light-sea-green lines represent chromosomes of C. sinensis, and thick cyan lines represent chromosomes of V. vinifera. Blue lines represent synteny events for ROS metabolism-associated genes. The triangles represent ROS metabolism-associated genes in C. sinensis.
3.4. Dynamic Expression Profiles of ROS Metabolism-Associated Genes in C. sinensis under CLas Infection
CLas infection significantly induces ROS accumulation in C. sinensis [8]. To analyze the expression changes of ROS metabolism-associated genes in response to CLas infection, we utilized transcriptome data to examine the expression patterns of ROS metabolism-associated genes in both CLas-free and CLas-infected samples. Eleven ROS metabolism-associated genes including four SOD genes, three GLR genes, two RBOH genes, and two PrxR genes showed no expression in either CLas-free or CLas-infected samples (Figure 4A). The remaining 80 genes exhibited varied expression patterns, with some showing a higher expression in CLas-free samples and others showing a higher expression in CLas-infected samples (Figure 4A). To further elucidate the differential genes induced by CLas infection, we identified 30 differentially expressed ROS-related genes, including 16 up-regulated and 14 down-regulated genes. These differentially expressed genes span various gene families, indicating diverse response patterns of ROS metabolism-associated genes to CLas infection (Figure 4B).
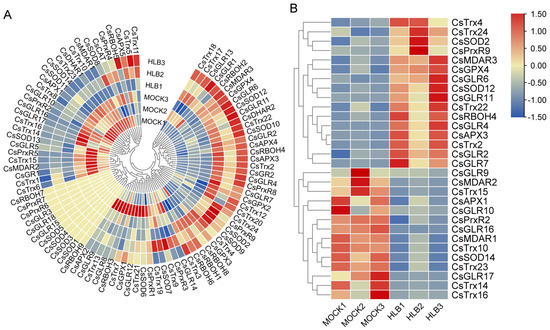
Figure 4.
Expression profiles of ROS metabolism-associated genes following infection by Candidatus Liberibacter asiaticus (CLas) in C. sinensis. (A) Expression profiles of all ROS metabolism-associated genes. (B) Expression profiles of differentially expressed ROS metabolism-associated genes. ‘MOCK’ represents CLas-free samples, ‘HLB’ represents CLas-infected samples. Numbers represent different biological replicates. Bar represents gradient colors to correspond with normalized expression values.
3.5. Dynamic Expression Profiles of ROS Metabolism-Associated Genes in C. sinensis under Xcc Infection
To clarify the expression changes of ROS metabolism-associated genes during Xcc infection, we also conducted transcriptome analysis to examine the expression patterns of ROS metabolism-associated genes at different time points post-inoculation with Xcc. Eight genes were found to be non-expressed in both the control and Xcc-inoculated samples (Figure 5A). These 8 genes, including CsSOD1/3/4/5, CsGLR3/15, CsPrxR6, and CsRBOH6, were part of the 11 genes identified earlier that were not expressed in both CLas-free and CLas-infection samples (Figure 4A). A subsequent analysis of differentially expressed genes revealed a total of 24 genes with altered expression levels 48 h post-inoculation, consisting of 12 up-regulated and 12 down-regulated genes (Figure 5B). It is worth noting the expression pattern of CsAPX3, which exhibited up-regulation at 24 h post-inoculation but down-regulation at 48 h post-inoculation when compared with the control samples (Figure 5B).
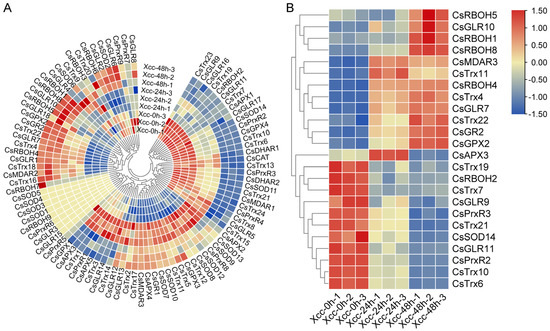
Figure 5.
Expression profiles of ROS metabolism-associated genes following incubation by Xanthomonas citri subspecies citri (Xcc) in C. sinensis. (A) Expression profiles of all ROS metabolism-associated genes. (B) Expression profiles of differentially expressed ROS metabolism-associated genes. Times ‘0 h’, ‘24 h’, and ‘48 h’ represent hours post incubation. Numbers represent different biological replicates. Bar represents gradient colors to correspond with normalized expression values.
3.6. Identification of ROS Metabolism-Associated Genes with Similar/Opposite Expression Patterns under CLas and Xcc Infection
To explore important genes involved in the response of C. sinensis to CLas and Xcc infection, we performed an analysis to pinpoint genes associated with ROS metabolism displaying comparable or contrasting expression profiles following CLas and Xcc infections. It was found that there were five common genes up-regulated in C. sinensis after CLas and Xcc infection, namely CsMDAR3, CsRBOH4, CsTrx4, CsGLR7, and CsTrx22 (Figure 6A). Additionally, there were two genes that were up-regulated in C. sinensis after CLas infection but down-regulated after Xcc infection, which were CsAPX3 and CsGLR11 (Figure 6B). Furthermore, four genes were identified to be down-regulated in C. sinensis after CLas and Xcc infections, namely CsGLR9, CsPrxR2, CsTrx10, and CsSOD14 (Figure 6C). Finally, there was one gene that was down-regulated after CLas infection but up-regulated after Xcc infection in C. sinensis, which was CsGLR10 (Figure 6D). The differentially expressed genes with similar expression patterns during CLas and Xcc infections may participate in common signal transduction pathways triggered by bacterial infections.
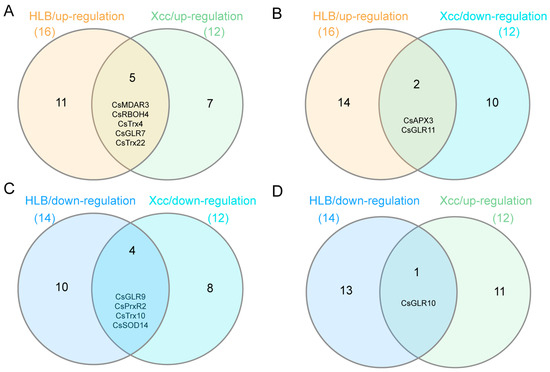
Figure 6.
Identification of ROS metabolism-associated genes with similar/opposite expression patterns under Candidatus Liberibacter asiaticus (CLas) and Xanthomonas citri subspecies citri (Xcc) infection in C. sinensis. (A) Common up-regulated genes under CLas and Xcc infection. (B) Common genes of up-regulated genes during CLas infection and down-regulated genes during Xcc infection. (C) Common down-regulated genes under CLas and Xcc infection. (D) Common genes of down-regulated genes during CLas infection and up-regulated genes during Xcc infection.
3.7. RT-qPCR Analysis
To validate the reliability of the RNA-seq analysis, RT-qPCR analysis was conducted to assess the expression profiles of four selected differentially expressed genes, including two up-regulated (CsMDAR3 and CsRBOH4) and two down-regulated (CsPrxR2 and CsTrx10) genes in response to CLas and Xcc infection. The RT-qPCR results demonstrated significantly higher expression levels of CsMDAR3 (Figure 7A) and CsRBOH4 (Figure 7B) in CLas-infected samples, along with significantly lower expression levels of CsPrxR2 (Figure 7C) and CsTrx10 (Figure 7D). Similarly, in Xcc-infected samples, CsMDAR3 (Figure 7E) and CsRBOH4 (Figure 7F) exhibited significantly elevated expression levels, while CsPrxR2 (Figure 7G) and CsTrx10 (Figure 7H) showed significantly reduced expression levels.
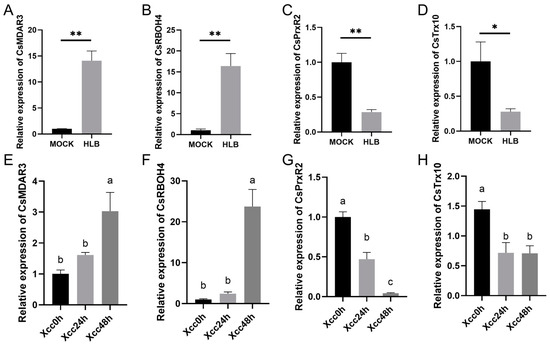
Figure 7.
RT-qPCR analysis of ROS metabolism-associated genes in C. sinensis under Candidatus Liberibacter asiaticus (CLas) and Xanthomonas citri subspecies citri (Xcc) infection in C. sinensis. Expression profiles of (A) CsMDAR3, (B) CsRBOH4, (C) CsPrxR2, and (D) CsTrx10 under CLas infection. Expression profiles of (E) CsMDAR3, (F) CsRBOH4, (G) CsPrxR2, and (H) CsTrx10 under Xcc infection. ‘*’ indicates significant difference with a p-value < 0.05. ‘**’ indicates significant difference with a p-value < 0.01. Lowercase letters annotated on bars indicate significant differences with a p-value < 0.05. ‘MOCK’ represents healthy plants, while ‘HLB’ represents plants infected with HLB. ‘Xcc0h’ represents control for citrus canker inoculation, and ‘Xcc24h’ and ‘Xcc48h’ represent samples collected at 24 h and 48 h post citrus canker inoculation, respectively. Each measurement was performed with three biological replicates.
3.8. Hydrogen Peroxide Content, CAT, and SOD Enzyme Activities during CLas and Xcc Infection
Pathogen invasion significantly alters the redox homeostasis within plant cells [21]. After CLas infection, we noted a significant accumulation of hydrogen peroxide (Figure 8A), and the activities of CAT (Figure 8B) and SOD (Figure 8C) were significantly increased in CLas-infected leaves of C. sinensis. Similarly, we found that Xcc infection also caused a significant increase in hydrogen peroxide levels (Figure 8D), as well as a significant elevation in CAT (Figure 8E) and SOD (Figure 8F) activities. However, we observed that the activities of CAT and SOD continued to significantly increase (Figure 8E,F), while the level of hydrogen peroxide did not show a significant change between samples of 24 h and 48 h post Xcc infection (Figure 8D).
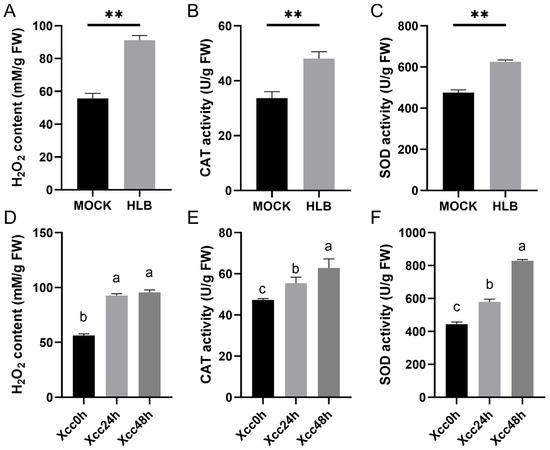
Figure 8.
Determination of hydrogen peroxide content, CAT, and SOD enzyme activities in C. sinensis under Candidatus Liberibacter asiaticus (CLas) and Xanthomonas citri subspecies citri (Xcc) infection in C. sinensis. (A) Hydrogen peroxide content, (B) CAT activities, and (C) SOD activities under CLas infection. (D) Hydrogen peroxide content, (E) CAT activities, and (F) SOD activities under Xcc infection. ‘**’ indicates significant difference with a p-value < 0.01. Lowercase letters annotated on bars indicate significant differences with a p-value < 0.05. ‘MOCK’ represents healthy plants, while ‘HLB’ represents plants infected with HLB. ‘Xcc0h’ represents control for citrus canker inoculation, and ‘Xcc24h’ and ‘Xcc48h’ represent samples collected at 24 h and 48 h post citrus canker inoculation, respectively. Each measurement was performed with three biological replicates.
3.9. PPI Network Analysis of Key Candidate ROS Metabolism-Associated Genes In Silico
In order to identify more cooperation factors involved in ROS metabolism, we performed a PPI network analysis of the common up-regulated and down-regulated genes during CLas and Xcc infections in silico (Figure 9). CsMDAR3 exhibited interactions with DAO domain-containing proteins, stress-response A/B barrel domain-containing proteins, and Rieske domain-containing proteins (Figure 9A). CsRBOH4 interacted with calcium-dependent protein kinases (Figure 9B). CsTrx4 and CsTrx22 primarily interacted with Thioredoxin domain-containing proteins and Peroxiredoxins (Figure 9C,D). CsGLR7 exhibited variable interaction partners such as Ribonucleoside-diphosphate reductases, Glutathione reductases, and Expansin (Figure 9E). CsGLR9 interacted with Ribonucleoside-diphosphate reductases and Glutathione reductases (Figure 9F). The interaction partners of CsPrxR2 included Tryptophan synthases and Peptide-methionine (R)-S-oxide reductases (Figure 9G). CsTrx10 interacted with Thioredoxin domain-containing proteins and Peroxiredoxins (Figure 9H). The interaction partners of CsSOD14 included DJ-1_PfpI domain-containing proteins and catalases (Figure 9I). Interacting partners play a crucial role in the catalytic activity of these enzymes, but their specific biological functions require further investigation.
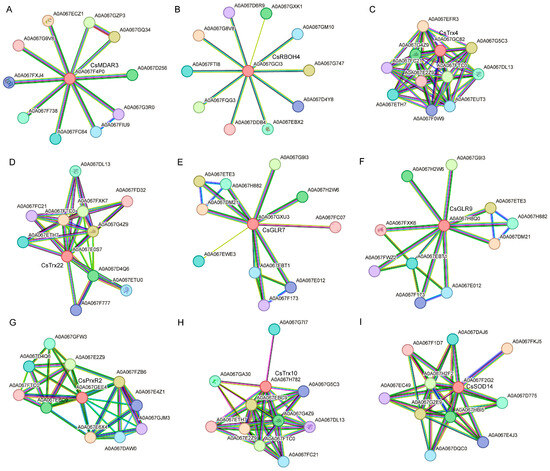
Figure 9.
Protein–protein interaction (PPI) network analysis of key ROS metabolism-associated genes under Candidatus Liberibacter asiaticus and Xanthomonas citri subspecies citri infection in C. sinensis in silico. PPI network analysis of (A) CsMADR3, (B) CsROBH4, (C) CsTrx4, (D) CsTrx22, (E) CsGLR7, (F) CsGLR9, (G) CsPrxR2, (H) CsTrx10, and (I) CsSOD14. Raw networks were downloaded from STRING database and modified using Cytoscape.
3.10. Cis-Acting Elements in the Promoter of ROS Metabolism-Associated Genes in C. Sinensis
To elucidate the potential expression regulatory mechanisms of ROS metabolism-associated genes, we analyzed the promoter sequences of these genes, focusing on cis-acting elements involved in the hormone signaling response and abiotic stress response. In the promoter region of CsSOD11, as many as 31 cis-acting elements are involved in hormone signaling and abiotic stress responses, while no such cis-acting elements were found in the promoter sequences of CsTrx1, CsTrx14, and CsTrx22 (Figure S1). Furthermore, the distribution of cis-acting elements is irregular in the promoter region of ROS metabolism-associated genes in C. sinensis, even within the same gene family, with completely different distributions of cis-acting elements (Figure S1). It is worth noting that although no cis-acting elements responding to hormone signals or abiotic stress were found in the promoter region of CsTrx22, CsTrx22 was up-regulated during CLas and Xcc infection (Figure 6A). This suggests that there are still deficiencies in predicting cis-acting elements using the current method, and how CsTrx22 responds to pathogen infection may involve new regulatory mechanisms.
3.11. Gene Gain and Loss Events in ROS Metabolism-Associated Genes between C. sinensis and P. trifoliata
Previous studies suggested that P. trifoliata is one of the germplasms resistant to HLB, while C. sinensis is more susceptible to the disease, although there are differences between different varieties [42,43,44]. Given the ROS accumulation observed in CLas-infected tissues, we systematically identified the occurrence of gene gain and loss events within ROS metabolism-associated genes between C. sinensis and P. trifoliata by analyzing their orthologous relationships (Table 1). Gene gain and loss events were found in the RBOH, SOD, PrxR, APX, and Trx gene families (Table 1, Figure 8 and Figures S2–S11). Specifically, one gene loss event was identified in the RBOH gene family, along with three gene gain events and five gene loss events in the SOD gene family. Furthermore, three gene gain events and three gene loss events were documented in the PrxR gene family, while one gene gain event was observed in the APX gene family. Lastly, one gene gain event and one gene loss event were found in the Trx gene family (Table 1).

Table 1.
Potential gain and loss events of ROS metabolism-associated genes between HLB-susceptible C. sinensis and HLB-tolerant P. trifoliata.
Taking the SOD gene family as an example, a sequence alignment and a phylogenetic analysis were performed on all SOD gene members in C. sinensis and P. trifoliata. The examination of individual evolutionary branches facilitated the identification of gene gain and loss events (Figure 10).
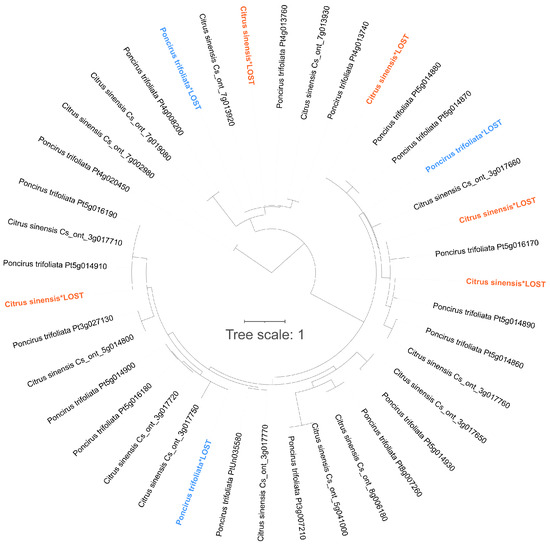
Figure 10.
Gene gain and loss events in SOD gene family between C. sinensis and P. trifoliata. Gene loss events in C. sinensis are highlighted in bold orange color, while gene loss events in P. trifoliata are highlighted in bold blue color.
4. Discussion
ROS are continuously produced as by-products of aerobic metabolism in plant cells, with their levels intricately regulated through a complex network of metabolic pathways [45]. These pathways, regulated by ROS metabolism-associated genes, are responsible for scavenging and detoxifying ROS, maintaining a balance between ROS production and elimination [46]. The balance between ROS production and scavenging is vital for the maintenance of cellular homeostasis [47]. The coordinated expression of ROS metabolism-associated genes ensures this balance, maintains cellular redox homeostasis, and allows for ROS to act as crucial signaling molecules in plant physiology [48].
There are significant differences in the number of ROS metabolism-associated genes among different species. Taking SOD as an example, there are 8 SOD genes in A. thaliana [22], 14 in C. sinensis, 29 in Brassica juncea [49], 18 in B. rapa [49], 25 in Musa acuminata [50], 18 in Gossypium raimondii and G. arboreum [51], and 8 in Salvia miltiorrhiza [52]. More surprising is that there are 18 SOD genes identified in P. trifoliata, a closely related species of C. sinensis. Since SOD functions as the initial step in catalyzing the conversion of O2− to hydrogen peroxide, the knockout mutant of AtSOD2 caused SOD activity to decline significantly in chloroplast and cytosol [53]. Nevertheless, functional overlap among SOD genes in A. thaliana has been noted, as some of them originated from gene duplication events [54]. Moreover, in C. sinensis, there is a single CAT gene responsible for eliminating hydrogen peroxide, whereas in A. thaliana, there are three gene loci encoding CAT [55]. Additionally, three CAT genes were identified in Oryza sativa [56], Zea mays [57] and Nicotiana plumbaginifolia [58]. Furthermore, two CAT genes were identified within P. trifoliata in this study. To determine the number of CAT genes in C. sinensis and potential evolutionary events, we conducted a homology search in genome versions 1 and 2 of C. sinensis, revealing two CAT genes located at adjacent genetic loci. Considering that the sequence of CAT in the genome of C. sinensis (version 3) is notably long and contains two conserved catalase domains, we have reason to suspect an annotation error of this gene in the genome of C. sinensis (version 3). Therefore, it is reasonable to conclude that C. sinensis contains two CAT genes. The APX gene family in C. sinensis comprises a total of 5 members, while A. thaliana has 9 members [22], and there are 11 APX genes in the genome of Populus trichocarpa [59]. In addition, while the genome of A. thaliana contained 27 GLR genes and 32 Trx genes [22], 18 GLR genes and 24 Trx genes were found in the genome of C. sinensis. This indicates that the quantity of ROS metabolism-associated genes in C. sinensis is generally lower than that in A. thaliana, with the exception of the SOD gene family.
Given that CLas infection significantly induces the accumulation of ROS in the phloem tissues, thereby triggering an exaggerated immune response in plants, our focus lies on determining the altered expression patterns of ROS metabolism-associated genes following CLas infection. Thirty differentially expressed genes, including 16 up-regulated genes and 14 down-regulated genes were observed in C. sinensis during CLas infection. Among them, 1 RBOH gene CsRBOH4 was up-regulated. A previous study indicated that RBOH genes are activated in the process of CLas infection, thus leading to ROS accumulation [1]. The homologous gene of CsRBOH4 in A. thaliana is AtRBOHF. AtRBOHF is a crucial factor involved not only in cell death associated with the hypersensitive response but also in modulating plant defense responses to pathogens [60]. Significantly, CLas infection leads to a notable increase in the accumulation of hydrogen peroxide and facilitates cell death in citrus plants [8]. Therefore, CsRBOH4 may serve as a key player in the interaction between CLas and citrus plants. Interestingly, the differentially expressed genes did not include genes from the CAT and GR gene families. One reason is that the number of genes in the CAT and GR gene families is small, and another reason is that these two genes are widely and importantly involved in biological processes within cells. Xcc infection also induced a differential expression of multiple genes, including 12 up-regulated and 12 down-regulated genes. Four RBOH genes were up-regulated, including CsRBOH4, while one RBOH gene, namely CsRBOH2, was down-regulated. The homologous gene of CsRBOH2 in A. thaliana is AtRBOHE. Research has demonstrated the involvement of AtRBOHE in the development of various tissues such as roots and pollen, as well as its participation in plant–Trichoderma interactions [61,62]. The negatively regulated expression of CsRBOH2 in C. sinensis during Xcc infection suggests an antagonistic relationship between plant development and stress responses. The identification of differentially expressed genes revealed that multiple genes exhibit similar or contrasting expression patterns in response to both CLas and Xcc infections. It is worthy to note that CsRBOH4 was up-regulated during both CLas and Xcc infections, suggesting that CsRBOH4 may play a central role in the C. sinensis response to pathogen invasion. Nevertheless, CsAPX3 and CsGLR11 were observed to be up-regulated during CLas infection but down-regulated during Xcc infection, highlighting distinct responses of C. sinensis to the infections caused by the two pathogens. Upon Pseudomonas syringae infection, APX was significantly up-regulated in Actinidia chinensis [63]. In contrast, upon Erwinia amylovora infection, the APX gene demonstrated down-regulation in Pyrus communis [64]. Therefore, this contrasting regulatory pattern of genes in response to different pathogens could provide novel clues for further elucidating pathogenic mechanisms.
In plant–pathogen interactions, ROS metabolism-associated genes play a dual role [12,21,45]. These genes play a role in triggering the oxidative burst, a quick and temporary increase in ROS levels that acts as an initial defense mechanism against pathogens [65]. However, some pathogens have developed mechanisms to control the generation of ROS and alter the functions of ROS metabolism enzymes, ultimately impacting the plant’s defense against these pathogens [66,67]. In this study, results showed a notable rise in hydrogen peroxide levels after CLas and Xcc infection, suggesting that C. sinensis relies on increased hydrogen peroxide to combat pathogen attacks post-infection. SOD facilitates the conversion of superoxide anions into hydrogen peroxide, while CAT plays a pivotal role in eliminating intracellular hydrogen peroxide [14]. Interestingly, enzymatic activity assessments in our study indicated a marked elevation in SOD and CAT activities of C. sinensis subsequent to being infected by CLas and Xcc. Nevertheless, RNA-seq analysis revealed no substantial alteration in CAT gene expression and a noteworthy decrease in CsSOD14 expression post CLas and Xcc infection. The findings imply inconsistencies between transcriptional and protein levels, emphasizing the complexity of the ROS metabolic pathway. Within this pathway, these genes cooperate to sustain the intricate balance of ROS.
5. Conclusions
In summary, this study systematically identified ROS metabolism-associated genes in the genome of C. sinensis using bioinformatics methods, including gene families such as RBOH, SOD, CAT, PrxR, APX, GPX, MDAR, DHAR, GR, Trx, and GLR. Through gene structure analysis, conserved motifs, and conserved domain prediction, sequence attributes of genes related to ROS metabolism were analyzed. A synteny analysis was used to explore the collinearity relationships of ROS metabolism-associated genes in sweet orange with those in Arabidopsis and grape. The changes in the expression patterns of genes related to ROS metabolism in sweet orange during CLas infection and Xcc infection processes were investigated, revealing multiple differentially expressed genes. Furthermore, five genes were found to be up-regulated in both CLas infection and Xcc infection processes, while four genes were down-regulated. A PPI network analysis was conducted to explore the protein interaction partners of these genes. An analysis of the promoter cis-elements of genes related to sweet orange ROS metabolism provided clues to their expression regulation patterns. Lastly, gene gain and loss analysis clarified events of gene gain and loss related to ROS metabolism genes in the sweet orange and trifoliate orange genomes. This study provides data support for investigating the reactive oxygen species burst during the sweet orange response to CLas infection and offers clues for further understanding the citrus–CLas interaction.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae10060625/s1, Figure S1. Identification of cis-acting elements in the promoter regions of ROS metabolism-associated genes in Citrus sinensis; Figure S2. Gene gain and loss events in the APX gene family between Citrus sinensis and Poncirus trifoliata; Figure S3. Gene gain and loss events in the CAT gene family between Citrus sinensis and Poncirus trifoliata; Figure S4. Gene gain and loss events in the DHAR gene family between Citrus sinensis and Poncirus trifoliata; Figure S5. Gene gain and loss events in the GLR gene family between Citrus sinensis and Poncirus trifoliata; Figure S6. Gene gain and loss events in the GPX gene family between Citrus sinensis and Poncirus trifoliata; Figure S7. Gene gain and loss events in the GR gene family between Citrus sinensis and Poncirus trifoliata; Figure S8. Gene gain and loss events in the MDAR gene family between Citrus sinensis and Poncirus trifoliata; Figure S9. Gene gain and loss events in the PrxR gene family between Citrus sinensis and Poncirus trifoliata; Figure S10. Gene gain and loss events in the RBOH gene family between Citrus sinensis and Poncirus trifoliata; Figure S11. Gene gain and loss events in the Trx gene family between Citrus sinensis and Poncirus trifoliata; Table S1. Primers used in this study; Table S2. Details of ROS metabolism-associated genes in Citrus sinensis; Table S3. Expression profiles of ROS metabolism-associated genes in Citrus sinensis during Candidatus Liberibacter asiaticus infection; Table S4. Expression profiles of ROS metabolism-associated genes in Citrus sinensis during Xanthomonas citri subspecies citri infection; Table S5. Annotation of proteins interacting with up-regulated ROS-associated members in Citrus sinensis; Table S6. Annotation of proteins interacting with down-regulated ROS-associated members in Citrus sinensis; Table S7. Gene gain and loss events of ROS metabolism-associated genes between Citrus sinensis and Poncirus trifoliata.
Author Contributions
Conceptualization, R.L. and Z.O.; methodology, G.H., F.L., Y.H. and R.L.; software, G.H., F.L., Y.H. and R.L.; validation, R.L.; formal analysis, R.L.; investigation, G.H., F.L., Y.H. and R.L.; resources, R.L.; data curation, G.H., F.L., Y.H. and R.L.; writing—original draft preparation, G.H., F.L., Y.H. and R.L.; writing—review and editing, G.H., Z.O. and R.L.; visualization, R.L.; supervision, R.L.; project administration, R.L.; funding acquisition, R.L, G.H. and F.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32160621, 32260659), the Innovation Program of the short-term Leader of Jiangxi Province (jxsq2020102129), and the Innovation Fund Designated for Graduate Students of Jiangxi Province (YC2023-S850).
Data Availability Statement
All data generated and analyzed during this study are included in this published article. The original RNA-seq data has been uploaded to the NCBI_SRA database (PRJNA953196 and PRJNA1088720).
Acknowledgments
The authors are grateful for the helpful discussions from the laboratory members.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ribeiro, C.; Xu, J.; Hendrich, C.; Pandey, S.S.; Yu, Q.; Gmitter, F.G.; Wang, N. Seasonal Transcriptome Profiling of Susceptible and Tolerant Citrus Cultivars to Citrus Huanglongbing. Phytopathology 2022, 113, 286–298. [Google Scholar] [CrossRef]
- Hu, Y.; Zhong, X.; Liu, X.; Lou, B.; Zhou, C.; Wang, X. Comparative transcriptome analysis unveils the tolerance mechanisms of Citrus hystrix in response to ‘Candidatus Liberibacter asiaticus’ infection. PLoS ONE 2017, 12, e0189229. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Shao, J.; Zhou, C.; Hartung, J.S. Transcriptome analysis of sweet orange trees infected with ‘Candidatus Liberibacter asiaticus’ and two strains of Citrus Tristeza Virus. BMC Genom. 2016, 17, 349. [Google Scholar] [CrossRef]
- Martinelli, F.; Uratsu, S.L.; Albrecht, U.; Reagan, R.L.; Phu, M.L.; Britton, M.; Buffalo, V.; Fass, J.; Leicht, E.; Zhao, W. Transcriptome profiling of citrus fruit response to huanglongbing disease. PLoS ONE 2012, 7, e38039. [Google Scholar] [CrossRef]
- Liu, C.; Chang, X.; Li, F.; Yan, Y.; Zuo, X.; Huang, G.; Li, R. Transcriptome analysis of Citrus sinensis reveals potential responsive events triggered by Candidatus Liberibacter asiaticus. Protoplasma 2024, 261, 499–512. [Google Scholar] [CrossRef]
- Tang, L.; Vashisth, T. New insight in Huanglongbing-associated mature fruit drop in citrus and its link to oxidative stress. Sci. Hortic. 2020, 265, 109246. [Google Scholar] [CrossRef]
- Gao, C.; Li, C.; Li, Z.; Liu, Y.; Li, J.; Guo, J.; Mao, J.; Fang, F.; Wang, C.; Deng, X.; et al. Comparative transcriptome profiling of susceptible and tolerant citrus species at early and late stage of infection by “Candidatus Liberibacter asiaticus”. Front. Plant Sci. 2023, 14, 1191029. [Google Scholar] [CrossRef]
- Ma, W.; Pang, Z.; Huang, X.; Xu, J.; Pandey, S.S.; Li, J.; Achor, D.S.; Vasconcelos, F.N.C.; Hendrich, C.; Huang, Y.; et al. Citrus Huanglongbing is a pathogen-triggered immune disease that can be mitigated with antioxidants and gibberellin. Nat. Commun. 2022, 13, 529. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, J.; Jia, H.; Sosso, D.; Li, T.; Frommer, W.B.; Yang, B.; White, F.F.; Wang, N.; Jones, J.B. Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA 2014, 111, E521–E529. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.W.; Loake, G.J.; Spoel, S.H. Transcription Dynamics in Plant Immunity. Plant Cell 2011, 23, 2809–2820. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef]
- Marino, D.; Dunand, C.; Puppo, A.; Pauly, N. A burst of plant NADPH oxidases. Trends Plant Sci. 2012, 17, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, A.; Sevilla, F.; Martí, M.C. Reactive oxygen species homeostasis and circadian rhythms in plants. J. Exp. Bot. 2021, 72, 5825–5840. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S.; Panda, P.; Sahoo, L.; Panda, S.K. Reactive oxygen species signaling in plants under abiotic stress. Plant Signal. Behav. 2013, 8, e23681. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Anee, T.I.; Parvin, K.; Nahar, K.; Mahmud, J.A.; Fujita, M. Regulation of Ascorbate-Glutathione Pathway in Mitigating Oxidative Damage in Plants under Abiotic Stress. Antioxidants 2019, 8, 384. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Xiao, J.; Zhao, Y.; Zhang, Y.; Jie, Y.; Shen, D.; Yue, C.; Huang, J.; Hua, Y.; Zhou, T. Comparative physiological and transcriptomic analyses reveal ascorbate and glutathione coregulation of cadmium toxicity resistance in wheat genotypes. BMC Plant Biol. 2021, 21, 459. [Google Scholar] [CrossRef] [PubMed]
- Paiva, C.N.; Bozza, M.T. Are Reactive Oxygen Species Always Detrimental to Pathogens? Antioxid. Redox Signal. 2013, 20, 1000–1037. [Google Scholar] [CrossRef] [PubMed]
- Vernon, P.J.; Tang, D. Eat-Me: Autophagy, Phagocytosis, and Reactive Oxygen Species Signaling. Antioxid. Redox Signal. 2012, 18, 677–691. [Google Scholar] [CrossRef]
- Lodde, V.; Morandini, P.; Costa, A.; Murgia, I.; Ezquer, I. cROStalk for Life: Uncovering ROS Signaling in Plants and Animal Systems, from Gametogenesis to Early Embryonic Development. Genes 2021, 12, 525. [Google Scholar] [CrossRef]
- Camejo, D.; Guzmán-Cedeño, Á.; Moreno, A. Reactive oxygen species, essential molecules, during plant–pathogen interactions. Plant Physiol. Biochem. 2016, 103, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Inupakutika, M.A.; Sengupta, S.; Devireddy, A.R.; Azad, R.K.; Mittler, R. The evolution of reactive oxygen species metabolism. J. Exp. Bot. 2016, 67, 5933–5943. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, X.; Liu, S.; Huang, Y.; Guo, Y.-X.; Xie, W.-Z.; Liu, H.; ul Qamar, M.T.; Xu, Q.; Chen, L.-L. Citrus Pan-Genome to Breeding Database (CPBD): A comprehensive genome database for citrus breeding. Mol. Plant 2022, 15, 1503–1505. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, 458–460. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Bolser, D.M.; Staines, D.M.; Perry, E.; Kersey, P.J. Ensembl Plants: Integrating Tools for Visualizing, Mining, and Analyzing Plant Genomic Data. Methods Mol. Biol. 2017, 1533, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Cheadle, C.; Vawter, M.P.; Freed, W.J.; Becker, K.G. Analysis of Microarray Data Using Z Score Transformation. J. Mol. Diagn. 2003, 5, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Bredeson, J.V.; Wu, G.A.; Shu, S.; Rawat, N.; Du, D.; Parajuli, S.; Yu, Q.; You, Q.; Rokhsar, D.S.; et al. A chromosome-scale reference genome of trifoliate orange (Poncirus trifoliata) provides insights into disease resistance, cold tolerance and genome evolution in Citrus. Plant J. 2020, 104, 1215–1232. [Google Scholar] [CrossRef] [PubMed]
- Rawat, N.; Kumar, B.; Albrecht, U.; Du, D.; Huang, M.; Yu, Q.; Zhang, Y.; Duan, Y.-P.; Bowman, K.D.; Gmitter, F.G., Jr.; et al. Genome resequencing and transcriptome profiling reveal structural diversity and expression patterns of constitutive disease resistance genes in Huanglongbing-tolerant Poncirus trifoliata and its hybrids. Hortic. Res. 2017, 4, 17064. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Roose, M.L.; Yu, Q.; Du, D.; Yu, Y.; Zhang, Y.; Deng, Z.; Stover, E.; Gmitter, F.G., Jr. Construction of High-Density Genetic Maps and Detection of QTLs Associated with Huanglongbing Tolerance in Citrus. Front. Plant Sci. 2018, 9, 1694. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, S.; Ali Wani, O.; Lone, J.K.; Manhas, S.; Kour, N.; Alam, P.; Ahmad, A.; Ahmad, P. Reactive Oxygen Species in Plants: From Source to Sink. Antioxidants 2022, 11, 225. [Google Scholar] [CrossRef] [PubMed]
- Janků, M.; Luhová, L.; Petřivalský, M. On the Origin and Fate of Reactive Oxygen Species in Plant Cell Compartments. Antioxidants 2019, 8, 105. [Google Scholar] [CrossRef] [PubMed]
- Decros, G.; Baldet, P.; Beauvoit, B.; Stevens, R.; Flandin, A.; Colombie, S.; Gibon, Y.; Pétriacq, P. Get the Balance Right: ROS Homeostasis and Redox Signalling in Fruit. Front. Plant Sci. 2019, 10, 1091. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Parvin, K.; Bardhan, K.; Nahar, K.; Anee, T.I.; Masud, A.A.; Fotopoulos, V. Biostimulants for the Regulation of Reactive Oxygen Species Metabolism in Plants under Abiotic Stress. Cells 2021, 10, 2537. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Lakhanpal, N.; Singh, K. Genome-wide identification and characterization of abiotic-stress responsive SOD (superoxide dismutase) gene family in Brassica juncea and B. rapa. BMC Genom. 2019, 20, 227. [Google Scholar] [CrossRef]
- Feng, X.; Lai, Z.; Lin, Y.; Lai, G.; Lian, C. Genome-wide identification and characterization of the superoxide dismutase gene family in Musa acuminata cv. Tianbaojiao (AAA group). BMC Genom. 2015, 16, 823. [Google Scholar] [CrossRef]
- Wang, W.; Xia, M.; Chen, J.; Deng, F.; Yuan, R.; Zhang, X.; Shen, F. Genome-wide analysis of superoxide dismutase gene family in Gossypium raimondii and G. arboreum. Plant Gene 2016, 6, 18–29. [Google Scholar] [CrossRef]
- Han, L.-M.; Hua, W.-P.; Cao, X.-Y.; Yan, J.-A.; Chen, C.; Wang, Z.-Z. Genome-wide identification and expression analysis of the superoxide dismutase (SOD) gene family in Salvia miltiorrhiza. Gene 2020, 742, 144603. [Google Scholar] [CrossRef] [PubMed]
- Cohu, C.M.; Abdel-Ghany, S.E.; Gogolin Reynolds, K.A.; Onofrio, A.M.; Bodecker, J.R.; Kimbrel, J.A.; Niyogi, K.K.; Pilon, M. Copper Delivery by the Copper Chaperone for Chloroplast and Cytosolic Copper/Zinc-Superoxide Dismutases: Regulation and Unexpected Phenotypes in an Arabidopsis Mutant. Mol. Plant 2009, 2, 1336–1350. [Google Scholar] [CrossRef] [PubMed]
- Shams, M.; Pokora, W.; Khadivi, A.; Aksmann, A. Superoxide dismutase in Arabidopsis and Chlamydomonas: Diversity, localization, regulation, and role. Plant Soil 2024. [Google Scholar] [CrossRef]
- Su, T.; Wang, P.; Li, H.; Zhao, Y.; Lu, Y.; Dai, P.; Ren, T.; Wang, X.; Li, X.; Shao, Q.; et al. The Arabidopsis catalase triple mutant reveals important roles of catalases and peroxisome-derived signaling in plant development. J. Integr. Plant Biol. 2018, 60, 591–607. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Mieulet, D.; Hubberten, H.-M.; Obata, T.; Hoefgen, R.; Fernie, A.R.; Fisahn, J.; San Segundo, B.; Guiderdoni, E.; Schippers, J.H.M.; et al. SALT-RESPONSIVE ERF1 Regulates Reactive Oxygen Species–Dependent Signaling during the Initial Response to Salt Stress in Rice. Plant Cell 2013, 25, 2115–2131. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Wei, Y.; Zhang, J.; Shao, W.; Li, J.; Ding, D.; Liu, M.; Xu, J. Bioinformatic analysis of catalase gene family of Arabidopsis and Maize. Mol. Plant Breed. 2021, 12, 34. [Google Scholar] [CrossRef]
- Willekens, H.; Villarroel, R.; Van Montagu, M.; Inzé, D.; Van Camp, W. Molecular identification of catalases from Nicotiana plumbaginifolia (L.). FEBS Lett. 1994, 352, 79–83. [Google Scholar] [CrossRef]
- Leng, X.; Wang, H.; Zhang, S.; Qu, C.; Yang, C.; Xu, Z.; Liu, G. Identification and Characterization of the APX Gene Family and Its Expression Pattern under Phytohormone Treatment and Abiotic Stress in Populus trichocarpa. Genes 2021, 12, 334. [Google Scholar] [CrossRef]
- Chaouch, S.; Queval, G.; Noctor, G. AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J. 2012, 69, 613–627. [Google Scholar] [CrossRef]
- Esparza-Reynoso, S.; Ávalos-Rangel, A.; Pelagio-Flores, R.; López-Bucio, J. Reactive oxygen species and NADPH oxidase-encoding genes underly the plant growth and developmental responses to Trichoderma. Protoplasma 2023, 260, 1257–1269. [Google Scholar] [CrossRef]
- Truskina, J.; Boeuf, S.; Renard, J.; Andersen, T.G.; Geldner, N.; Ingram, G. Anther development in Arabidopsis thaliana involves symplastic isolation and apoplastic gating of the tapetum-middle layer interface. Development 2022, 149, dev200596. [Google Scholar] [CrossRef] [PubMed]
- Nunes da Silva, M.; Vasconcelos, M.W.; Gaspar, M.; Balestra, G.M.; Mazzaglia, A.; Carvalho, S.M.P. Early Pathogen Recognition and Antioxidant System Activation Contributes to Actinidia arguta Tolerance against Pseudomonas syringae Pathovars actinidiae and actinidifoliorum. Front. Plant Sci. 2020, 11, 1022. [Google Scholar] [CrossRef] [PubMed]
- Azarabadi, S.; Abdollahi, H.; Torabi, M.; Salehi, Z.; Nasiri, J. ROS generation, oxidative burst and dynamic expression profiles of ROS-scavenging enzymes of superoxide dismutase (SOD), catalase (CAT) and ascorbate peroxidase (APX) in response to Erwinia amylovora in pear (Pyrus communis L.). Eur. J. Plant Pathol. 2017, 147, 279–294. [Google Scholar] [CrossRef]
- Singh, Y.; Nair, A.M.; Verma, P.K. Surviving the odds: From perception to survival of fungal phytopathogens under host-generated oxidative burst. Plant Commun. 2021, 2, 100142. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, Q.; Liu, T.; Liu, L.; Shen, D.; Zhu, Y.; Liu, P.; Zhou, J.-M.; Dou, D. Two Cytoplasmic Effectors of Phytophthora sojae Regulate Plant Cell Death via Interactions with Plant Catalases. Plant Physiol. 2015, 167, 164. [Google Scholar] [CrossRef]
- Yuan, H.; Jin, C.; Pei, H.; Zhao, L.; Li, X.; Li, J.; Huang, W.; Fan, R.; Liu, W.; Shen, Q.-H. The Powdery Mildew Effector CSEP0027 Interacts with Barley Catalase to Regulate Host Immunity. Front. Plant Sci. 2021, 12, 733237. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).