Abstract
Self-incompatibility (SI) systems in plants prevent self-pollination and mating among relatives, enhancing genetic diversity in nature but posing challenges in almond production and breeding. S-allele composition alongside the flowering periods of these cultivars enables the anticipation of cross-compatibility and optimal cultivar combinations for the allocation of pollinating trees in production. In the current study, 65 materials containing 61 almond (Prunus dulcis) germplasm resources, of which two were hybrids and the remaining four were peach (Prunus persica) germplasms, were used for the S-RNase genotype. The results showed that 55 genomic samples were amplified by PCR to obtain double-banded types, which identified their complete S-RNase genotypes, while the rest of the samples amplified only a single band, identifying one S-RNase gene in the S gene. A total of 30 S-RNase genes were identified in Prunus dulcis, Prunus webbii, Prunus persica, Prunus armeniaca, Prunus salicina, and Prunus cerasifera. Sequence analysis revealed polymorphisms spanning from 313 to 2031 bp within the amplified fragment sequence. The S57-RNase gene exhibited the highest frequency at 31.75% among the identified materials, with S1S57, S10S57, and S7S57 being the predominant S genotypes. A new S-RNase gene, named S65, was identified with a sequencing length of 1483 bp. Its deduced amino acid sequence shared 98.24% similarity with the amino acid sequence of the S-RNase gene on GenBank, with the highest homology. Furthermore, according to the findings, 65 materials belong to eight S genotype cross-incompatibility groups (CIG) and one semi-compatibility or compatibility group (0). Among them, most of the seven main almond germplasm resources and 35 cultivars can be cross-pollinated. The results of the study can lay the foundation for pollinator tree allocation and breeding hybrid parent selection in almond production.
1. Introduction
The almond [Prunus dulcis (Mill.) D.A. Webb] is a cultivated species classified within the almond group of the subgenus Amygdalus L. within the genus Prunus L. of the Rosaceae family. The almond is renowned globally as a dry fruit and as a woody oil-bearing tree species. The seed kernels of almonds hold a distinguished status as one of the four primary dry fruits worldwide, alongside walnut, hazelnut, and cashew. The almond group includes plants such as Prunus tangutica Batal., Prunus pedunculata Maxim., Prunus mongolica Maxim., Prunus ledebouriana Schleche., Prunus dulcis (Mill.) D.A. Webb (cultivated almonds), and Prunus triloba Lindl. []. Almonds are believed to have originated in the Eastern Mediterranean and Central Asian regions and were then distributed to other parts of the world. The Xinjiang region is the only main production area of almonds suitable for cultivation in China, with a cultivation history of more than 1300 years. Yarkant County, Yengisar County, and Shufu County in the Kashgar region of Xinjiang are mostly cultivated. The main cultivars are ‘Zhipi’, ‘Shuangguo’, ‘Shuangruan’, ‘Aifeng’, ‘Wanfeng’, ‘Yingzui’, and ‘Yeerqiang’. Most of these seven local cultivars are of the early-flowering, early- to mid-maturing type. As of the end of 2021, the planting area of almonds in Xinjiang reached 977,100 mu (=65,140 ha), yielding a total output of approximately 45,000 tons (seed kernels).
Almonds are cross-pollinated plants and are a typical gametophytic self-incompatibility (GSI) tree species. Cross-pollination is required during flowering to achieve a certain fruit-setting rate []. The failure of fruit setting after cross-pollination of two different varieties is a common phenomenon in almonds []. At present, the almonds used in cultivation and production in many central Asian regions are self-incompatible varieties. The unstable fruit-setting rate and low yield of almonds in production are caused by factors such as sandstorm weather during the flowering period, low temperatures in spring, and possible unreasonable allocation of pollinating trees, which hinder economic benefits from almond production. Among them, the scientific planning of pollination trees is closely related to the S genotype encoding self-incompatibility. The gametophytic self-incompatibility phenotype is controlled by multiple alleles (S-alleles) at a single locus (S-locus) containing at least two genes, the pistil S-gene (S-RNase) and the pollen S-gene (SFB/SLF) []. Mutations in the S-RNase gene of the style or SFB/SLF gene of the pollen can affect the mechanism of gamete incompatibility, resulting in the formation of self-compatible materials []. Under normal circumstances, when the S alleles of both parents are different, they pollinate each other, and the fruit-setting rate is higher, which is manifested as cross compatibility, and vice versa. When the parents have one of the same S genes, the fruit-setting rate is low, which is manifested as semi-cross-compatibility []. Therefore, the identification of the germplasm S genotype is very important for the allocation of pollinating trees in production and the design of effective cross combinations in breeding. In the past, electron microscopy was used to observe the growth of pollen tubes after pollination and to count the fruit-setting rate of ripe fruits for determining the difference in the S genotype, but this method was time-consuming, laborious, and inaccurate. At present, genomic DNA extraction, sequence amplification, and sequencing based on S gene fragments have become mainstream technologies for determining the S genotype of materials, and are more accurate, which can efficiently clarify the S genotype of research materials. He et al. [] identified 84 S genotypes of native Chinese pear accessions by allele-specific PCR using one pair of consensus primers and 29 pairs of S-allele-specific primers, including 49 new pear varieties (P. pyrifolia, P. sinkiangensis, and inter-specific hybrids) and 35 wild pear varieties (P. ussuriensis, P. pseudopashia, P. betulifolia, P. calleryana, and landraces). Jiang et al. [] used three pairs of Rosaceae universal primer combinations for PCR amplification of specific alleles of leaf genomic DNA of 27 apricot varieties to obtain four registered S-RNase genes of the genus Apricot, as well as 15 new S-RNase genes. Liu et al. [] identified the S genotype of 88 Malus germplasm resources using seven pairs of S-RNase gene-specific primers, and the results showed that 70 materials obtained complete S-RNase genotypes.
In terms of almonds, more than 180 almond cultivars have been identified for S gene research in foreign countries [,]. Researchers grouped varieties or germplasms with the same S genotype into a hybrid incompatibility group (CIG), and currently 48 CIGs have been established [,]. Chinese researchers have also conducted related studies on the identification of the S genotype of almonds. Ma et al. [] designed primers based on the conserved sequences of the S gene of Rosaceae species and amplified 10 S gene fragments of different sizes from the genome of eight Xinjiang almond varieties. Jiang et al. [] identified the S genotype of 10 almond varieties using two pairs of universal primers of Rosaceae. Guo et al. [] PCR-amplified almond varieties with multiple pairs of universal primers from Rosaceae and identified the S genotypes of 15 materials.
As one of the origin centers of almonds in the world, Xinjiang’s topography and climate are very suitable for the growth of almonds. There is abundant phenotypic diversity of almond germplasm resources in the woodland. At present, research on the S genotype of almonds and close relatives in China is not comprehensive. Although the S genotypes of some germplasms have been determined, this has been mainly limited to more than ten local cultivars and no CIG has been established for domestic almonds. More materials for almonds and close relatives need to be identified and systematically analyzed for the S genotype. The phenomenon of the same variety name but completely different genetic backgrounds in almond production requires further clarification and identification. Therefore, in this study, PCR combined with DNA sequencing technology was used to identify the S-RNase genotypes of 65 almond germplasm resources and close relatives in Xinjiang, establish an almond CIG in China for the first time, and analyze the compatibility between seven main almond cultivars and 35 cultivars in Xinjiang. This will provide a reference for enriching the S genotype and genetic information of almonds, studying the self-incompatibility mechanism, selecting suitable pollinating trees in production, designing new hybrid combinations, and breeding new cultivars.
2. Materials and Methods
2.1. Materials
A total of 65 materials were used in the current study, which includes 61 almond (Prunus dulcis) germplasm resources such as Prunus communis Fritsch, Prunus triloba Lindl., Prunus Mongolica Maxim, and almond × peach natural hybrids available in Xinjiang, China. Additionally, 4 peach (Prunus persica) germplasms, including Prunus ferganensis Kost. et Riab and Prunus davidiana Franch. Most of the materials (62) were collected from the almond germplasm resource garden in Yarkant county, Xinjiang, China, while 3 Jinbian varieties (‘Jinbian 1’, ‘Jinbian 2’, and ‘Jinbian 3’) were obtained from Shanxi Agricultural University. Tender leaves of the mentioned materials were collected in spring 2022, frozen in liquid nitrogen, and stored in a −80 °C refrigerator for future use.
2.2. Extraction of Leaf Genomic DNA
Leaf genomic DNA was extracted using a plant genomic DNA rapid extraction kit (Aidlab Biotechnologies Co.,Ltd., Beijing, China). An appropriate amount of genomic DNA was taken to test its quality and concentration. After 1.5% agarose gel electrophoresis and nucleic acid protein analyzer detection, it was stored at −80 °C for standby.
2.3. Primer
S-RNase genotype identification was performed using Rosaceae consensus primer pairs 1 to 3 (with minor adjustments), as reported in the literature [,,,]. For materials that did not amplify or only amplified a single band, primers C1-F and C5-R were designed using Primer Premier 5.0 based on the highly conserved amino acid sequence of the S-RNase gene of Prunus L. in the Rosaceae family. In addition, primer pair 5 “PaConsI-F/C5-R” and primer pair 6 “C1-F/PruC4R” were combined for further identification (Table 1).

Table 1.
Primers used for S-RNase genotype identification and sequences.
2.4. PCR Amplification of the S-RNase Gene
The leaf genomic DNA was used as a template to amplify the S-RNase allele of the test genotypes. The reaction volume was 25 μL and contained 12.5 μL of 2×Taq Master Mix (Vazyme, Nanjing, China), 1 μL of primers (10 μmol/L), 1 μL of DNA template (100 ng), and 9.5 μL of RNase-free ddH2O. Amplification was performed in the Mastercycler nexus GSX1 (Eppendorf, Germany) gradient PCR instrument using the following procedure: predenaturation at 95 °C for 3 min; denaturation at 95 °C for 30 s, annealing at 53 °C for 1 min, extension at 72 °C for 1 min, 32 cycles; finally, extension at 72 °C for 5 min and storage at 4 °C. The amplified product was detected in 1.5% agarose gel in 1×TAE buffer, and the stained gel was photographed under UV light with a UVitec gel recorder.
2.5. Cloning and Sequencing of the S-RNase Gene
After the PCR expansion product was imaged by 1.5% agarose electrophoresis and gel, the purified product was ligated to the pCE2 TA/Blunt-Zero vector, the recombinant vector was transferred to E. coli DH5α-competent cells, and the positive clones were sequenced at least three times to confirm the correct sequence.
2.6. Sequence Analysis
Sample sequencing was conducted by the Zhengzhou Branch of Shenggong Biotechnology (Shanghai) Co., Ltd. (Shanghai, China) using the ABI 3730XL gene sequencer. The process involved utilizing Bio XM 2.7 to eliminate vector sequences, Chromas for correction, SeqManII for integrating three duplicate sequencing results, and finally, employing the Blast program to conduct homology sequence retrieval with the S-RNase sequences already registered in the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 19 April 2024).
Nucleotide and amino acid sequence analysis of the S-RNase gene was carried out using DNAMAN software (Version V6; Lynnon Biosoft, San Ramon, CA, USA). If the amino acid sequence consistency reaches 99%, it is considered to be the same gene. Conversely, if there is inconsistency, it is regarded as a new S-RNase gene. This process aids in determining the S-RNase allele information of the test material.
2.7. Construction of the Phylogenetic Tree of the S Gene
The amino acid sequence of Prunus L. S-RNase was downloaded from the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/, accessed on 19 April 2024). With MEGA 11 software, the Clustal W method was used to conduct multiple sequence comparisons between the S-RNase obtained from the experiment and the downloaded amino acid sequence, and the phylogenetic tree was constructed using the neighbor-joining (NJ) method. The bootstrap values were kept 1000 times.
2.8. Construction of a Database of Hybridization Combinations
According to the principle of S genotype incompatibility, if the hybrid parent shares the same S genotype, the hybrid is incompatible. If the S genotype of the hybrid parent contains one identical S gene, the hybrid is semi-compatible. When the S genotype of the hybrid parent is completely different, the hybrid is compatible. A database of almond hybrid incompatibility can be constructed based on this principle. Additionally, an analysis of hybrid incompatibility/compatibility between seven main almond varieties planted in Xinjiang and 35 other varieties was conducted. This analysis provides a reference basis for selecting parents in production and breeding.
2.9. Field Validation of Cross-Compatibility of Some Almond Cultivars
From late March to early April 2022–2023, according to the hybrid combination in Table 2, the balloon-stage flower buds were collected, the anthers were peeled, dried in the shade, completely pollinated, and then loaded into dry small tubes, which were placed at −20 °C for later use. No less than 300 large bud-stage flowers were selected for pollination in each combination. To improve the accuracy of the results, the unopened and already blooming flowers on the hybrid flower branches were removed, and finally, the hybrid branches were bagged. The bagging was removed after 2 weeks of pollination, and the number of fruits were counted two months later.

Table 2.
Cross-pollination and fruit setting of some almond cultivars in 2022–2023.
According to Audergon et al. [], a fruit-setting rate higher than 5% indicates cross-compatibility, whereas a rate lower than 5% suggests cross-incompatibility. This criterion is utilized to determine the cross-compatibility between different almond cultivars.
3. Results
3.1. Identification of S-RNase Alleles
PCR amplification of leaf DNA using consensus primers and self-designed degenerate primers showed that two different fragments were detected in 55 samples, and only one fragment was amplified in the remaining 10 samples (Figure 1). The size of the product fragments showed a certain degree of polymorphism, and the sequencing results showed that the sequence fragments ranged from 313 to 2031 bp. Akram et al. [] also showed that the size of S-RNase allele fragments in 22 Iranian almond cultivars ranged from 600 to 2400 bp. The sequences and sizes of introns in different S genes were different and showed high differences. The coding region of each S-RNase was almost separated by introns at the same position. Therefore, it causes high variability in fragment sizes.
After multiple sequence alignment of all nucleotide sequences obtained through DNAMAN, it was found that the bands of different varieties located at the same position have more than 99% similarity in the nucleic acid sequence, which can be considered as the same gene fragment. A total of 30 nucleotide sequences of different sizes were isolated from 65 germplasm: 313 bp (‘Jinbian 3’), 347 bp (‘XX3’), 350 bp (Prunus triloba), 408 bp (‘Yeerqiang holy Fruit’), 419 bp (‘Jinbian 3’, etc.), 591 bp (‘Amannisha’, etc.), 621 bp (Prunus triloba), 624 bp (‘Paizhui’), 653 bp (‘Aifeng’, etc.), 722 bp (Prunus mongolica), 754 bp (‘Badanwang’, etc.), 772 bp (‘Shuangruan’, etc.), 797 bp (‘Shache 29’), 797 bp (‘Shuangruan’, etc.), 811 bp (‘Dabadan’, etc.), 821 bp (‘Wanhua Taobadan’), 822 bp (‘Jinbian 2’, etc.), 857 bp (‘Jinsha Fruit’, etc.), 982 bp (‘Hanfeng’), 993 bp (‘Jinbian 2’), 994 bp (‘Baishuang’, etc.), 1086 bp (‘A1’, etc.), 1130 bp (‘Tianren Taobadan’, etc.), 1254 bp (‘Yuanruan’, etc.), 1274 bp (‘Baiboke’, etc.), 1483 bp (‘Shuangbo’, etc.), 1592 bp (‘Shache 29’, etc.), 1634 bp (‘Huangshuang’, etc.), 1650 bp (‘Shuangguo’, etc.), and 2031 bp (‘Shitou Almond’, etc.).
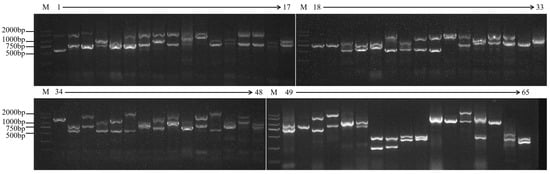


Figure 1.
S-RNase gene amplification in 65 almond germplasm resources and close relatives. M: 2000 bp marker; The materials numbered 1–65 are shown in Table 3; Test materials 1–48 were amplified using primer pair 1, test materials 49–54, 60 using primer pair 4, test materials 55–59, 65 using primer pair 2, test materials 63–64 using primer pair 3, test material 61 using primer pair 5, and test material 62 using primer pair 6.

Table 3.
S-RNase genotypes from 65 Xinjiang almond germplasms and close relatives.
Table 3.
S-RNase genotypes from 65 Xinjiang almond germplasms and close relatives.
| No. | Sample | Fragment Length/bp | Genotype | No. | Sample | Fragment Length/bp | Genotype |
|---|---|---|---|---|---|---|---|
| 1 | Shuangbo | 591/1483 | S5S65 | 34 | Kexi | 1274 | S54 |
| 2 | Huangshuang | 1634/811 | S22S57 | 35 | Yeerqiang | 591/811 | S5S57 |
| 3 | Shitou Almond | 2031/811 | S7S57 | 36 | Shache 29 | 1592/797 | S1S55 |
| 4 | Ku 1 | 977/747 | S10S63 | 37 | A1 | 591/1086 | S5S16 |
| 5 | Ydl 1 | 977/811 | S10S57 | 38 | Tianren Taobadan | 591/1130 | S5S24 |
| 6 | Shache 28 | 754/1650 | S6S35 | 39 | Jianzuihuang | 591/2031 | S5S7 |
| 7 | Jinsha Fruit × XX3 | 1592/1086 | S1S16 | 40 | Badanwang | 591/754 | S5S6 |
| 8 | Dabadan × XX3 | 1592/811 | S1S57 | 41 | Baiboke | 653/1274 | Sn3S54 |
| 9 | Taobadan | 1592/811 | S1S57 | 42 | Aitelaisi Badan | 977/1634 | S10S22 |
| 10 | Xiaokuren Taobadan | 1592/1086 | S1S16 | 43 | Paizhui | 624/1086 | SgS16 |
| 11 | Threeling Taobadan | 1592/1130 | S1S24 | 44 | Wanhua Taobadan | 1592/821 | S1S2 |
| 12 | Badanxing | 994/747 | S23S63 | 45 | Wanfeng | 591/2031 | S5S7 |
| 13 | Shache 79 | 977/811 | S10S57 | 46 | Aifeng | 653/994 | Sn3S23 |
| 14 | Xinjiang Peach 1 | 1592/821 | S1S2 | 47 | XX1 | 1592/1086 | S1S16 |
| 15 | Arele Taobadan | 1592/811 | S1S57 | 48 | Houke Taobadan | 977/811 | S10S57 |
| 10 | Ydl 2 | 977/811 | S10S57 | 49 | Zhipi | 632/811 | Sn3S57 |
| 17 | Jinsha Fruit | 857/1086 | S6S16 | 50 | Prunus mongolica | 722 | S3 |
| 18 | Duoguo | 747 | S63 | 51 | Yingzui | 632/1576 | Sn3S1 |
| 19 | Dabadan | 811 | S57 | 52 | XX2 | 2021/811 | S7S57 |
| 20 | Amannisha | 591/977 | S5S10 | 53 | Shuangruan | 797/772 | Sn5S63 |
| 21 | Gongbadan | 591/811 | S5S57 | 54 | Jinbian 2 | 822/993 | S49Sf |
| 22 | Xiaoshuangren | 1086/811 | S16S57 | 55 | Ku Taobadan | 455/190 | S2S57 |
| 23 | Shuangguo | 591/1650 | S5S35 | 56 | XX3 | 200/347 | S1S27 |
| 24 | Kubadan | 591/977 | S5S10 | 57 | Jinbian 1 | 335/419 | S49Sf′ |
| 25 | Baichang Badan | 591/1274 | S5S54 | 58 | Yeerqiang holy Fruit | 355/408 | S7S8 |
| 26 | Shache 27 | 1592/591 | S1S5 | 59 | Yuanruan | 1254 | S10 |
| 27 | Xinjiang Peach 2 | 1592 | S1 | 60 | Make | 1212 | S24 |
| 28 | Tianrenxiao Taobadan | 1592/811 | S1S57 | 61 | Shache 48 | 1967/1008 | S7S57 |
| 29 | Taohuaguazi Badan | 977/1130 | S10S24 | 62 | Prunus davidiana | 1231/375 | S1S57 |
| 30 | Xinjiang Peach 3 | 1592/977 | S1S10 | 63 | Hanfeng | 982 | S61 |
| 31 | Guazi Badan | 811/1483 | S57S65 | 64 | Prunus triloba | 350/621 | S7S11 |
| 32 | Changying | 811 | S57 | 65 | Jinbian 3 | 313/419 | Sn1Sf′ |
| 33 | Baishuang | 994 | S23 |
Note: The underline indicates that the gene is not Prunus dulcis S-RNase.
Homology sequence retrieval was performed on 30 nucleotide sequences of different fragment sizes in NCBI, and the results showed that the nucleotide sequences of the S-RNase genes highly homologous to the sequencing sequence were all S-RNase genes of the Prunus L. in the Rosaceae family, including 20 Prunus dulcis-style S gene sequences. Prunus webbii, Prunus persica, Prunus armeniaca, Prunus salicina, and Prunus cerasifera sequences were 4, 3, 1, 1, and 1, respectively (Table 4).

Table 4.
Comparison results of 30 gene fragments in GenBank.
For nucleotide sequences with different fragment sizes, the amino acid sequences of nucleotide sequences with various fragment sizes were deduced by removing introns, utilizing the cDNA of the S-RNase gene with the highest homology found on GenBank. The amino acid sequences of the S genes with the highest homology rate in GenBank were compared for consistency by DNAMAN (V6) software. The results showed that the deduced amino acid sequences with amplified fragment sizes of 1483 bp and 1634 bp differed from the amino acid sequences of Prunus armeniaca S2 (AY587562.1) and Prunus dulcis S22 (AM231671.1) by three and two amino acids, respectively, with consistencies of 98.24% and 98.57%, respectively. Among them, a sequence of 1483 bp was identified as a new S-RNase gene named S65. The amino acid sequences of 722 bp, 772 bp, 1592 bp, and 1650 bp had consistencies of over 99% with the gene of the highest homology (the only difference in amino acid sequence). The remaining fragments were 100% consistent with the amino acid sequence of the gene with the highest homology. Based on the above results, the S-RNase genotypes of the test materials were determined (Table 3).
3.2. Structural Analysis of the Amino Acid Sequences of Different S-RNase Genes
Except for the sequence regions of Sf’, S8, S27, and Sn1 alleles obtained from the signal peptide region to the C1 region, when comparing the deduced amino acid sequence structures, it was found that the amino acid sequences of 25 registered S-RNases and one new S-RNase all exhibit typical structural features of the S-RNase of Prunus L. Downstream of the N-terminal SP of the sequence are five conserved regions, namely C1, C2, C3, RC4, and C5. There is a high-variability region (RHV) between C2 and C3, as well as two histidine residues necessary for RNase activity and four conserved cysteine residues (Figure 2).
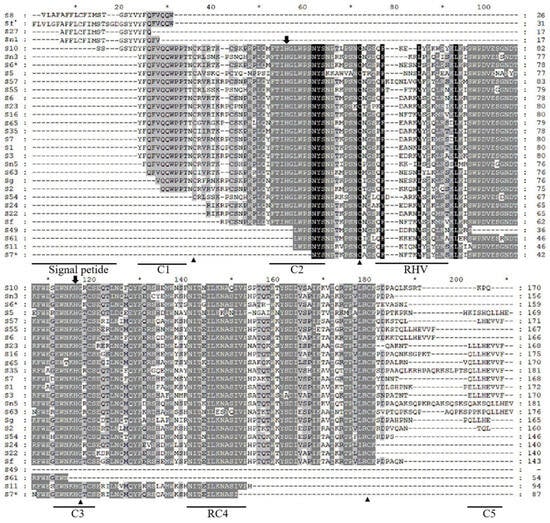
Figure 2.
Amino acid sequence structures of 30 S-RNase genes. Conserved residues are located in the black area and dashes indicate gaps. The asterisk represents a difference of 10 residues from the previous number. The signal peptide region (SP), conserved regions (C1, C2, C3, RC4, and C5), and the hypervariable region (RHV) are underlined. The two histidine residues essential for RNase activity are labeled with the symbol ↓, and the four conserved cysteine residues are labeled with the symbol ▲. To distinguish the two S6s, S6* stands for Prunus webbii S6.
3.3. Phylogenetic Analysis of the S Gene in Rosaceae Prunoideae
We removed four shorter gene amino acid sequences that cannot meet the requirements for constructing a phylogenetic tree, including Sf′, S8, S27, and Sn1. The neighbor-joining (NJ) feature of MEGA 11 software was used to obtain 26 S-RNase gene amino acid sequences, combined with 19 registered S-RNase gene amino acid sequences of Prunus webbii, Prunus salicina, Prunus armeniaca, Prunus avium, Prunus simonii, and Prunus mume in the NCBI database to construct a phylogenetic tree of Prunoideae (Figure 3). Cluster analysis showed that 25 identified S-RNase genes and one new almond S-RNase gene clustered to varying degrees with 19 registered plum genes in six branches of the evolutionary tree, and their evolutionary relationships with Prunus armeniaca, Prunus webbii, and Prunus avium were relatively close in different branches. This result is consistent with the research results of Jiang et al. []. In addition, most of the S-RNase genes obtained from the experiment have close genetic relationships.
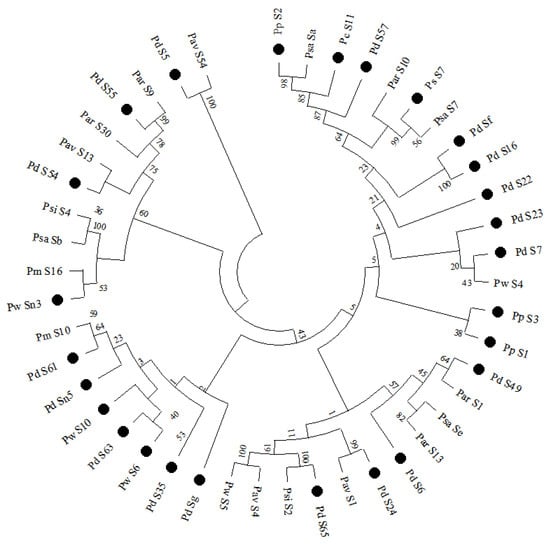
Figure 3.
Phylogenetic tree of 50 S-RNase genes in the Prunus L. genus. Pd: Prunus dulcis; Pw: Prunus webbii; Pp: Prunus persica; Pc: Prunus cerasifera; Psa: Prunus salicina; Par: Prunus armeniaca; Pav: Prunus avium; Psi: Prunus simonii; Pm: Prunus mume. Those marked with circles are the S-RNase genes in the test.
3.4. Analysis of S-RNase Genes and Gene Frequencies
Among the 30 S-RNase gene fragments amplified from 65 germplasm resources, the highest gene frequency of S57-RNase was 31.75%, which was amplified from 20 cultivars. The frequencies of S1-, S5-, S7-, S10-, and S16-RNase are relatively high, ranging from 9.52% to 23.81%. The gene frequency of the remaining S-RNases ranged from 1.59% to 6.35%, appearing 1–4 times (Figure 4). The results also showed that high-frequency genes were commonly present in the seven main almond cultivars in Xinjiang, including ‘Zhipi’ Sn3S57, ‘Shuangguo’ S5S35, ‘Shuangruan’ Sn5S63, ‘Aifeng’ Sn3S23, ‘Wanfeng’ S5S7, ‘Yingzui’ Sn3S1, and ‘Yeerqiang’ S5S57. This resulted in different cultivars having the same S-RNase gene or even the same S genotype, which may be due to long-term natural selection or purposeful breeding by humans.
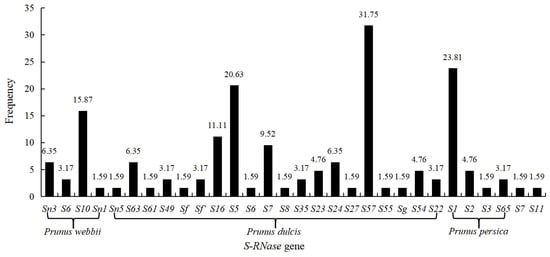
Figure 4.
Gene frequencies of 30 S-RNase genes in the germplasm resources.
3.5. Establishment of Almond Hybrid Incompatibility Groups
In this study, the S genotype results of 55 out of 65 materials were successfully identified. Based on the theory that materials with the same S genotype are incompatible for cross-pollination, and there is semi-compatibility or compatibility for materials with one identical S-RNase gene [], eight CIGs (I-VIII) and one semi-compatibility or compatibility group (0) were initially established (Table 5). Additionally, cross-incompatibility/compatibility between seven main cultivars of Xinjiang almond and 35 other cultivars was analyzed. The results showed that most varieties could pollinate each other, with only a few showing cross-incompatibility (Figure 5). In the later stages, further verification was needed through cross-pollination in the field.

Table 5.
Identification results of almond cross-incompatibility groups and their S genotypes.
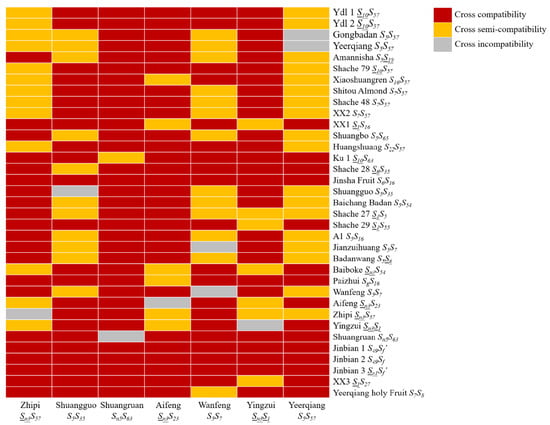
Figure 5.
Heat map of cross-(non)compatibility between 7 local almond cultivars of the Xinjiang region and different almond cultivars.
3.6. Field Identification and Analysis of Some Almond Cultivars
Among the eight hybrid combinations from 2022 to 2023, all exhibited fruit-setting rates higher than 5%. Notably, the combination ‘Jinsha Fruit’ × ‘Taohuaguazi Badan’ yielded the highest fruit-setting rate at 29.70% (Table 2). Following the criteria outlined by Audergon et al. [], all eight combinations demonstrated cross-compatibility. The identification results of parental genotypes in each cross-combination revealed that the S genotype of the parents differed across combinations, and these identifications were consistent with field observations.
4. Discussion
4.1. Analysis of the Results of S-RNase Allele Identification
PCR amplification combined with DNA sequencing has been widely used to identify S-RNase alleles in almonds and other Prunus L. plants [,,]. It is only necessary to collect tissues such as the young leaves of plants, without long-term pollination observation. The S1S57 and S1S16 genotypes of hybrid offspring “Dabadan × XX3” and “Jinsha Fruit × XX3”, as well as the genotypes of their parents ‘Dabadan’ S57, ‘Jinsha Fruit’ (S6S16), and ‘XX3’ (S1S27), conform to Mendelian genetic inheritance rules, which to some extent indicates the reliability of this identification method. In addition, the field verification results of eight different hybrid combinations in this study also supported the identification of S genotypes, that is, the fruit-setting rates of all combinations were higher than 5%, and the S-RNase genotypes were different among the hybrid parents. Researchers conducted pollination experiments on some Xinjiang almond varieties, pointing out that when the flowering periods of ‘Zhipi’, ‘Shuangguo’, and ‘Shuangruan’ meet, they can become pollinating trees for each other []. In this study, the S-RNase genotypes of these three cultivars were different. In addition, a pollination experiment was conducted using ‘Zhipi’ and ‘Yeerqiang’ as male parents, and the results showed that ‘Zhipi’ had good fruit-setting rates with female parents ‘Wanfeng’, ‘Shuangguo’, ‘Shuangbo’, and ‘Amannisha’, while ‘Yeerqiang’ had better fruit-setting rates with female parents ‘Zhipi’, ‘Shuangguo’, ‘Shuangbo’, and ‘Shuangruan’ []. In this study, the S-RNase genotype of ‘Zhipi’ was completely different from the above maternal genotype, and the S-RNase genotype of ‘Yeerqiang’ had one identical S-RNase gene as the above maternal parent, which also showed that the identification method was accurate and reliable. In this study, the S-RNase genotypes of 85% of the materials were successfully identified using the consensus primers of Rosaceae and degeneracy primers designed on the basis of the conserved amino acid sequence of the S-allele. The electrophoretic map of ‘Shuangruan’ showed a single band, but the cloning and sequencing results revealed two different S-RNases with the genotype Sn5S63 (797/772). For the 10 materials that only amplified single bands, it is speculated that the reason may be due to large introns in the target fragment causing amplification failure, or that the two S-RNase allele fragments of the cultivars were too close, and another S-RNase gene was not connected during cloning and sequencing. It is also possible that the variety carried a homozygous S-RNase gene, necessitating the use of specific primers for identification.
The nucleotide sequences of the 30 S-RNase genes obtained by amplification had 95.95~100% homology with the S-RNase genes registered in GenBank. The consistency of amino acid sequences was analyzed according to the GT/AG conserved features of the intron/exon splice site. The amino acid sequences of 29 S-RNase genes were more than 99% consistent with that of homologous genes, while the amino acid sequence of the 1483 bp amplified fragment was three amino acids different from that of homologous genes, and was identified as a new S-RNase gene named S65. The discovery of the new S-RNase gene will help to expand the diversity of the local almond gene information database and enrich the hybrid parental material. At the same time, studies have shown that for SI populations, pollens carrying the new S gene are more likely to be successfully fertilized, thereby increasing their population size [].
In the current research, it was found that Taobadan is a natural hybrid of almonds and peaches in Xinjiang, China [], and was integrated into Prunus persica S1 (AB252415.1) in the S genotype of most Taobadan resources. Although the names of the resources are similar, the S genotype is not completely consistent, indicating that the genetic background of the materials is different. In practical production applications, it is necessary to understand the S genotype of the materials. In addition, the identification results of the genotypes of ‘Zhipi’ and ‘Yeerqiang’ were Sn3S57 and S5S57, respectively, while previous research results were S50S61 and S15S16 []. Sequence analysis showed that the nucleotide sequence consistency between Prunus dulcis cultivar S50 RNase (AY613337) and Sn3, and Prunus dulcis S15 (AM231664.1) and S5, was 98.92% and 99.33%, respectively. When the S genotype identification results of the same variety are different, it is often due to sampling error caused by different species having the same name, or the different ways researchers evaluate the sequencing results. Ortega et al. [] proposed that the sequences of the Prunus dulcis alleles S5 and S15 are identical, as are the sequences of S4 and S20 and S13 and S19. In the early studies, the researchers repeated the naming due to a lack of unified regulations and lack of communication. If the identification results are wrong, it will not only lead to the occurrence of different varieties with the same name or different names for the same variety, which will affect the evaluation and utilization of varieties and the research process but will also mislead production and utilization, resulting in serious economic losses. It can be seen that the identification of genotypes needs to be repeatedly compared and verified to be error-free before the accuracy of S-RNase allele identification and naming results can be guaranteed.
4.2. Analysis of the S Genotype
In the constructed phylogenetic tree, the S-RNase genes of Prunus dulcis, Prunus salicina, Prunus armeniaca, Prunus avium, Prunus webbii, Prunus simonii, Prunus mume, and Prunus cerasifera did not cluster into a single subclass, nor did they show intra-specific similarity higher than inter-specific similarity. The evolution of S-RNase genes may be out of sync with the development of plant systems within Prunoideae. In this study, the consistency between the ‘Baiboke’ 653 bp sequence, Prunus webbii Sn3 (AM690354.1), the Prunus mongolica 722 bp sequence, and the Prunus persica S3 (AB537563.1) amino acid sequence is higher than 99%. This suggests that the S-RNase gene has evolved across species. These findings align with previous studies indicating that the inter-specific homology of S-RNase genes in Rosaceae is often greater than intra-specific homology []. The evolutionary tree reveals relatively close phylogenetic relationships between tree species, particularly evident in the proximity of Prunus dulcis, Prunus webbii, and Prunus avium. Correspondingly, the genetic relationship among S-RNase genes obtained from experiments is also relatively close, narrowing the genetic range of almonds and potentially hindering the inheritance of S genes in crossbreeding parents. Therefore, it is imperative to identify the S genotype of almond germplasm resources and update the information base to broaden the genetic background of almonds, identify excellent genes, and provide precise guidance continuously and accurately for practical production.
The results of the pollination test performed by Heng et al. [] showed that both self-pollination and cross-pollination among varieties of the same S genotype produced low fruiting rates (0%), except ‘Hongding’ which yielded 0.64%. Further, the results of Liang [] showed that there was one identical S-RNase gene. The style S genotype of the two varieties showed semi-affinity, and the offspring only inherited 1/2 of the pollen of the parents. When the parent S genotype is completely different, four genotypes of offspring can be produced. Similarly, foreign researchers have placed varieties with the same S genotype in a cross-incompatibility group (CIG) based on this characteristic, and currently, 48 CIGs have been established [,]. Based on the identification results, this study established, for the first time, eight almond CIGs (I-VIII) and one semi-compatible or compatible group (0) in China. Materials within the same CIG group should be excluded from almond production and hybrid breeding. The cross-(in)compatibility between seven main almond cultivars and 35 other cultivars in Xinjiang was analyzed, and the heat map showed that most of the varieties could pollinate each other, with only a few showing hybridization incompatibilities. Further field cross-pollination is needed to verify this later. Understanding the affinity relationship between CIG and materials is very necessary for breeding programs. This understanding can provide a reference for enriching the genetic information of varieties, studying the mechanism of self-incompatibility, selecting suitable pollination trees, designing new hybrid combinations, and selecting and breeding new cultivars.
5. Conclusions
This study utilized 65 germplasm resources from 61 almond and four peach germplasms to investigate the genotype composition of S-RNase and its impact on the cross-compatibility of almond cultivars. Through PCR-based amplification and sequence analysis, 55 of the identified materials obtained a complete S-RNase genotype, while only one S-RNase gene was found in 10 of them. A total of 30 S-RNase genes were identified, including the discovery of a new S-RNase named S65. Furthermore, this study classified the materials into eight S-genotype cross-incompatibility groups (CIG) and one semi-compatibility or compatibility group (0). The research results will contribute to pollinator tree allocation and breeding hybrid parent selection in almond production, providing valuable insights into the potential cross-pollination compatibility among almond cultivars and providing practical applications for improving almond production and breeding strategies.
Author Contributions
Conceptualization, P.X. and M.A.; methodology, P.X. and X.W.; software, Y.A.; investigation, Y.X.; resources, L.W. and M.A.; data curation, Y.A.; writing—original draft preparation, P.X.; writing—review and editing, P.X. and X.W.; supervision, L.W.; project administration, Y.X.; funding acquisition, P.X., Y.X., C.G. and M.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Youth Science and Technology Backbone Project of Xinjiang Academy of Agricultural Sciences, grant number xjnkq-2023009; Natural Science Foundation of Xinjiang Uygur Autonomous Region Sponsored Programs, grant number 2022D01A90; National Region Fund Science Project of China, grant number 32260738; Central Government Guide Local Science and Technology development special fund project “Characteristic Fruit Tree Germplasm Innovation and Breeding Capacity Enhancement”; Basic Research Business of Public Welfare Research Institutes in Xinjiang Uygur Autonomous Region, grant number KY2022039.
Data Availability Statement
The datasets supporting the results presented in this manuscript are included within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Wang, L. Chinese Peach Genetic Resources, 1st ed.; China Agriculture Press: Beijing, China, 2012. [Google Scholar]
- Martinez-Gomez, P.; Sanchez-Perez, R.; Dicenta, F.; Howard, W.; Arus, P.; Gradziel, T.M. Fruits and Nuts. In Genome Mapping and Molecular Breeding in Plants, 1st ed.; Kole, C., Ed.; Springer: Berlin, Germany, 2007; Volume 3, pp. 229–242. [Google Scholar]
- Boskovic, R.; Tobutt, K.R.; Batlle, I.; Duval, H.; Martinez-Gomez, P.; Gradziel, T.M. Stylar ribonucleases in almond: Correlation with and prediction of incompatibility genotypes. Plant Breed. 2003, 122, 70–76. [Google Scholar] [CrossRef]
- He, M.; Gu, C.; Wu, J.Y.; Zhang, S.L. Recent advances on self-incompatibility mechanism in fruit trees. Acta Hortic. Sin. 2021, 48, 759–777. [Google Scholar]
- Tao, R.; Watari, A.; Hanada, T.; Habu, T.; Yaegaki, H.; Yamaguchi, M.; Yamane, H. Self-compatible peach (Prunus persica) has mutant versions of the S haplotypes found in self-incompatible Prunus species. Plant Mol. Biol. 2007, 63, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Liu, M. Study on Pollination Characteristic and S Genotype Identification of Self-Incompatibility of Kernel-Apricot. Ph.D. Thesis, Chinese Academy of Forestry, Beijing, China, 2015. [Google Scholar]
- He, M.; Li, L.; Xu, Y.; Mu, J.; Xie, Z.; Gu, C.; Zhang, S. Identification of S-genotypes and a novel S-RNase in 84 native Chinese pear accessions. Hortic. Plant J. 2022, 8, 713–726. [Google Scholar] [CrossRef]
- Jiang, X.; Cao, X.; Wang, D.; Feng, J.; Liu, Y.; Fan, X. Identification of self-incompatibility S-RNase genotypes for apricot cultivars in South of Xinjiang area. J. Fruit Sci. 2012, 29, 569–576. [Google Scholar]
- Liu, Z.; Gao, Y.; Wang, K.; Feng, J.; Sun, S.; Lu, X.; Wang, L.; Tian, W.; Wang, G.; Li, Z.; et al. Identification of S-RNase genotype and analysis of its origin and evolutionary patterns in Malus plants. J. Integr. Agric. 2024, 23, 1205–1221. [Google Scholar] [CrossRef]
- Lopez, M.; Vargas, F.J.; Batlle, I. Self-(in)compatibility almond genotypes: A review. Euphytica 2006, 150, 1–16. [Google Scholar] [CrossRef]
- Kodad, O.; Alonso, J.M.; Sanchez, A.; Oliveira, M.; Company, R.S.I. Evaluation of genetic diversity of S-alleles in an almond germplasm collection. J. Hortic. Sci. Biotech. 2008, 83, 603–608. [Google Scholar] [CrossRef]
- Akram, H.; Behrouz, S.; Bahram, M.; Ali, I.; Bojana, B. Identification of new S-RNase self-incompatibility alleles and characterization of natural mutations in Iranian almond cultivars. Trees 2013, 27, 497–510. [Google Scholar]
- Gomez, E.M.; Dicenta, F.; Batlle, I.; Romero, A.; Ortega, E. Significance of S-genotype determination in the conservation of genetic resources and breeding of almond. Ciheam 2016, 119, 83–86. [Google Scholar]
- Ma, Y.; Ma, R. Analysis on polymorphism of S gene in almond. Acta Hortic. Sin. 2006, 33, 137–139. [Google Scholar]
- Jiang, J.; Feng, J.; Cao, X.; Li, W.; Wang, X.; Chen, L. Identification of self-Incompatibility S-RNase genotypes for almond cultivars in Xinjiang. Xinjiang Agric. Sci. 2011, 48, 1177–1182. [Google Scholar]
- Guo, C.; Yang, B.; Tudi, M.; Tang, Y.; Li, N.; Wang, J.; Gong, P. Comparison of amplification effects on almond resources in China using different primer combinations. Xinjiang Agric. Sci. 2016, 53, 641–646. [Google Scholar]
- Tamura, M.; Ushijima, K.; Sassa, H.; Hirano, H.; Tao, R.; Gradziel, T.M.; Dandekar, A.M. Identification of self-incompatibility genotypes of almond by allele-specific PCR analysis. Theor. Appl. Genet. 2000, 101, 344–349. [Google Scholar] [CrossRef]
- Sonneveld, T.; Tobutt, K.R.; Robbins, T.P. Allele-specific PCR detection of sweet cherry self-incompatibility (S) alleles S1 to S16 using consensus and allele-specific primers. Theor. Appl. Genet. 2003, 107, 1059–1070. [Google Scholar] [CrossRef]
- Ortega, E.; Sutherland, B.G.; Dicenta, F.; Boskovic, R.; Tobutt, K.R. Determination of incompatibility genotypes in almond using first and second intron consensus primers: Detection of new S alleles and correction of reported S genotypes. Plant Breed. 2005, 124, 188–196. [Google Scholar] [CrossRef]
- Tao, R.; Yamane, H.; Sugiura, A. Molecular Typing of S-alleles through Identification, characterization and cDNA cloning for S-RNases in sweet cherry. J. Am. Soc. Hortic. Sci. 1999, 24, 224–233. [Google Scholar] [CrossRef]
- Audergon, J.M. Conribution to the study of inheritance of the character self incompatibility in apricot. Acta Hortic. 1999, 488, 275–279. [Google Scholar] [CrossRef]
- Sharafi, Y.; Hajilou, J.; Mohammadi, S.A.; Dadpour, M.R.; Eskandari, S. Pollen-pistil compatibility relationships in some Iranian almond (Prunus dulcis, Batch) genotypes as revealed by PCR analysis. Afr. J. Biotechnol. 2010, 9, 3338–3341. [Google Scholar]
- Zeng, B.; Gao, Q.; Tian, J.; Li, J. Research Progress on Self-Incompatibility of Almond Plants. In Proceedings of the Dried Fruit Branch of Chinese Horticultural Society, Shijiazhuang, China, 29 November 2013. [Google Scholar]
- Tian, J. China Almond, 1st ed.; China Agriculture Press: Beijing, China, 2008. [Google Scholar]
- Charlesworth, D.; Guttman, D.S. Seeing selection in S allele sequence. Curr. Biol. 1997, 7, 34–37. [Google Scholar] [CrossRef]
- Ortega, E.; Bonkovi, R.I.; Sargent, D.J.; Tobutt, K.R. Analysis of S-RNase alleles of almond (Prunus dulcis): Characterization of new sequences, resolution of synonyms and evidence of intragenic recombination. Mol. Gen. Genomics. 2006, 276, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Heng, W.; Wu, H.Q.; Chen, Q.X.; Wu, J.; Huang, S.; Zhang, S.L. Identification of S-genotypes and novel S-RNase alleles in Prunus mume. J. Hortic. Sci. Biotech. 2008, 83, 689–694. [Google Scholar] [CrossRef]
- Liang, M. Identification and Evolution of Citrus Self-Incompatibility Related Genes. Master’s Thesis, Huazhong Agricultural University, Wuhan, China, 2019. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).