The Complete Chloroplast Genome of an Epiphytic Leafless Orchid, Taeniophyllum complanatum: Comparative Analysis and Phylogenetic Relationships
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, DNA Extraction and Sequencing
2.2. Chloroplast Genome Assembly and Annotation
2.3. Genome Comparison and Analysis, IR Border and Divergence Analyses
2.4. Codon Usage Frequency and Amino Acid Abundance
2.5. Repeat Sequence Analysis
2.6. Reconstruction of Phylogenetic Relationship
3. Results
3.1. Characteristics of the Chloroplast
3.2. Comparative Genome Analysis
3.3. Repeated Analysis
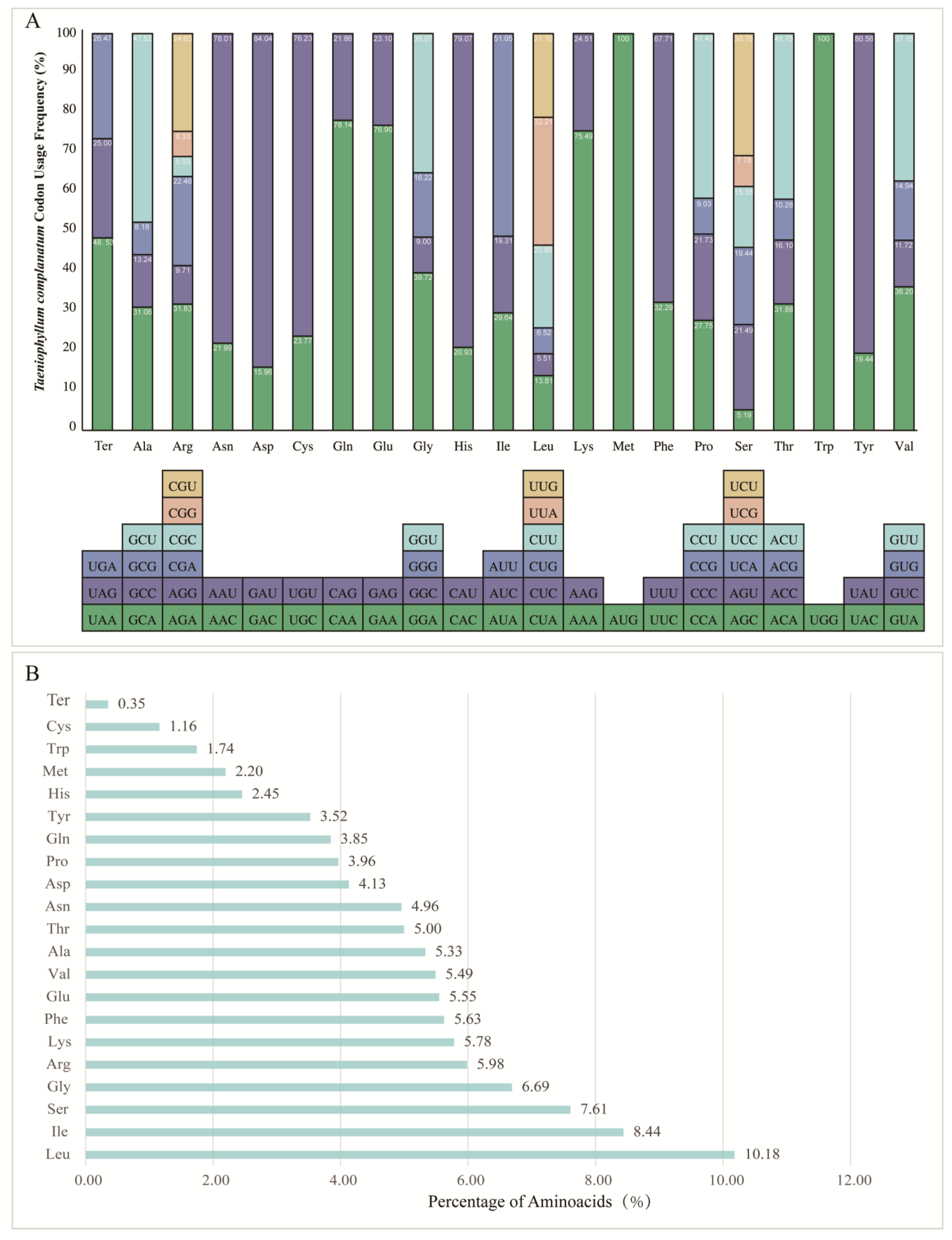
3.4. Codon Usage Frequency and Aminoacid Abundance
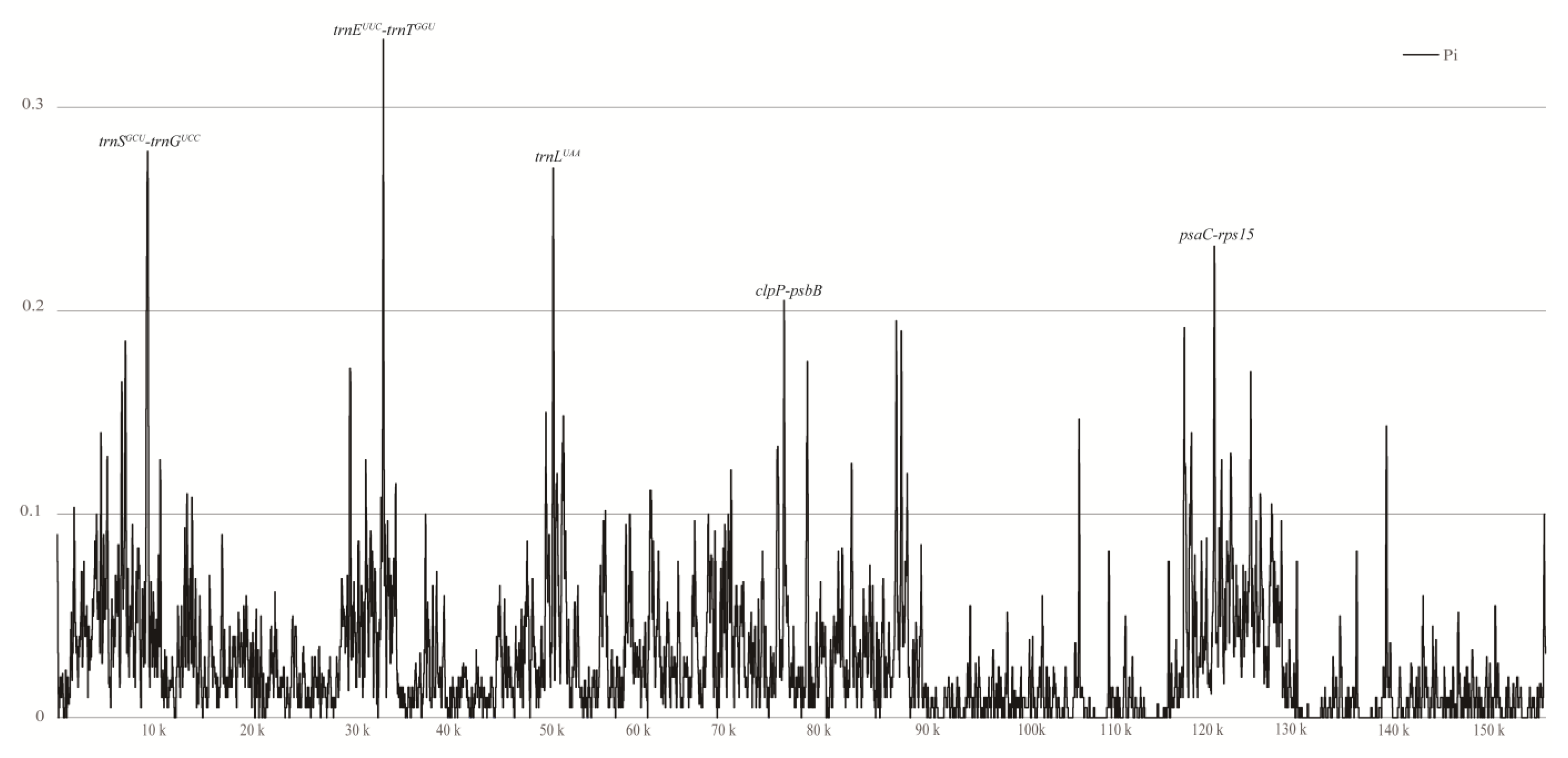
3.5. Sequence Divergence and Barcoding Investigation
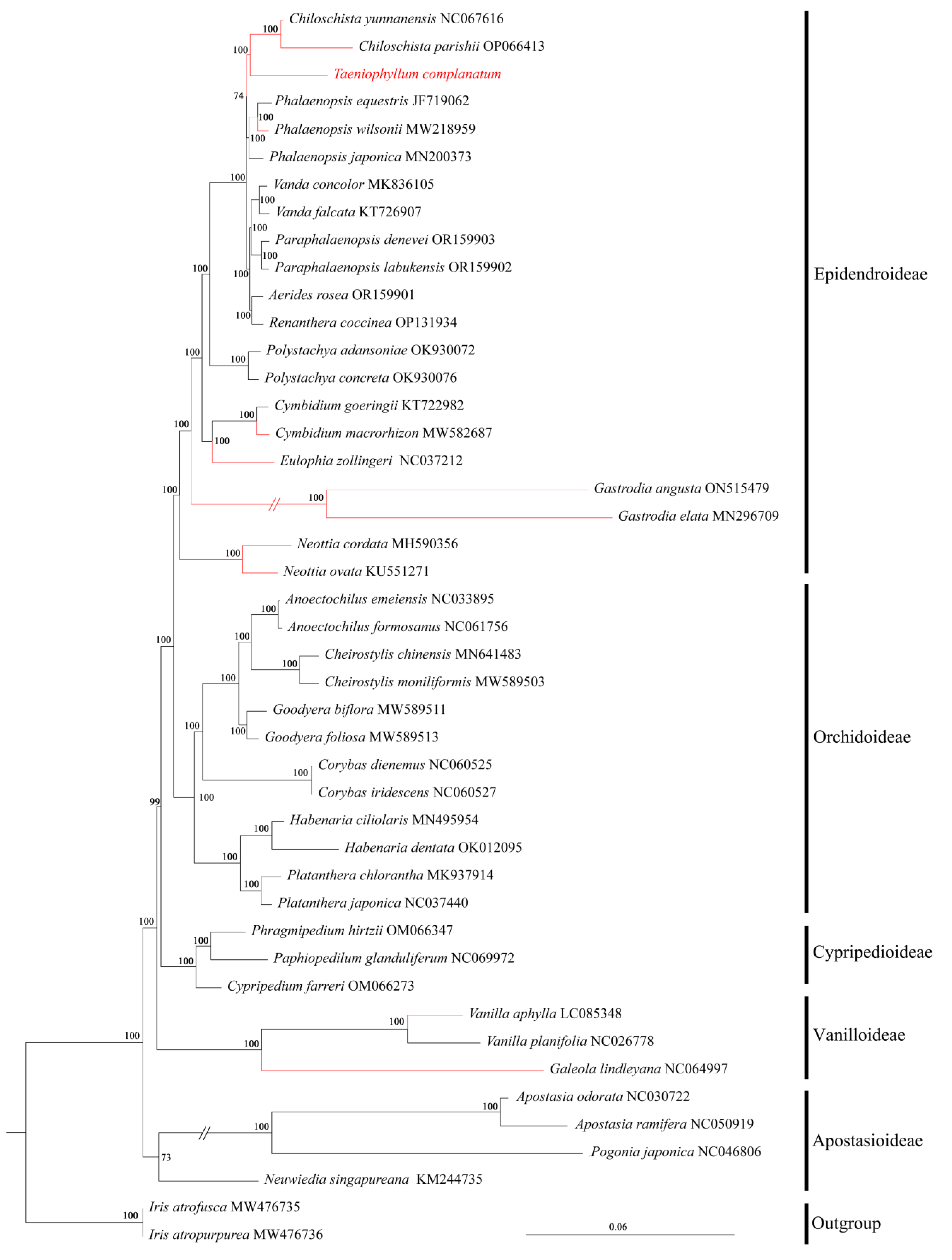
3.6. Phylogenetic Reconstruction
4. Discussion
4.1. Chloroplast Genome Organization and Gene Content of T. complanatum and Comparison among Other Orchidaceae
4.2. Comparative Analysis of Sequence Comparison and Gene Order
4.3. SSRs and Repetitive Region Analysis
4.4. Codon Usage Frequency Analysis and Amino Acid Abundance
4.5. Divergent Hot Spot Analysis
4.6. Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hernández-Hernández, T.; Hernández, H.M.; De-Nova, J.A.; Puente, R.; Eguiarte, L.E.; Magallón, S. Phylogenetic relationships and evolution of growth form in Cactaceae (Caryophyllales, Eudicotyledoneae). Am. J. Bot. 2011, 98, 44–61. [Google Scholar] [CrossRef] [PubMed]
- Majure, L.C.; Achá, S.; Baker, M.A.; Puente-Martínez, R.; Köhler, M.; Fehlberg, S. Phylogenomics of One of the World’s Most Intriguing Groups of CAM Plants, the Opuntioids (Opuntioideae: Cactaceae): Adaptation to Tropical Dry Forests Helped Drive Prominent Morphological Features in the Clade. Diversity 2023, 15, 570. [Google Scholar] [CrossRef]
- Zhong, C.; Mansour, S.; Nambiar-Veetil, M.; Bogusz, D.; Franche, C. Casuarina glauca: A model tree for basic research in actinorhizal symbiosis. J. Biosci. 2013, 38, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wei, Y.; Meng, J.; Wang, Y.; Nie, S.; Zhang, Z.; Wang, H.; Yang, Y.; Gao, Y.; Wu, J.; et al. Chromosome-scale de novo genome assembly and annotation of three representative Casuarina species: C. equisetifolia, C. glauca, and C. cunninghamiana. Plant J. 2023, 114, 1490–1505. [Google Scholar] [CrossRef]
- Chen, X.; Fang, D.; Xu, Y.; Duan, K.; Yoshida, S.; Yang, S.; Sahu, S.K.; Fu, H.; Guang, X.; Liu, M.; et al. Balanophora genomes display massively convergent evolution with other extreme holoparasites and provide novel insights into parasite-host interactions. Nat. Plants 2023, 9, 1627–1642. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Hu, X.; Li, J.; Shao, B.; Wang, Y.; Wang, L.; Li, K.; Lin, D.; Wang, H.; Gao, Z.; et al. The Sapria himalayana genome provides new insights into the lifestyle of endoparasitic plants. BMC Biol. 2023, 21, 134. [Google Scholar] [CrossRef]
- Sun, G.; Xu, Y.; Liu, H.; Sun, T.; Zhang, J.; Hettenhausen, C.; Shen, G.; Qi, J.; Qin, Y.; Li, J.; et al. Large-scale gene losses underlie the genome evolution of parasitic plant Cuscuta australis. Nat. Commun. 2018, 9, 2683. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Yu, W.B.; Tan, Y.; Liu, B.; Yao, X.; Jin, J.; Padmanaba, M.; Yang, J.B.; Corlett, R.T. Evolutionary Comparisons of the Chloroplast Genome in Lauraceae and Insights into Loss Events in the Magnoliids. Genome Biol. Evol. 2017, 9, 2354–2364. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Read, D. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: San Diego, CA, USA, 2008. [Google Scholar] [CrossRef]
- Young, B.W.; Massicotte, H.B.; Tackaberry, L.E.; Baldwin, Q.F.; Egger, K.N. Monotropa uniflora: Morphological and molecular assessment of mycorrhizae retrieved from sites in the sub-boreal spruce biogeoclimatic zone in central British Columbia. Mycorrhiza 2002, 12, 75–82. [Google Scholar] [CrossRef]
- Bidartondo, M.I.; Burghardt, B.; Gebauer, G.; Bruns, T.D.; Read, D.J. Changing partners in the dark: Isotopic and molecular evidence of ectomycorrhizal liaisons between forest orchids and trees. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2004, 271, 1799–1806. [Google Scholar] [CrossRef]
- Cameron, D.D.; Leake, J.R.; Read, D.J. Mutualistic mycorrhiza in orchids: Evidence from plant-fungus carbon and nitrogen transfers in the green-leaved terrestrial orchid Goodyera repens. New Phytol. 2006, 171, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Christenhusz, M.J.M.; Byng, J.W. The number of known plants species in the world and its annual increase. Phytotaxa 2016, 261, 201–217. [Google Scholar] [CrossRef]
- Zimmer, K.; Hynson, N.A.; Gebauer, G.; Allen, E.B.; Allen, M.F.; Read, D.J. Wide geographical and ecological distribution of nitrogen and carbon gains from fungi in pyroloids and monotropoids (Ericaceae) and in orchids. New Phytol. 2007, 175, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Motomura, H.; Selosse, M.A.; Martos, F.; Kagawa, A.; Yukawa, T. Mycoheterotrophy evolved from mixotrophic ancestors: Evidence in Cymbidium (Orchidaceae). Ann. Bot. 2010, 106, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Selosse, M.A.; Faccio, A.; Scappaticci, G.; Bonfante, P. Chlorophyllous and Achlorophyllous Specimens of Epipactis microphylla (Neottieae, Orchidaceae) Are Associated with Ectomycorrhizal Septomycetes, including Truffles. Microb. Ecol. 2004, 47, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-H.; Liu, K.-W.; Li, Z.; Lu, H.-C.; Ye, Q.-L.; Zhang, D.; Wang, J.-Y.; Li, Y.-F.; Zhong, Z.-M.; Liu, X.; et al. Genomes of leafy and leafless Platanthera orchids illuminate the evolution of mycoheterotrophy. Nat. Plants 2022, 8, 373–388. [Google Scholar] [CrossRef] [PubMed]
- Carlsward, B.S.; Whitten, W.M.; Williams, N.H.; Bytebier, B. Molecular phylogenetics of Vandeae (Orchidaceae) and the evolution of leaflessness. Am. J. Bot. 2006, 93, 770–786. [Google Scholar] [CrossRef] [PubMed]
- Benzing, D.H.; Friedman, W.E.; Peterson, G.; Renfrow, A. Shootlessness, velamentous roots and the pre-eminence of Orchidaceae in the epiphytic biotope. Am. J. Bot. 1983, 70, 121–133. [Google Scholar] [CrossRef]
- Pridgeon, A.M.; Cribb, P.J.; Chase, M.W.; Rasmussen, F.N.; Pridgeon, A.M.; Cribb, P.J.; Chase, M.W.; Rasmussen, F.N. Genera Orchidacearum Volume 6: Epidendroideae (Part 3); Oxford University Press: Oxford, UK, 2014. [Google Scholar]
- Jinlu, L.; Shuo, W.; Jing, Y.; Ling, W.; Shiliang, Z. A Modified CTAB Protocol for Plant DNA Extraction. Chin. Bull. Bot. 2013, 48, 72–78. [Google Scholar] [CrossRef]
- Liu, D.-K.; Tu, X.-D.; Zhao, Z.; Zeng, M.-Y.; Zhang, S.; Ma, L.; Zhang, G.-Q.; Wang, M.-M.; Liu, Z.-J.; Lan, S.-R.; et al. Plastid phylogenomic data yield new and robust insights into the phylogeny of Cleisostoma-Gastrochilus clades (Orchidaceae, Aeridinae). Mol. Phylogenetics Evol. 2020, 145, 106729. [Google Scholar] [CrossRef]
- Luo, C.; Huang, W.; Yer, H.; Kamuda, T.; Li, X.; Li, Y.; Rong, Y.; Yan, B.; Wen, Y.; Wang, Q.; et al. Complete Chloroplast Genomes and Comparative Analyses of Three Ornamental Impatiens Species. Front. Genet. 2022, 13, 816123. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-K.; Jo, S.; Cheon, S.-H.; Joo, M.-J.; Hong, J.-R.; Kwak, M.; Kim, K.-J. Plastome Evolution and Phylogeny of Orchidaceae, with 24 New Sequences. Front. Plant Sci. 2020, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.Y.; Zeng, M.Y.; Gao, X.; Zhao, Z.; Li, R.; Wu, Y.; Liu, Z.J.; Zhang, D.; Li, M.H. Characteristics and Comparative Analysis of Seven Complete Plastomes of Trichoglottis s.l. (Aeridinae, Orchidaceae). Int. J. Mol. Sci. 2023, 24, 14544. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; de Pamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Poczai, P.; Hyvonen, J.; Tang, J.; Amiryousefi, A. Chloroplot: An Online Program for the Versatile Plotting of Organelle Genomes. Front. Genet. 2020, 11, 576124. [Google Scholar] [CrossRef]
- Brudno, M.; Malde, S.; Poliakov, A.; Do, C.B.; Couronne, O.; Dubchak, I.; Batzoglou, S. Glocal alignment: Finding rearrangements during alignment. Bioinformatics 2003, 19 (Suppl. S1), i54–i62. [Google Scholar] [CrossRef]
- Rissman, A.I.; Mau, B.; Biehl, B.S.; Darling, A.E.; Glasner, J.D.; Perna, N.T. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics 2009, 25, 2071–2073. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvonen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zhou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. DAMBE7: New and Improved Tools for Data Analysis in Molecular Biology and Evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef] [PubMed]
- Mehmetoglu, E.; Kaymaz, Y.; Ates, D.; Kahraman, A.; Tanyolac, M.B. The complete chloroplast genome sequence of Cicer echinospermum, genome organization and comparison with related species. Sci. Hortic. 2022, 296, 110912. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef]
- Ye, B.J.; Zhang, S.; Tu, X.D.; Liu, D.K.; Li, M.H. The complete plastid genome of Thrixspermum tsii (Orchidaceae, Aeridinae). Mitochondrial DNA Part B 2020, 5, 384–385. [Google Scholar] [CrossRef]
- Xia, K.; Liu, D.K.; Wang, J.Y. The complete chloroplast genome sequence of Phalaenopsis wilsoniii (Orchidaceae). Mitochondrial DNA B Resour. 2021, 6, 3303–3305. [Google Scholar] [CrossRef]
- Xiao, T.; He, L.; Yue, L.; Zhang, Y.; Lee, S.Y. Comparative phylogenetic analysis of complete plastid genomes of Renanthera (Orchidaceae). Front. Genet. 2022, 13, 998575. [Google Scholar] [CrossRef]
- Chen, J.; Wang, F.; Zhou, C.; Ahmad, S.; Zhou, Y.; Li, M.; Liu, Z.; Peng, D. Comparative Phylogenetic Analysis for Aerides (Aeridinae, Orchidaceae) Based on Six Complete Plastid Genomes. Int. J. Mol. Sci. 2023, 24, 12473. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, F.; Zhao, Z.; Li, M.; Liu, Z.; Peng, D. Complete Chloroplast Genomes and Comparative Analyses of Three Paraphalaenopsis (Aeridinae, Orchidaceae) Species. Int. J. Mol. Sci. 2023, 24, 11167. [Google Scholar] [CrossRef]
- Lin, C.-S.; Chen, J.J.W.; Huang, Y.-T.; Chan, M.-T.; Daniell, H.; Chang, W.-J.; Hsu, C.-T.; Liao, D.-C.; Wu, F.-H.; Lin, S.-Y.; et al. The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Rep. 2015, 5, 9040. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, M.J.; Copetti, D.; Búrquez, A.; Bustamante, E.; Charboneau, J.L.M.; Eguiarte, L.E.; Kumar, S.; Lee, H.O.; Lee, J.; McMahon, M.; et al. Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): Loss of the ndh gene suite and inverted repeat. Am. J. Bot. 2015, 102, 1115–1127. [Google Scholar] [CrossRef]
- Bock, R. Structure, Function, and Inheritance of Plastid Genomes. In Cell and Molecular Biology of Plastids; Springer: Berlin/Heidelberg, Germany, 2007; pp. 29–63. [Google Scholar] [CrossRef]
- Palmer, J.D.; Thompson, W.F. Chloroplast DNA rearrangements aremore frequent when a large inverted repeat sequence is lost. Cell 1982, 29, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Shi, C.; Liu, Y.; Mao, S.-Y.; Gao, L.-Z. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: Genome structure and phylogenetic relationships. BMC Evol. Biol. 2014, 14, 151. [Google Scholar] [CrossRef]
- Raubeson, L.A.; Peery, R.; Chumley, T.W.; Dziubek, C.; Fourcade, H.M.; Boore, J.L.; Jansen, R.K. Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom. 2007, 8, 174. [Google Scholar] [CrossRef]
- Guo, Y.Y.; Yang, J.X.; Bai, M.Z.; Zhang, G.Q.; Liu, Z.J. The chloroplast genome evolution of Venus slipper (Paphiopedilum): IR expansion, SSC contraction, and highly rearranged SSC regions. BMC Plant Biol. 2021, 21, 248. [Google Scholar] [CrossRef]
- Guo, Y.-Y.; Yang, J.-X.; Li, H.-K.; Zhao, H.-S. Chloroplast Genomes of Two Species of Cypripedium: Expanded Genome Size and Proliferation of AT-Biased Repeat Sequences. Front. Plant Sci. 2021, 12, 609729. [Google Scholar] [CrossRef] [PubMed]
- Chi, X.; Zhang, F.; Dong, Q.; Chen, S. Insights into Comparative Genomics, Codon Usage Bias, and Phylogenetic Relationship of Species from Biebersteiniaceae and Nitrariaceae Based on Complete Chloroplast Genomes. Plants 2020, 9, 1605. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T. Chloroplast evolution secondary symbiogenesis and multiple losses chloroplasts originated from cyanobacteria only. Curr. Biol. 2002, 12, R62–R64. [Google Scholar] [CrossRef] [PubMed]
- Munyao, J.N.; Dong, X.; Yang, J.X.; Mbandi, E.M.; Wanga, V.O.; Oulo, M.A.; Saina, J.K.; Musili, P.M.; Hu, G.W. Complete Chloroplast Genomes of Chlorophytum comosum and Chlorophytum gallabatense: Genome Structures, Comparative and Phylogenetic Analysis. Plants 2020, 9, 296. [Google Scholar] [CrossRef] [PubMed]
- Mo, Z.; Lou, W.; Chen, Y.; Jia, X.; Zhai, M.; Guo, Z.; Xuan, J. The chloroplast genome of Carya illinoinensis: Genome structure, adaptive evolution, and phylogenetic analysis. Forests 2020, 11, 207. [Google Scholar] [CrossRef]
- Huo, X.; Zhao, Y.; Qian, Z.; Liu, M. Characterization of the complete chloroplast genome of Eulophia zollingeri, an endangered orchid in China. Conserv. Genet. Resour. 2017, 10, 817–819. [Google Scholar] [CrossRef]
- Niu, Z.; Xue, Q.; Zhu, S.; Sun, J.; Liu, W.; Ding, X. The Complete Plastome Sequences of Four Orchid Species: Insights into the Evolution of the Orchidaceae and the Utility of Plastomic Mutational Hotspots. Front. Plant Sci. 2017, 8, 715. [Google Scholar] [CrossRef] [PubMed]
- Zong, D.; Gan, P.; Zhou, A.; Li, J.; Xie, Z.; Duan, A.; He, C. Comparative analysis of the complete chloroplast genomes of seven Populus species: Insights into alternative female parents of Populus tomentosa. PLoS ONE 2019, 14, e0218455. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zeng, M.Y.; Wu, Y.W.; Li, J.W.; Zhou, Z.; Liu, Z.J.; Li, M.H. Characterization and Comparative Analysis of the Complete Plastomes of Five Epidendrum (Epidendreae, Orchidaceae) Species. Int. J. Mol. Sci. 2023, 24, 14437. [Google Scholar] [CrossRef]
- Ding, S.; Dong, X.; Yang, J.; Guo, C.; Cao, B.; Guo, Y.; Hu, G. Complete Chloroplast Genome of Clethra fargesii Franch, an Original Sympetalous Plant from Central China: Comparative Analysis, Adaptive Evolution, and Phylogenetic Relationships. Forests 2021, 12, 441. [Google Scholar] [CrossRef]
- Abdullah; Mehmood, F.; Shahzadi, I.; Waseem, S.; Mirza, B.; Ahmed, I.; Waheed, M.T. Chloroplast genome of Hibiscus rosasinensis (Malvaceae): Comparative analyses and identification of mutational hotspots. Genomics 2020, 112, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.-H.; Huang, J.-X.; Zhang, G.-Q.; Liu, Z.-J.; Zhuang, X.-Y. A molecular phylogeny of Aeridinae (Orchidaceae: Epidendroideae) inferred from multiple nuclear and chloroplast regions. Mol. Phylogenetics Evol. 2015, 85, 247–254. [Google Scholar] [CrossRef] [PubMed]
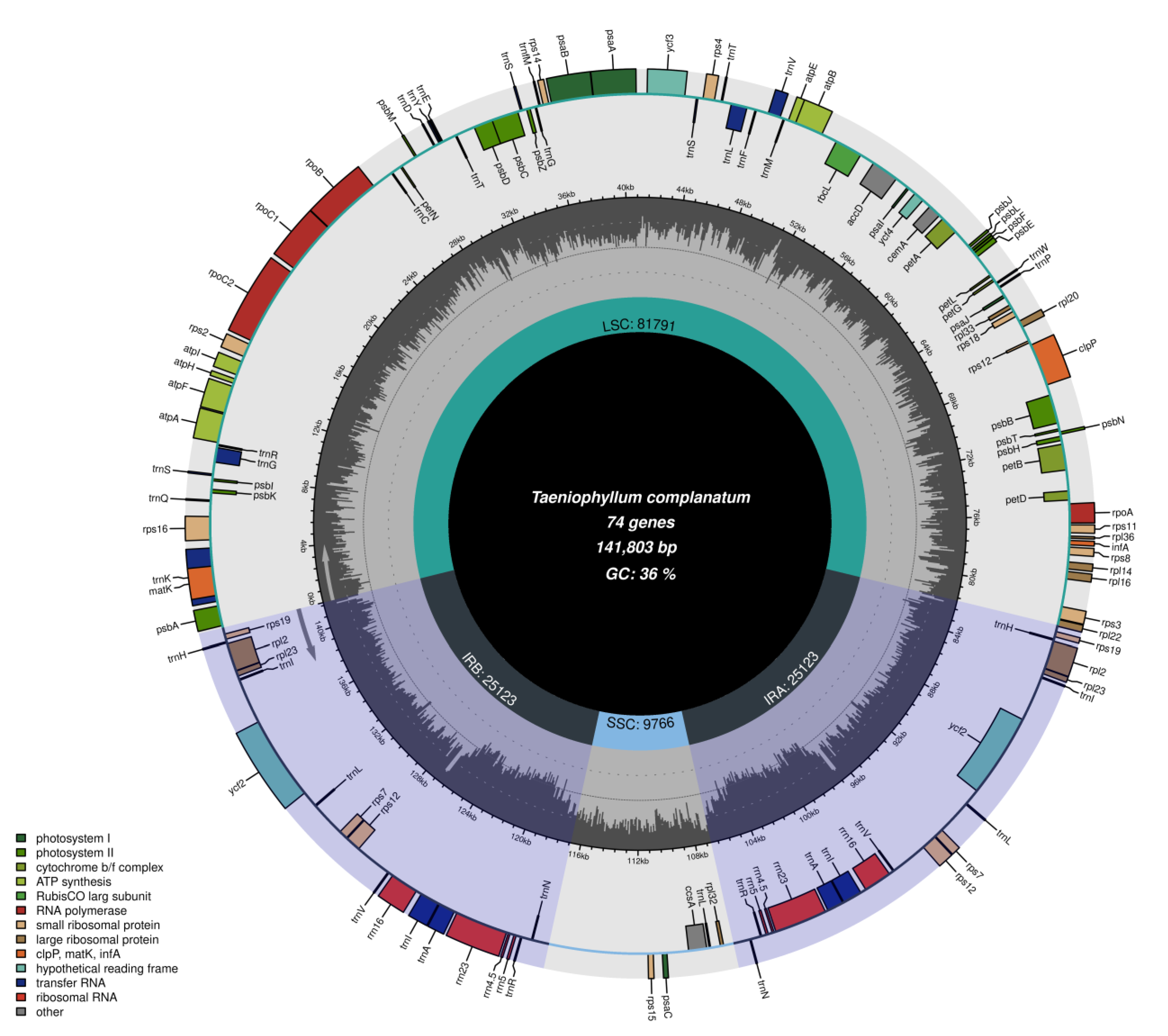
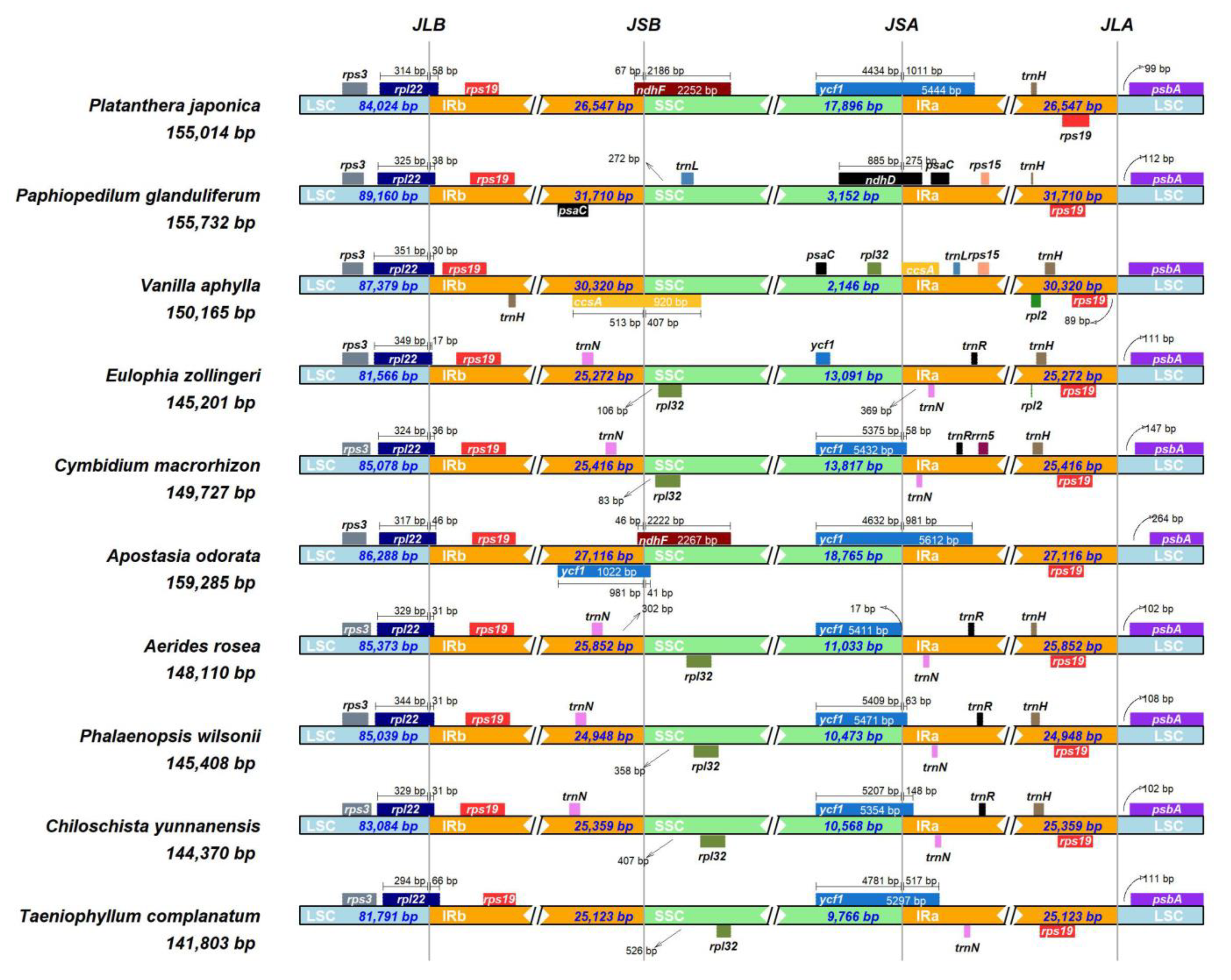
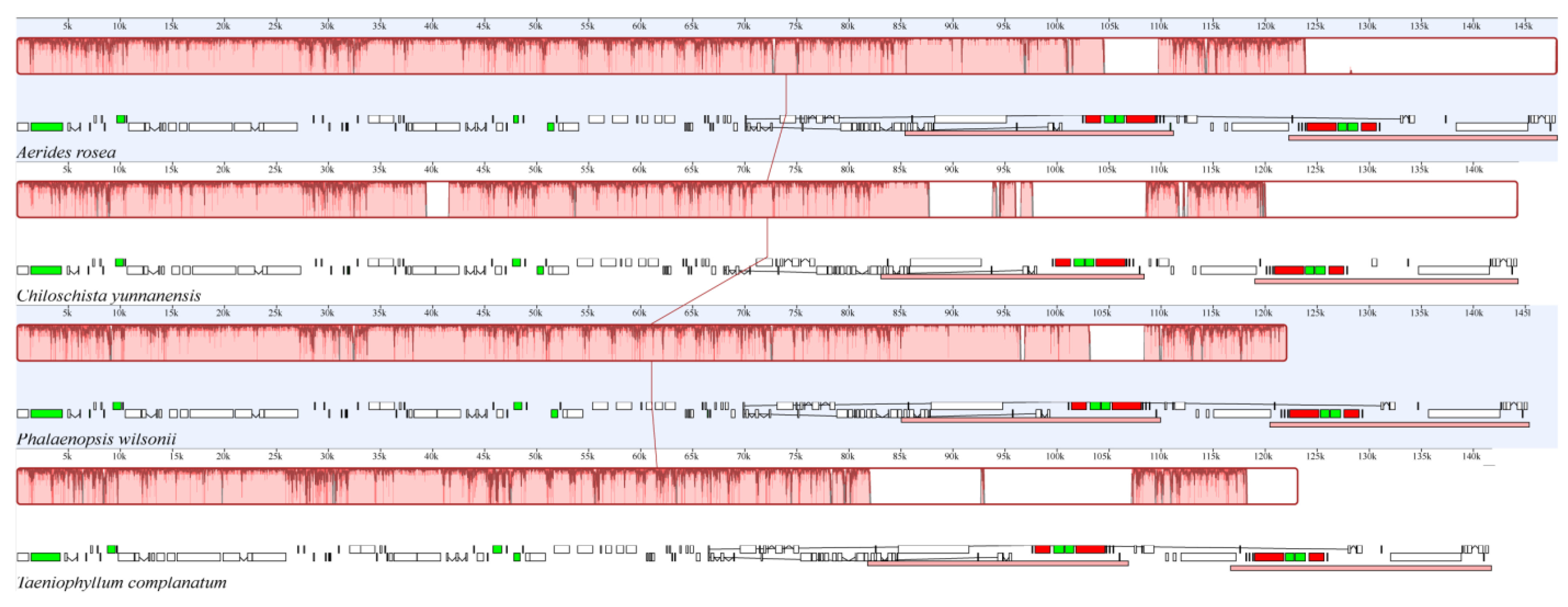
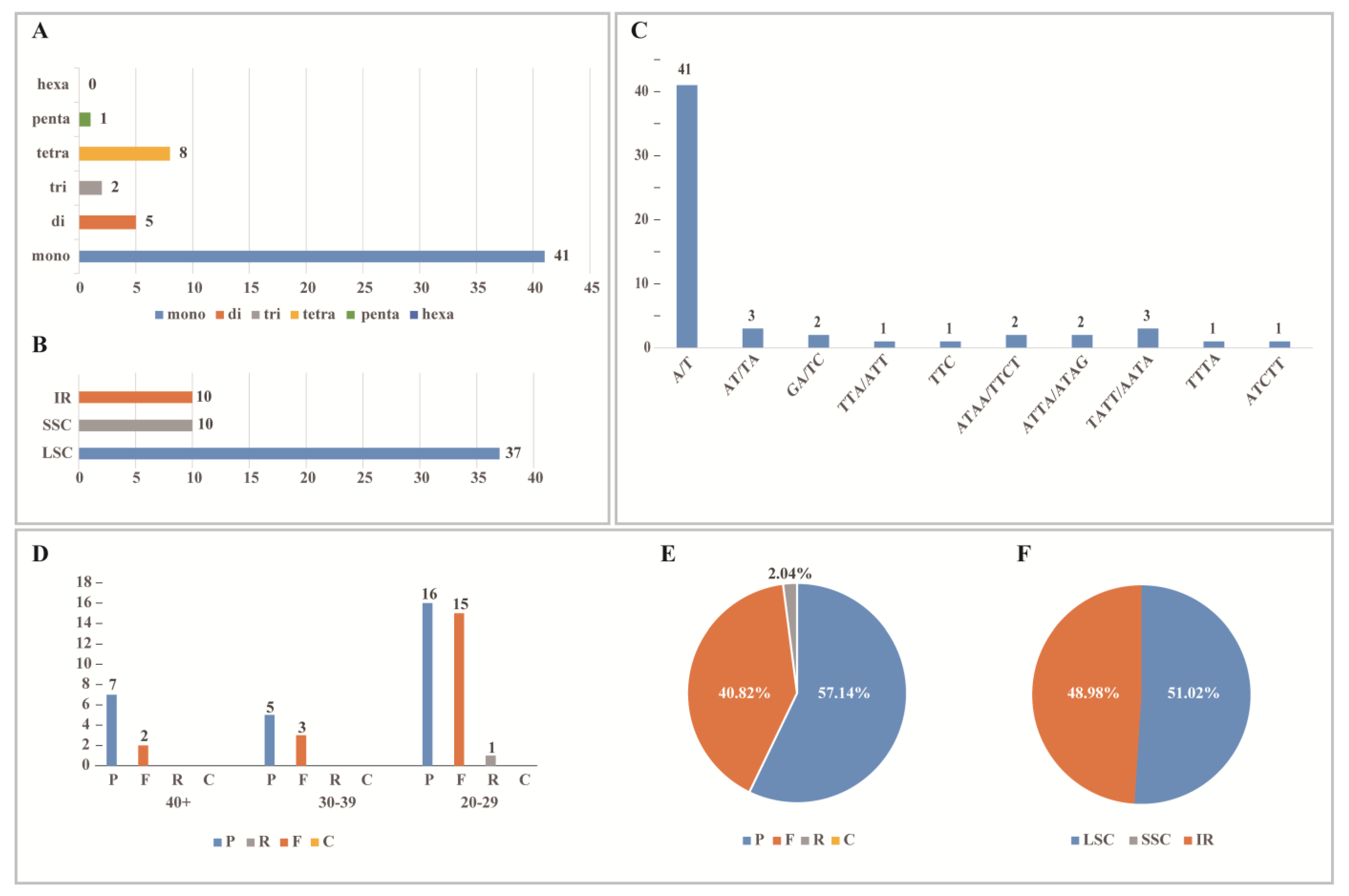
| Species | Size (bp) | LSC (bp) | SSC (bp) | IRs (bp) | Number of Genes | Protein-Coding Genes | tRNA Genes | rRNA Genes | Total GC (%) | LSC GC (%) | SSC GC (%) | IR GC (%) | Number of ndh Gene Loss/Pseudogenization |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T. complanatum | 141,174 | 81,791 | 9766 | 25,123 | 120 | 74 | 38 | 8 | 36.3 | 33.6 | 26.9 | 42.2 | 9 (3) |
| Aerides rosea | 148,110 | 85,373 | 11,033 | 25,852 | 120 | 74 | 38 | 8 | 36.7 | 34.0 | 28.3 | 43.1 | 7 (5) |
| Chiloschista yunnanensis | 144,370 | 83,084 | 10,568 | 25,359 | 119 | 73 | 38 | 8 | 36.9 | 33.7 | 27.6 | 43.2 | 7 (5) |
| Phalaenopsis wilsonii | 145,408 | 85,039 | 10,473 | 24,948 | 120 | 74 | 38 | 8 | 36.8 | 34.1 | 28.2 | 43.3 | 6 (6) |
| Category | Group of Genes | Name of Genes |
|---|---|---|
| Self-replication | Proteins of large ribosomal subunit | rpl2(×2) a, rpl14, rpl16 a, rpl20, rpl22, rpl23(×2), rpl32, rpl33, rpl36 |
| Proteins of small ribosomal subunit | rps2, rps3, rps4, rps7(×2), rps8, rps11, rps12(×2) b, rps14, rps15, rps16 a, rps18, rps19(×2) | |
| Subunits of RNA polymerase | rpoA, rpoB, rpoC1 a, rpoC2 | |
| Ribosomal RNA genes | rrn4.5(×2), rrn5(×2), rrn16(×2), rrn23(×2) | |
| Transfer RNA genes | trnA-UGC(×2) a, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC a, trnH-GUG(×2), trnI-CAU(×2), trnI-GAU(×2) a, trnK-UUU a, trnL-CAA(×2), trnL-UAA a, trnL-UAG, trnM-CAU, trnN-GUU(×2), trnP-UGG, trnQ-UUG, trnR-ACG(×2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(×2), trnV-UAC a, trnW-CCA, trnY-GUA, trnfM-CAU | |
| Genes for photosynthesis | Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of NADH dehydrogenase | atpA, atpB, atpE, atpF a, atpH, atpI | |
| Subunits of cytochrome b/f complex | ndhB (×2), ndhD | |
| Subunits of ATP synthase | petA, petB a, petD a, petG, petL, petN | |
| Large subunit of rubisco | rbcL | |
| Other genes | Protease | clpP b |
| Maturase | matK | |
| Envelope membrane protein | cemA | |
| Translation initiation factor | infA | |
| C-type cytochrome synthesis gene | ccsA | |
| Subunit of acetyl-CoA-carboxylase | accD | |
| Genes of unknown function | Conserved hypothetical chloroplast | ycf1, ycf(×2), ycf3 b, ycf4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Z.; Chen, J.; Wang, F.; Wu, X.; Liu, Z.; Peng, D.; Lan, S. The Complete Chloroplast Genome of an Epiphytic Leafless Orchid, Taeniophyllum complanatum: Comparative Analysis and Phylogenetic Relationships. Horticulturae 2024, 10, 660. https://doi.org/10.3390/horticulturae10060660
Zhou Z, Chen J, Wang F, Wu X, Liu Z, Peng D, Lan S. The Complete Chloroplast Genome of an Epiphytic Leafless Orchid, Taeniophyllum complanatum: Comparative Analysis and Phylogenetic Relationships. Horticulturae. 2024; 10(6):660. https://doi.org/10.3390/horticulturae10060660
Chicago/Turabian StyleZhou, Zhuang, Jinliao Chen, Fei Wang, Xiaopei Wu, Zhongjian Liu, Donghui Peng, and Siren Lan. 2024. "The Complete Chloroplast Genome of an Epiphytic Leafless Orchid, Taeniophyllum complanatum: Comparative Analysis and Phylogenetic Relationships" Horticulturae 10, no. 6: 660. https://doi.org/10.3390/horticulturae10060660
APA StyleZhou, Z., Chen, J., Wang, F., Wu, X., Liu, Z., Peng, D., & Lan, S. (2024). The Complete Chloroplast Genome of an Epiphytic Leafless Orchid, Taeniophyllum complanatum: Comparative Analysis and Phylogenetic Relationships. Horticulturae, 10(6), 660. https://doi.org/10.3390/horticulturae10060660






