Identification and Functional Analysis of the Flower Development-Related TCP Genes in Erycina pusilla
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. TCP Family Members Identification and Phylogenetic Analysis
2.3. Relative Expression Analysis of EpTCPs
2.4. Arabidopsis Transformation
2.5. Statistical Analysis
3. Results
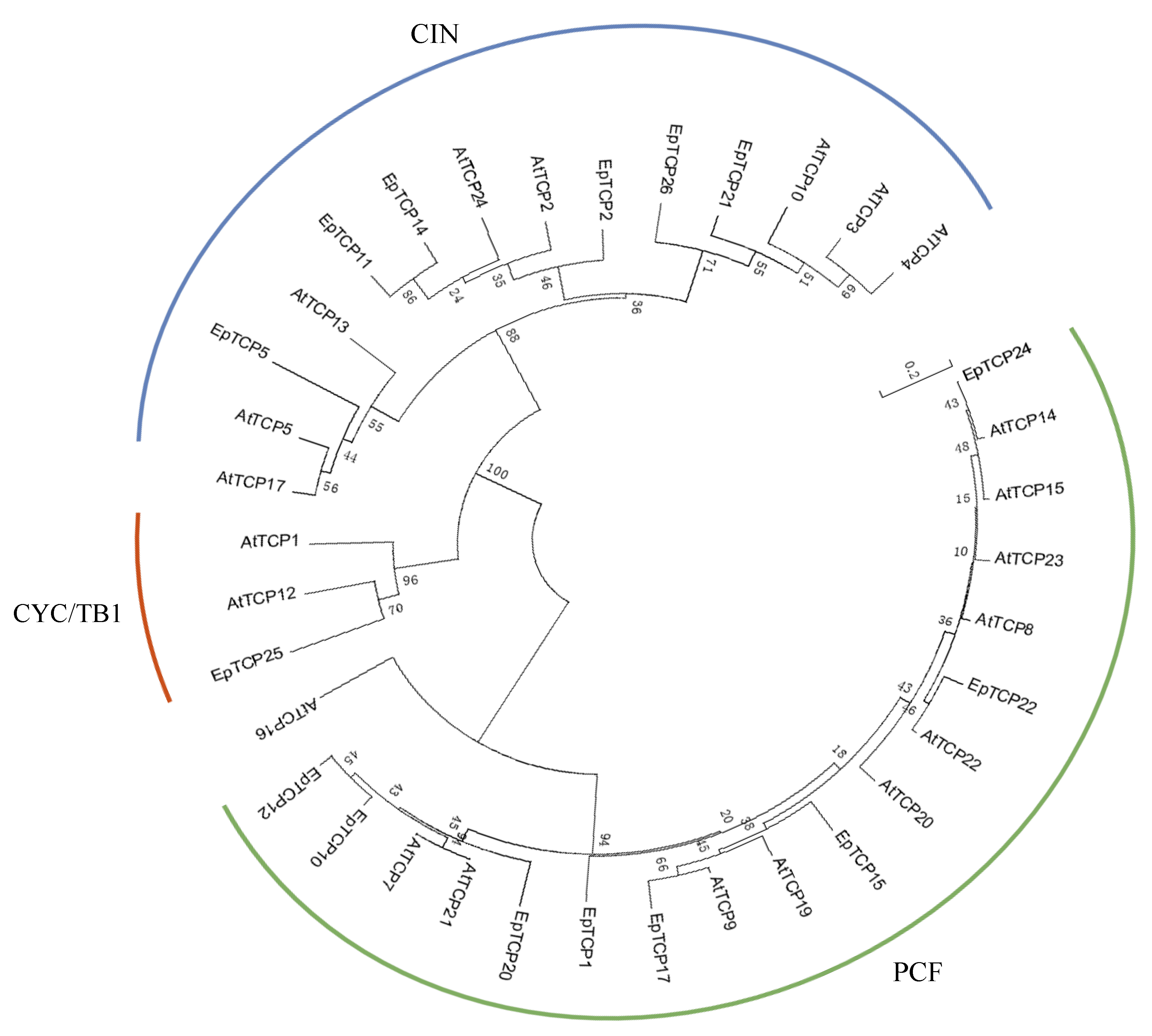
3.1. Phylogeny of TCP Transcript Factors from Erycina pusilla
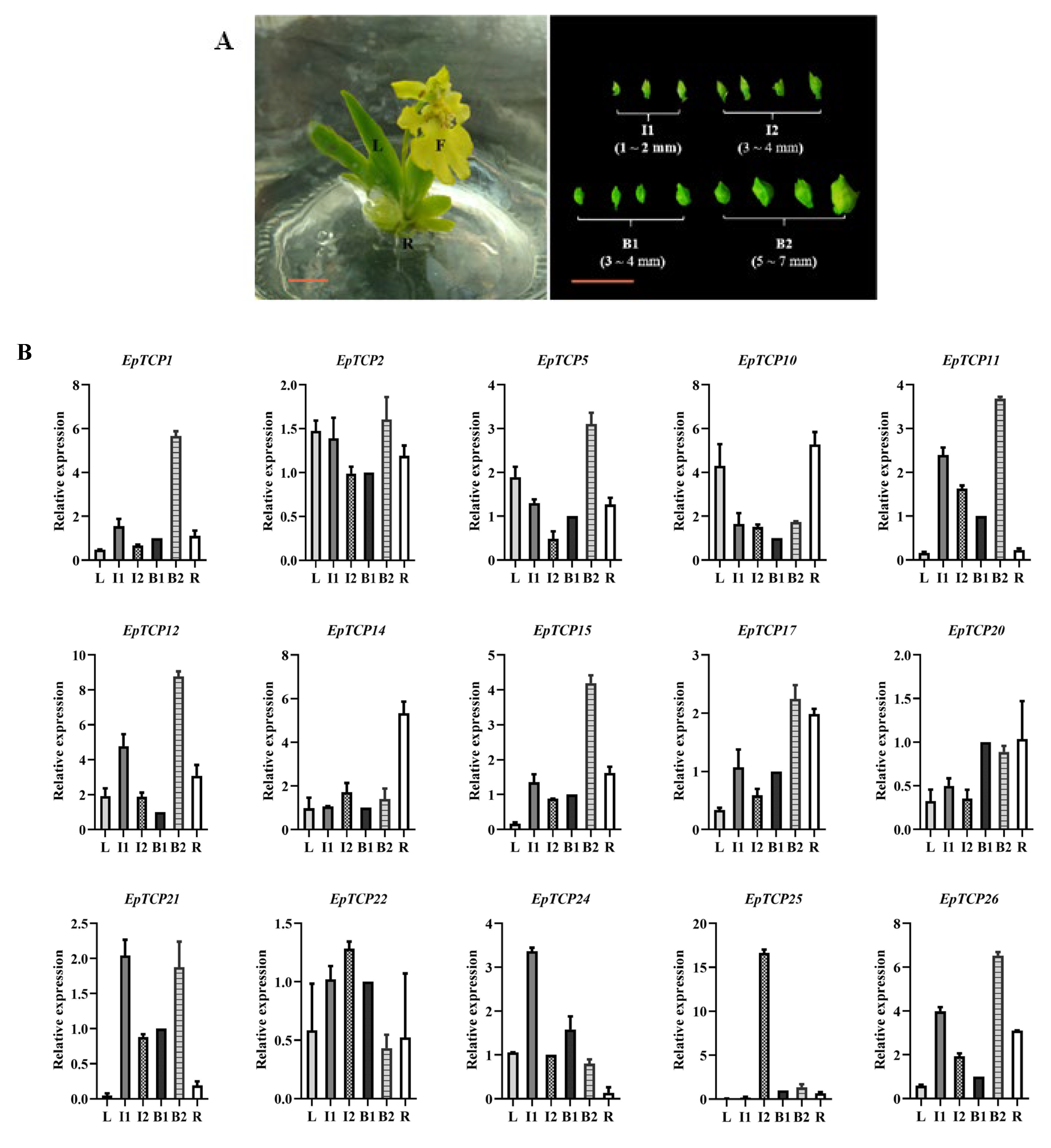
3.2. Expression Patterns of the EpTCP Genes in Different Tissues
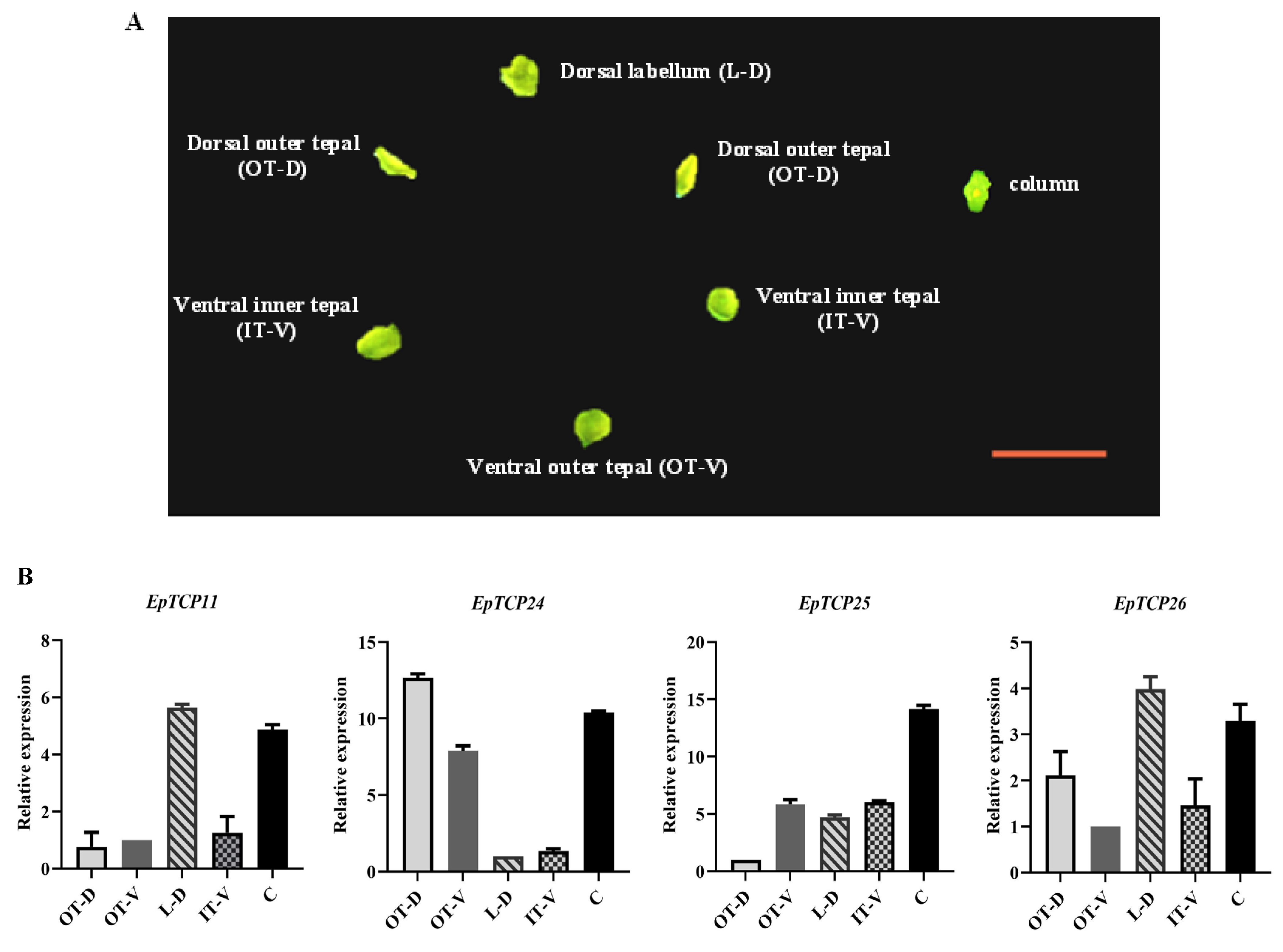
3.3. Expression Patterns of the EpTCP Genes in Different Floral Organs
3.4. Phenotypes of EpTCP11, EpTCP25 or EpTCP26 Overexpression in Arabidopsis
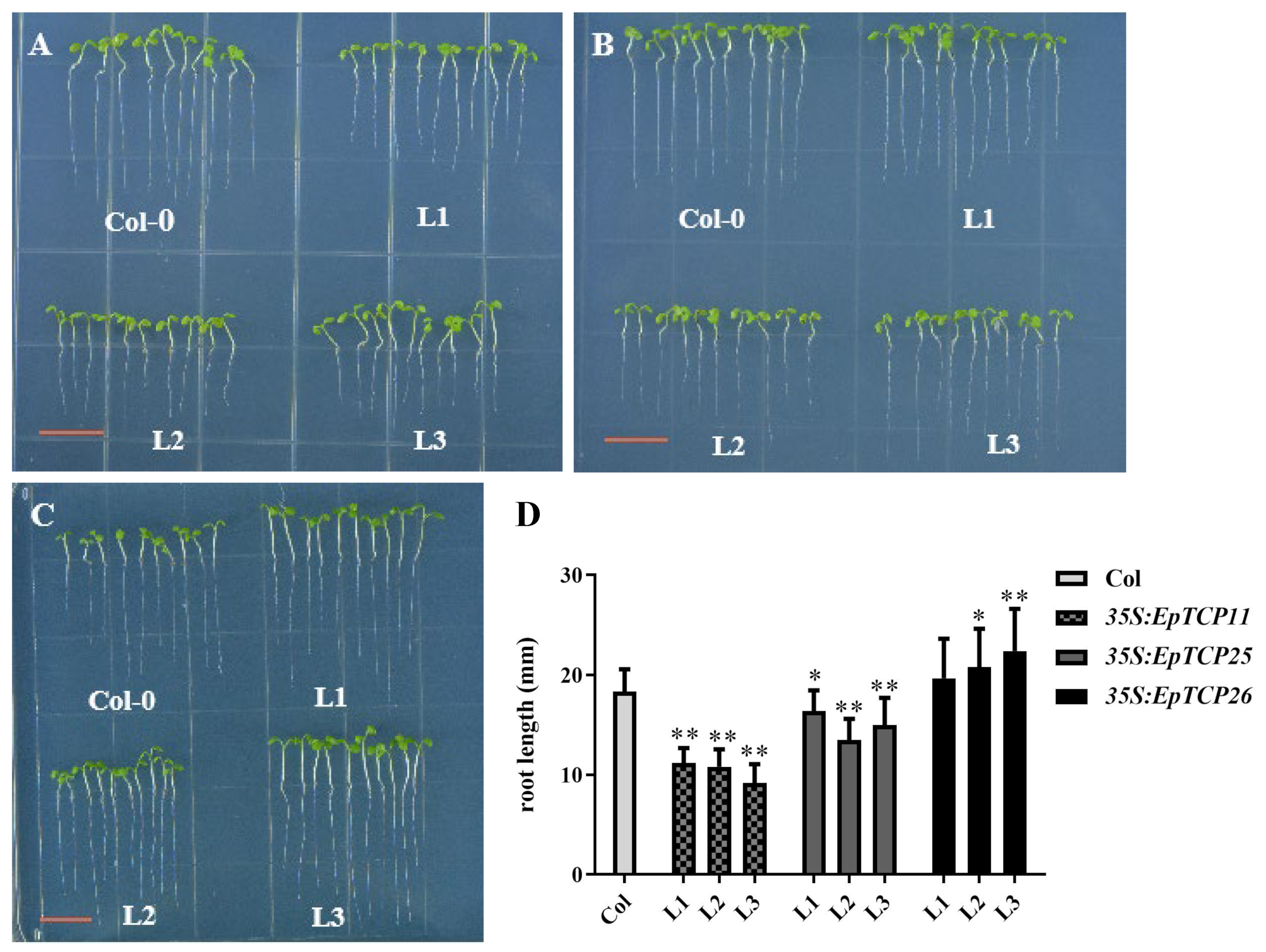
3.4.1. Effects of EpTCP11, EpTCP25 or EpTCP26 Overexpression on the Primary Root Growth of Arabidopsis
3.4.2. Effects of EpTCP11, EpTCP25 or EpTCP26 Overexpression on the Plant Growth and Development of Arabidopsis
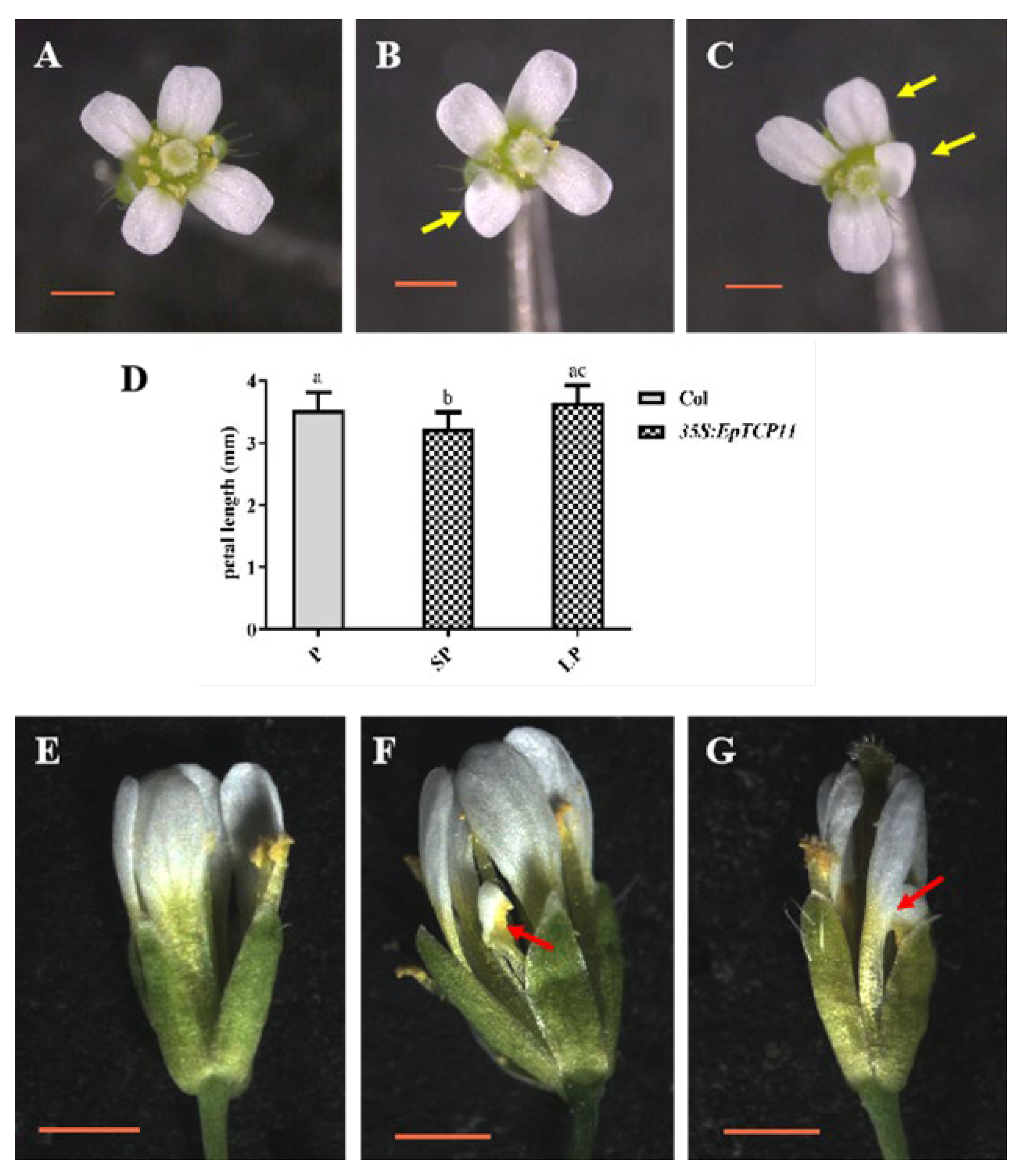
3.4.3. Effects of EpTCP11, EpTCP25 or EpTCP26 Overexpression on the Floral Organs Development of Arabidopsis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Aceto, S.; Gaudio, L. The MADS and the beauty: Genes involved in the development of orchid Flowers. Curr. Genom. 2011, 12, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Goh, C.J. Identification and characterization of three orchid MADS-box genes of the AP1/AGL9 subfamily during floral transition. Plant Physiol. 2000, 123, 1325–1336. [Google Scholar] [CrossRef]
- Hsu, H.F.; Huang, C.H.; Chou, L.T.; Yang, C.H. Ectopic expression of an orchid (Oncidium Gower Ramsey) AGL6-like gene promotes flowering by activating flowering time genes in Arabidopsis thaliana. Plant Cell Physiol. 2003, 44, 783–794. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.C.; Kuoh, C.S.; Chuang, M.H.; Chen, W.H.; Chen, H.H. Four DEF-like MADS box genes displayed distinct floral morphogenetic roles in Phalaenopsis orchid. Plant Cell Physiol. 2004, 45, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Teo, L.L.; Zhou, J.; Kumar, P.P.; Yu, H. Floral organ identity genes in the orchid Dendrobium crumenatum. Plant J. 2006, 46, 54–68. [Google Scholar] [CrossRef]
- Tsai, W.C.; Pan, Z.J.; Hsiao, Y.Y.; Jeng, M.F.; Wu, T.F.; Chen, W.H.; Chen, H.H. Interactions of B-class complex proteins involved in tepal development in Phalaenopsis orchid. Plant Cell Physiol. 2008, 49, 814–824. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.Y.; Chiu, Y.F.; Wu, J.W.; Yang, C.H. Four orchid (Oncidium Gower Ramsey) AP1/AGL9-like MADS box genes show novel expression patterns and cause different effects on floral transition and formation in Arabidopsis thaliana. Plant Cell Physiol. 2009, 50, 1425–1438. [Google Scholar] [CrossRef] [PubMed]
- Mondragón-Palomino, M.; Theissen, G. Conserved differential expression of paralogous DEFICIENS- and GLOBOSA-like MADS-box genes in the flowers of Orchidaceae: Refining the ‘orchid code’. Plant J. 2011, 66, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Shao, X.; Zhu, C.; Xu, J.; Lu, H.; Tang, Y.; Jiao, K.; Guo, W.; Xiao, W.; Liu, Z.; et al. Transcriptome-Wide Analysis Reveals the Origin of Peloria in Chinese Cymbidium (Cymbidium sinense). Plant Cell Physiol. 2018, 59, 2064–2074. [Google Scholar] [CrossRef]
- Mondragón-Palomino, M.; Theissen, G. Why are orchid flowers so diverse? Reduction of evolutionary constraints by paralogues of class B floral homeotic genes. Ann. Bot. 2009, 104, 583–594. [Google Scholar] [CrossRef]
- Hsu, H.F.; Hsu, W.H.; Lee, Y.I.; Mao, W.T.; Yang, J.Y.; Li, J.Y.; Yang, C.H. Model for perianth formation in orchids. Nat. Plants 2015, 1, 15046. [Google Scholar] [CrossRef]
- Wei, Y.; Jin, J.; Yao, X.; Lu, C.; Zhu, G.; Yang, F. Transcriptome analysis reveals clues into leaf-like flower mutant in Chinese orchid Cymbidium ensifolium. Plant Divers. 2020, 42, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.X.; Gao, J.; Wei, Y.L.; Ren, R.; Zhang, G.Q.; Lu, C.Q.; Jin, J.P.; Ai, Y.; Wang, Y.Q.; Chen, L.J.; et al. The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 2021, 19, 2501–2516. [Google Scholar] [CrossRef] [PubMed]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Horn, S.; Pabón-Mora, N.; Theuß, V.S.; Busch, A.; Zachgo, S. Analysis of the CYC/TB1 class of TCP transcription factors in basal angiosperms and magnoliids. Plant J. 2015, 81, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Carpenter, R.; Vincent, C.; Copsey, L.; Coen, E. Origin of floral asymmetry in Antirrhinum. Nature 1996, 383, 794–799. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Carpenter, R.; Copsey, L.; Vincent, C.; Clark, J.; Coen, E. Control of organ asymmetry in flowers of Antirrhinum. Cell 1999, 99, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Zhao, Z.; Tian, Z.; Xu, S.; Luo, Y.; Cai, Z.; Wang, Y.; Yang, J.; Wang, Z.; Weng, L.; et al. Control of petal shape and floral zygomorphy in Lotus japonicus. Proc. Natl. Acad. Sci. USA 2006, 103, 4970–4975. [Google Scholar] [CrossRef] [PubMed]
- Busch, A.; Zachgo, S. Control of corolla monosymmetry in the Brassicaceae Iberis amara. Proc. Natl. Acad. Sci. USA 2007, 104, 16714–16719. [Google Scholar] [CrossRef]
- Zhang, W.; Kramer, E.M.; Davis, C.C. Floral symmetry genes and the origin and maintenance of zygomorphy in a plant-pollinator mutualism. Proc. Natl. Acad. Sci. USA 2010, 107, 6388–6393. [Google Scholar] [CrossRef]
- Yang, X.; Pang, H.B.; Liu, B.L.; Qiu, Z.J.; Gao, Q.; Wei, L.; Dong, Y.; Wang, Y.Z. Evolution of double positive autoregulatory feedback loops in CYCLOIDEA2 clade genes is associated with the origin of floral zygomorphy. Plant Cell 2012, 24, 1834–1847. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Luo, Y.; Cai, Z.; Cao, X.; Hu, X.; Yang, J.; Luo, D. Functional diversity of CYCLOIDEA-like TCP genes in the control of zygomorphic flower development in Lotus japonicus. J. Integr. Plant Biol. 2013, 55, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Liu, J.; Li, P.W.; Li, C.Q.; Lü, T.F.; Yang, X.; Wang, Y.Z. Evolution of Darwin’s Peloric Gloxinia (Sinningia speciosa) is caused by a null mutation in a pleiotropic TCP Gene. Mol. Biol. Evol. 2018, 35, 1901–1915. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sun, M.; Jia, Y.; Qiu, D.; Peng, Q.; Zhuang, L. Genetic control of the lateral petal shape and identity of asymmetric flowers in mungbean (Vigna radiata L.). Front. Plant Sci. 2022, 13, 996239. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Knox, E.B.; Morden, C.W.; Cellinese, N.; Mossolem, F.; Zubair, A.S.; Howarth, D.G. Duplication and expression patterns of CYCLOIDEA-like genes in Campanulaceae. EvoDevo 2022, 13, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Liao, H.; Li, S.; Zhang, R.; Dai, J.; Ma, P.; Wang, T.; Wang, M.; Yuan, Y.; Fu, X.; et al. Delphinieae flowers originated from the rewiring of interactions between duplicated and diversified floral organ identity and symmetry genes. Plant Cell 2023, 35, 994–1012. [Google Scholar] [CrossRef] [PubMed]
- De Paolo, S.; Gaudio, L.; Aceto, S. Analysis of the TCP genes expressed in the inflorescence of the orchid Orchis italica. Sci. Rep. 2015, 5, 16265. [Google Scholar] [CrossRef]
- Liao, X.Z. Bioinformatic Analysis of TCP Genes in Orchidaceae and Cloning and Functional Study of CYC-like Genes in Phalaenopsis. Master’s Thesis, Nanjing Agricultural University, Nanjing, China, 2017. [Google Scholar]
- Lin, Y.F.; Chen, Y.Y.; Hsiao, Y.Y.; Shen, C.Y.; Hsu, J.L.; Yeh, C.M.; Mitsuda, N.; Ohme-Takagi, M.; Liu, Z.J.; Tsai, W.C. Genome-wide identification and characterization of TCP genes involved in ovule development of Phalaenopsis equestris. J. Exp. Bot. 2016, 67, 5051–5066. [Google Scholar] [CrossRef]
- Chou, M.L.; Shih, M.C.; Chan, M.T.; Liao, S.Y.; Hsu, C.T.; Haung, Y.T.; Chen, J.J.; Liao, D.C.; Wu, F.H.; Lin, C.S. Global transcriptome analysis and identification of a CONSTANS-like gene family in the orchid Erycina pusilla. Planta 2013, 237, 1425–1441. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Zhu, G.; Yang, F.; Shi, S.; Li, D.; Wang, Z.; Liu, H.; Huang, D.; Wang, C. Transcriptome characterization of Cymbidium sinense ‘dharma’ using 454 pyrosequencing and its application in the identification of genes associated with leaf color variation. PLoS ONE 2015, 10, e128592. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Martínez, J.A.; Poza-Carrión, C.; Cubas, P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 2007, 19, 458–472. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.M.; Fox, S.; Hanna, A.I.; Baxter, C.; Coen, E. Evolution of regulatory interactions controlling floral asymmetry. Development 2005, 132, 5093–5101. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, M.E.; Specht, C.D. Changes in expression pattern of the teosinte branched1-like genes in the Zingiberales provide a mechanism for evolutionary shifts in symmetry across the order. Am. J. Bot. 2011, 98, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Preston, J.C.; Hileman, L.C. Parallel evolution of TCP and B-class genes in Commelinaceae flower bilateral symmetry. EvoDevo 2012, 3, 6. [Google Scholar] [CrossRef] [PubMed]
- Hileman, L.C. Trends in flower symmetry evolution revealed through phylogenetic and developmental genetic advances. Philos. Trans. R. Soc. Lond B Biol. Sci. 2014, 369, 20130348. [Google Scholar] [CrossRef] [PubMed]
- Masuda, H.P.; Cabral, L.M.; De Veylder, L.; Tanurdzic, M.; de Almeida Engler, J.; Geelen, D.; Inzé, D.; Martienssen, R.A.; Ferreira, P.C.; Hemerly, A.S. ABAP1 is a novel plant Armadillo BTB protein involved in DNA replication and transcription. EMBO J. 2008, 27, 2746–2756. [Google Scholar] [CrossRef] [PubMed]
- Ballester, P.; Navarrete-Gómez, M.; Carbonero, P.; Oñate-Sánchez, L.; Ferrándiz, C. Leaf expansion in Arabidopsis is controlled by a TCP-NGA regulatory module likely conserved in distantly related species. Physiol. Plant 2015, 155, 21–32. [Google Scholar] [CrossRef]
- Palatnik, J.F.; Allen, E.; Wu, X.; Schommer, C.; Schwab, R.; Carrington, J.C.; Weigel, D. Control of leaf morphogenesis by microRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 2010, 22, 3574–3588. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Sato, F.; Ohme-Takagi, M. Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol. 2017, 175, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Qin, G. The Regulation of CIN-like TCP Transcription Factors. Int. J. Mol. Sci. 2020, 21, 4498. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Furutani, M.; Tasaka, M.; Ohme-Takagi, M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 2007, 19, 473–484. [Google Scholar] [CrossRef]
- Tanaka, Y.; Yamamura, T.; Oshima, Y.; Mitsuda, N.; Koyama, T.; Ohme-Takagi, M.; Terakawa, T. Creating ruffled flower petals in Cyclamen persicum by expression of the chimeric cyclamen TCP repressor. Plant Biotechnol. 2011, 28, 141–147. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, Y.-H.; Zhong, Y.-Y.; Huang, X. Identification and Functional Analysis of the Flower Development-Related TCP Genes in Erycina pusilla. Horticulturae 2024, 10, 534. https://doi.org/10.3390/horticulturae10060534
Tang Y-H, Zhong Y-Y, Huang X. Identification and Functional Analysis of the Flower Development-Related TCP Genes in Erycina pusilla. Horticulturae. 2024; 10(6):534. https://doi.org/10.3390/horticulturae10060534
Chicago/Turabian StyleTang, Yu-Huan, Ying-Yin Zhong, and Xia Huang. 2024. "Identification and Functional Analysis of the Flower Development-Related TCP Genes in Erycina pusilla" Horticulturae 10, no. 6: 534. https://doi.org/10.3390/horticulturae10060534
APA StyleTang, Y.-H., Zhong, Y.-Y., & Huang, X. (2024). Identification and Functional Analysis of the Flower Development-Related TCP Genes in Erycina pusilla. Horticulturae, 10(6), 534. https://doi.org/10.3390/horticulturae10060534





