Identification of Flavonoid 3′-Hydroxylase Genes from Red Chinese Sand Pear (Pyrus pyrifolia Nakai) and Their Regulation of Anthocyanin Accumulation in Fruit Peel
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Experimental Treatments
2.2. Research Methodology
2.2.1. Extraction of RNA and Synthesis of Complementary DNA (cDNA)
2.2.2. Isolation and Analysis of Gene Sequences
2.2.3. Extraction and Measurement of Total Anthocyanin
2.2.4. Real-Time Quantitative PCR (Q-PCR)
2.2.5. Yeast One-Hybrid (Y1H) Assay
2.2.6. Dual-Luciferase Assay
2.2.7. Statistical Analysis
3. Results
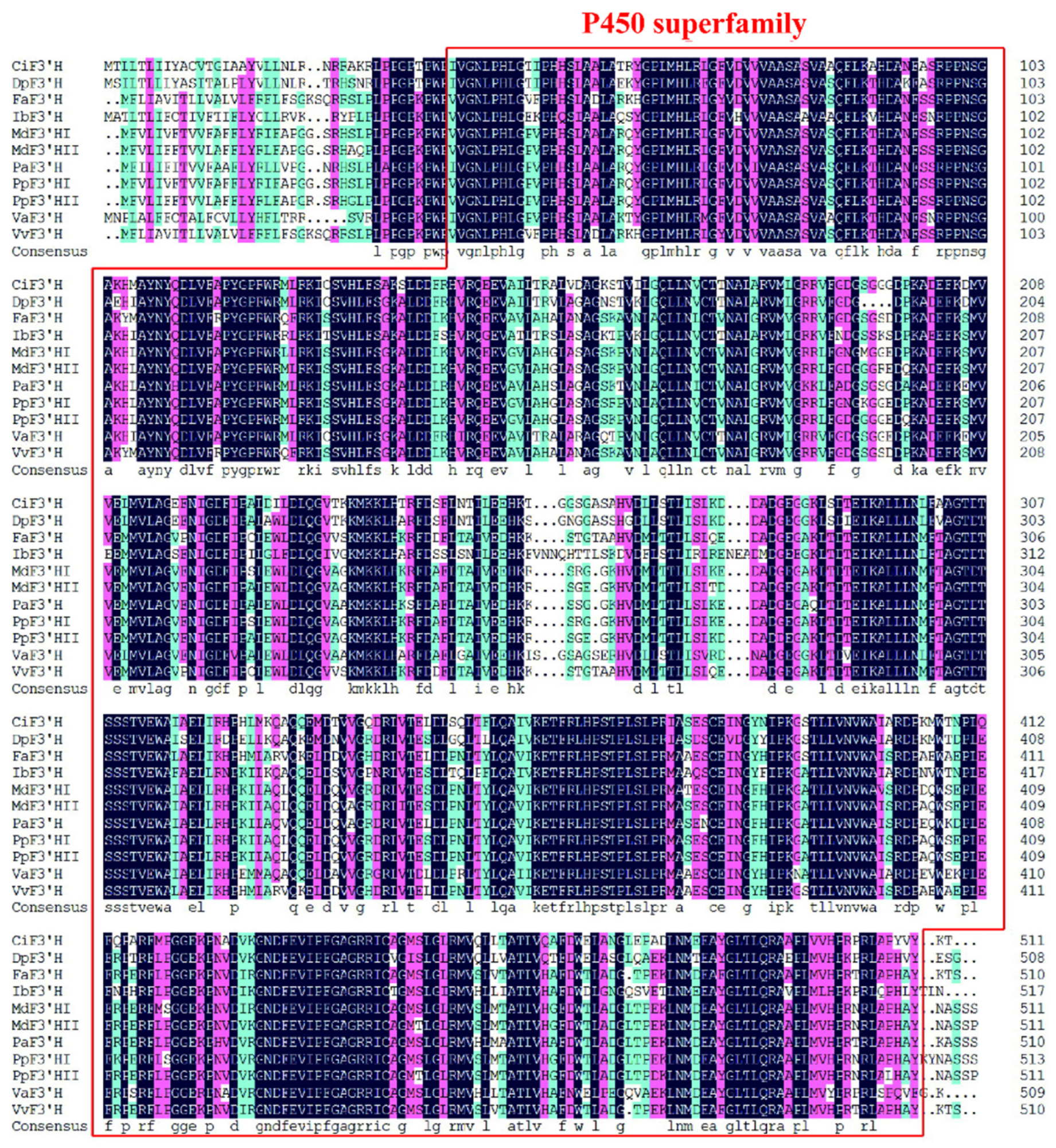
3.1. Isolation and Sequence Analysis of Pear PpF3′H Genes
3.2. Anthocyanin Concentration and Expression of PpF3′HI and PpF3′HII in Different Pear Tissues
3.3. Anthocyanin Concentration and Expression of PpF3′HI and PpF3′HII in Different Development Stages of Pear Fruit
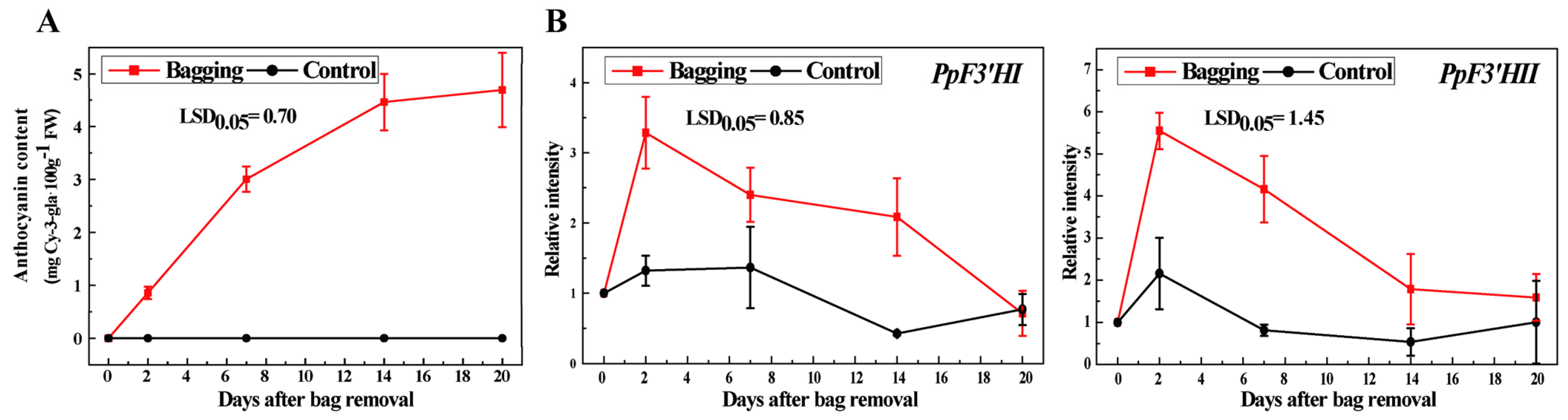
3.4. Anthocyanin Accumulation and Expression of PpF3′HI and PpF3′HII under Bagging Treatment Conditions in ‘Mantianhong’
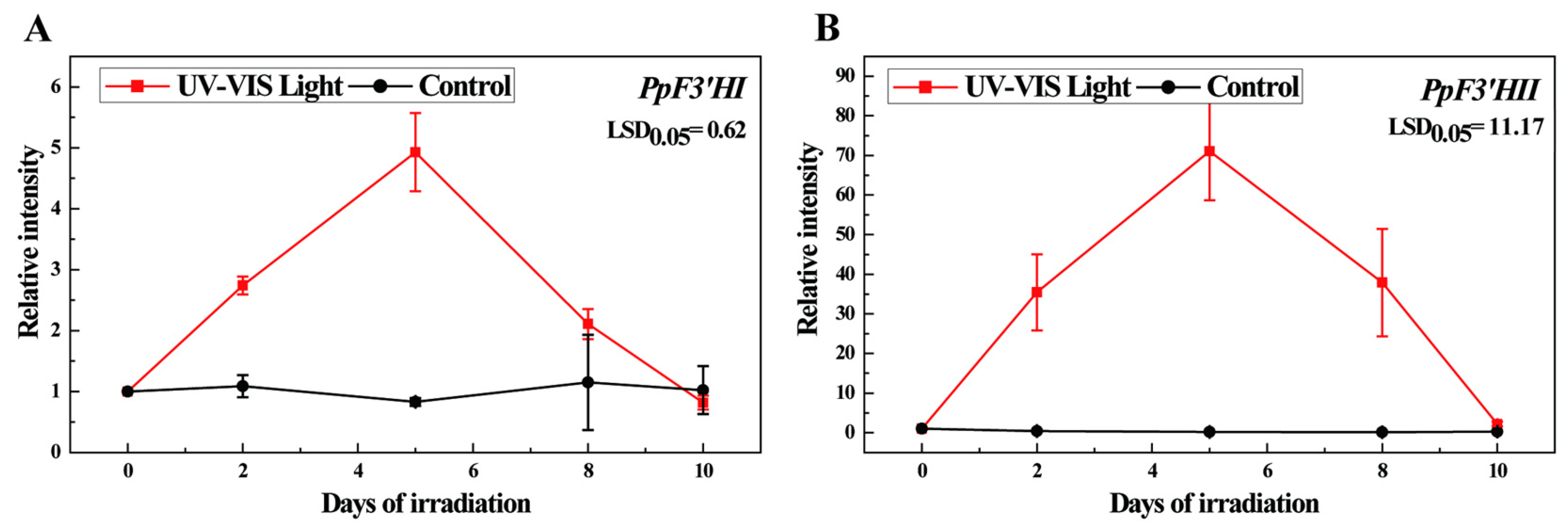
3.5. Anthocyanin Accumulation and Expression of PpF3′HI and PpF3′HII under Postharvest UV-B/Visible Irradiation Conditions in ‘Mantianhong’
3.6. PpMYB10 and PpHY5 Promote PpF3′HI Transcription
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dussi, M.C.; Sugar, D.; Wrolstad, R.E. Characterizing and Quantifying Anthocyanins in Red Pears and the Effect of Light Quality on Fruit Color. J. Am. Soc. Hortic. Sci. 1995, 120, 785–789. [Google Scholar] [CrossRef]
- Coombe, B.G. In the Regulation of Set and Development of the Grape Berry. Acta Hortic. 1973, 34, 261–274. [Google Scholar] [CrossRef]
- Steyn, W.J.; Wand, S.J.E.; Holcroft, D.M.; Jacobs, G. Red colour development and loss in pears. Int. Soc. Hortic. Sci. 2005, 1, 79–85. [Google Scholar] [CrossRef]
- Qian, M.J.; Zhang, D.; Yue, X.Y.; Wang, S.K.; Li, X.G.; Teng, Y.W. Analysis of different pigmentation patterns in ‘Mantianhong’ (Pyrus pyrifolia Nakai) and ‘Cascade’ (Pyrus communis L.) under bagging treatment and postharvest UV-B/visible irradiation conditions. Sci. Hortic. 2013, 151, 75–82. [Google Scholar] [CrossRef]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Holton, T.A.; Cornish, E.C. Genetics and Biochemistry of Anthocyanin Biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Forkmann, G. Flavonoids as Flower Pigments: The Formation of the Natural Spectrum and its Extension by Genetic Engineering. Plant Breed. 1991, 106, 1–26. [Google Scholar] [CrossRef]
- Bogs, J.; Ebadi, A.; McDavid, D.; Robinson, S.P. Identification of the flavonoid hydroxylases from grapevine and their regulation during fruit development. Plant Physiol. 2006, 140, 279–291. [Google Scholar] [CrossRef]
- Seitz, C.; Eder, C.; Deiml, B.; Kellner, S.; Martens, S.; Forkmann, G. Cloning, functional identification and sequence analysis of flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase cDNAs reveals independent evolution of flavonoid 3′,5′-hydroxylase in the Asteraceae family. Plant Mol. Biol. 2006, 61, 365–381. [Google Scholar] [CrossRef]
- Schoenbohm, C.; Martens, S.; Eder, C.; Forkmann, G.; Weisshaar, B. Identification of the Arabidopsis thaliana flavonoid 3’-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol. Chem. 2000, 381, 749–753. [Google Scholar] [CrossRef] [PubMed]
- Brugliera, F.; Barri-Rewell, G.; Holton, T.A.; Mason, J.G. Isolation and characterization of a flavonoid 3′-hydroxylase cDNA clone corresponding to the Ht1 locus of Petunia hybrida. Plant J. 1999, 19, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.P.; Vimolmangkang, S.; Soria-Guerra, R.E.; Rosales-Mendoza, S.; Zheng, D.M.; Lygin, A.V.; Korban, S.S. Ectopic Expression of Apple F3′H Genes Contributes to Anthocyanin Accumulation in the Arabidopsis tt7 Mutant Grown Under Nitrogen Stress. Plant Physiol. 2010, 153, 806–820. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Wang, Y.; Yang, S.; Xu, Y.; Chen, X. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 2010, 232, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Espley, R.V.; Brendolise, C.; Chagné, D.; Kutty-Amma, S.; Green, S.; Volz, R.; Putterill, J.; Schouten, H.J.; Gardiner, S.E.; Hellens, R.P. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell Online 2009, 21, 168–183. [Google Scholar] [CrossRef]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef]
- Kobayashi, S.; Goto-Yamamoto, N.; Hirochika, H. Retrotransposon-Induced Mutations in Grape Skin Color. Science 2004, 304, 982. [Google Scholar] [CrossRef]
- Azuma, A.; Kobayashi, S.; Goto-Yamamoto, N.; Shiraishi, M.; Mitani, N.; Yakushiji, H.; Koshita, Y. Color recovery in berries of grape (Vitis vinifera L.) ‘Benitaka’, a bud sport of ‘Italia’, is caused by a novel allele at the VvmybA1 locus. Plant Sci. 2009, 176, 470–478. [Google Scholar] [CrossRef]
- Wang, Z.; Meng, D.; Wang, A.; Li, T.; Jiang, S.; Cong, P.; Li, T. The methylation of PcMYB10 promoter is associated with Green skinned sport in ‘Max Red Bartlett’ pear. Plant Physiol. 2013, 162, 885–896. [Google Scholar] [CrossRef]
- Kui, L.-W.; Bolitho, K.; Grafton, K.; Kortstee, A.; Karunairetnam, S.; McGhie, T.K.; Espley, R.V.; Hellens, R.P.; Allan, A.C. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010, 10, 50. [Google Scholar]
- Lai, B.; Li, X.-J.; Hu, B.; Qin, Y.-H.; Huang, X.-M.; Wang, H.-C.; Hu, G.-B. LcMYB1 Is a Key Determinant of Differential Anthocyanin Accumulation among Genotypes, Tissues, Developmental Phases and ABA and Light Stimuli in Litchi chinensis. PLoS ONE 2014, 9, e86293. [Google Scholar] [CrossRef]
- Niu, S.-S.; Xu, C.-J.; Zhang, W.-S.; Zhang, B.; Li, X.; Lin-Wang, K.; Ferguson, I.B.; Allan, A.C.; Chen, K.-S. Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry (Myrica rubra) fruit by a R2R3 MYB transcription factor. Planta 2010, 231, 887–899. [Google Scholar] [CrossRef]
- Bai, S.; Sun, Y.; Qian, M.; Yang, F.; Ni, J.; Tao, R.; Li, L.; Shu, Q.; Zhang, D.; Teng, Y. Transcriptome analysis of bagging-treated red Chinese sand pear peels reveals light-responsive pathway functions in anthocyanin accumulation. Sci. Rep. 2017, 7, 63. [Google Scholar] [CrossRef]
- Ravaglia, D.; Espley, R.V.; Henry-Kirk, R.A.; Andreotti, C.; Ziosi, V.; Hellens, R.P.; Costa, G.; Allan, A.C. Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 2013, 13, 68. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Ren, L.; Lian, H.; Liu, Y.; Chen, H. Novel insight into the mechanism underlying light-controlled anthocyanin accumulation in eggplant (Solanum melongena L.). Plant Sci. 2016, 249, 46–58. [Google Scholar] [CrossRef]
- Binkert, M.; Kozma-Bognár, L.; Terecskei, K.; De Veylder, L.; Nagy, F.; Ulm, R. UV-B-Responsive Association of the Arabidopsis bZIP Transcription Factor ELONGATED HYPOCOTYL5 with Target Genes, Including Its Own Promoter. Plant Cell 2014, 26, 4200–4213. [Google Scholar] [CrossRef]
- Peng, T.; Saito, T.; Honda, C.; Ban, Y.; Kondo, S.; Liu, J.-H.; Hatsuyama, Y.; Moriguchi, T. Screening of UV-B-induced genes from apple peels by SSH: Possible involvement of MdCOP1-mediated signaling cascade genes in anthocyanin accumulation. Physiol. Plant. 2012, 148, 432–444. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Graham, S.E.; Peterson, J.A. How Similar Are P450s and What Can Their Differences Teach Us? Arch. Biochem. Biophys. 1999, 369, 24–29. [Google Scholar] [CrossRef]
- Schuler, M.A.; Werck-Reichhart, D. Functional Genomics of P450S. Annu. Rev. Plant Biol. 2003, 54, 629–667. [Google Scholar] [CrossRef]
- Werck-Reichhart, D.; Feyereisen, R. Cytochromes P450: A success story. Genome Biol. 2000, 1, 3003. [Google Scholar] [CrossRef]
- Nelson, D.R.; Schuler, M.A.; Paquette, S.M.; Werck-Reichhart, D.; Bak, S. Comparative Genomics of Rice and Arabidopsis. Analysis of 727 Cytochrome P450 Genes and Pseudogenes from a Monocot and a Dicot. Plant Physiol. 2004, 135, 756–772. [Google Scholar] [CrossRef]
- Xiao, C.C.; Li, J.M.; Yao, G.F.; Liu, J.; Hu, H.J.; Cao, Y.F.; Zhang, S.L.; Wu, J. Characteristics of components and contents of anthocyanin in peel of red-skinned pear fruits from different species. J. Nanjing Agric. Univ. 2014, 37, 60–66. [Google Scholar]
- Huang, C.H.; Yu, B.; Su, Q.; Shu, J.; Teng, Y.W. A study on coloration physiology of fruit in two red Chinese sand pear cultivars ‘Meirensu’ and ‘Yunhongli No.1’. Sci. Agric. Sin. 2010, 43, 1433–1440. [Google Scholar]
- Arakawa, O. Characteristics of color development in some apple cultivars: Changes in anthocyanin synthesis during maturation as affected by bagging and light quality. J. Jpn. Soc. Hortic. Sci. 1988, 57, 373–380. [Google Scholar] [CrossRef]
- Bai, S.; Tuan, P.A.; Saito, T.; Honda, C.; Hatsuyama, Y.; Ito, A.; Moriguchi, T. Epigenetic regulation of MdMYB1 is associated with paper bagging-induced red pigmentation of apples. Planta 2016, 244, 573–586. [Google Scholar] [CrossRef]
- Ubi, B.E.; Honda, C.; Bessho, H.; Kondo, S.; Wada, M.; Kobayashi, S.; Moriguchi, T. Expression analysis of anthocyanin biosynthetic genes in apple skin: Effect of UV-B and temperature. Plant Sci. 2006, 170, 571–578. [Google Scholar] [CrossRef]
- Sun, Y.; Qian, M.; Wu, R.; Niu, Q.; Teng, Y.; Zhang, D. Postharvest pigmentation in red Chinese sand pears (Pyrus pyrifolia Nakai) in response to optimum light and temperature. Postharvest Biol. Technol. 2014, 91, 64–71. [Google Scholar] [CrossRef]
- Li, X.D.; Wu, B.H.; Wang, L.J.; Zheng, X.B.; Yan, S.T.; Li, S.H. Changes in trans-resveratrol and other phenolic compounds in grape skin and seeds under low temperature storage after post-harvest UV-irradiation. J. Hortic. Sci. Biotechnol. 2009, 84, 113–118. [Google Scholar] [CrossRef]
- Basiouny, F.M. Effects of UV-B irradiance on storability and quality of blackberry. Proc. Fla. State Hortic. Soc. 1998, 35, 283–284. [Google Scholar]
- Kataoka, I.; Sugiyama, A.; Beppu, K. Involvement of UV rays in sweet cherry fruit coloration during maturation. Proc. IVth Int. Cherry Symp. 2005, 2, 461–466. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Tao, R.; Ni, J.; Qian, M.; Teng, Y. Identification of Flavonoid 3′-Hydroxylase Genes from Red Chinese Sand Pear (Pyrus pyrifolia Nakai) and Their Regulation of Anthocyanin Accumulation in Fruit Peel. Horticulturae 2024, 10, 535. https://doi.org/10.3390/horticulturae10060535
Zhou Y, Tao R, Ni J, Qian M, Teng Y. Identification of Flavonoid 3′-Hydroxylase Genes from Red Chinese Sand Pear (Pyrus pyrifolia Nakai) and Their Regulation of Anthocyanin Accumulation in Fruit Peel. Horticulturae. 2024; 10(6):535. https://doi.org/10.3390/horticulturae10060535
Chicago/Turabian StyleZhou, Yi, Ruiyan Tao, Junbei Ni, Minjie Qian, and Yuanwen Teng. 2024. "Identification of Flavonoid 3′-Hydroxylase Genes from Red Chinese Sand Pear (Pyrus pyrifolia Nakai) and Their Regulation of Anthocyanin Accumulation in Fruit Peel" Horticulturae 10, no. 6: 535. https://doi.org/10.3390/horticulturae10060535
APA StyleZhou, Y., Tao, R., Ni, J., Qian, M., & Teng, Y. (2024). Identification of Flavonoid 3′-Hydroxylase Genes from Red Chinese Sand Pear (Pyrus pyrifolia Nakai) and Their Regulation of Anthocyanin Accumulation in Fruit Peel. Horticulturae, 10(6), 535. https://doi.org/10.3390/horticulturae10060535








