Signaling of Plant Defense Mediated by Receptor-like Kinases, Receptor-like Cytoplasmic Protein Kinases and MAPKs Triggered by Fungal Chitin in Horticultural Crops
Abstract
1. Introduction
2. Chitin as an Inducer of the Defense Mechanism
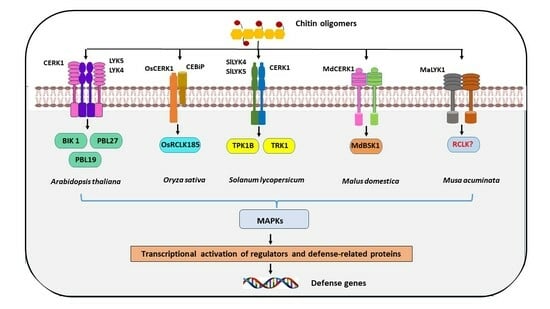
3. Plant Membrane Receptors That Perceive Chitin and Fungal Chitin Oligosaccharides
4. Receptor-Like Cytoplasmic Kinase (RLCK) Serving as a Signal Carrier
5. MAP Kinases Are Activated in Response to Chitin Stimulation
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Aune, D.; Giovannucci, E.; Boffetta, P.; Fadnes, L.T.; Keum, N.; Norat, T.; Greenwood, D.C.; Riboli, E.; Vatten, L.J.; Tonstad, S. Fruit and vegetable intake and the risk of cardiovascular disease, total cancer and all-cause mortality-a systematic review and dose-response meta-analysis of prospective studies. Int. J. Epidemiol. 2017, 46, 1029–1056. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.N.; Meng, X.; Li, Y.; Li, S.; Liu, Q.; Tang, G.Y.; Li, H.B. Fruits for Prevention and Treatment of Cardiovascular Diseases. Nutrients 2017, 9, 598. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Zhu, B.; Luo, Y.; Fu, D. Unraveling the protein network of tomato fruit in response to necrotrophic phytopathogenic Rhizopus nigricans. PLoS ONE 2013, 8, e73034. [Google Scholar] [CrossRef]
- Manzo, D.; Ferriello, F.; Puopolo, G.; Zoina, A.; D’Esposito, D.; Tardella, L.; Ferrarini, A.; Ercolano, M.R. Fusarium oxysporum f.sp. radicis-lycopersici induces distinct transcriptome reprogramming in resistant and susceptible isogenic tomato lines. BMC Plant Biol. 2016, 16, 53. [Google Scholar] [CrossRef] [PubMed]
- Alkan, N.; Fortes, A.M. Insights into molecular and metabolic events associated with fruit response to post-harvest fungal pathogens. Front. Plant Sci. 2015, 6, 889. [Google Scholar] [CrossRef] [PubMed]
- Troncoso-Rojas, R.; Tiznado-Hernández, M. Alternaria alternata (black rot, black spot). In Postharvest Decay of Fruits and Vegetables: Control Strategies; Bautista-Baños, S., Ed.; Elsevier, Inc.: Philadelphia, PA, USA, 2014; pp. 147–187. [Google Scholar]
- Takao, K.; Akagi, Y.; Tsuge, T.; Kodama, M. Functional characterization of putative G protein-coupled receptors in the tomato pathotype of Alternaria alternata. J. Gen. Plant Pathol. 2016, 82, 82–88. [Google Scholar] [CrossRef]
- Malerba, M.; Cerana, R. Recent Applications of Chitin- and Chitosan-Based Polymers in Plants. Polymers 2019, 11, 839. [Google Scholar] [CrossRef]
- Henry, G.Y.; Zamora, O.R.; Troncoso-Rojas, R.; Tiznado-Hernández, M.E.; Báez-Flores, M.E.; Carvajal-Millan, E.; Rascón-Chu, A. Toward Understanding the Molecular Recognition of Fungal Chitin and Activation of the Plant Defense Mechanism in Horticultural Crops. Molecules 2021, 26, 6513. [Google Scholar] [CrossRef]
- Abdul Malik, N.A.; Kumar, I.S.; Nadarajah, K. Elicitor and Receptor Molecules: Orchestrators of Plant Defense and Immunity. Int. J. Mol. Sci. 2020, 21, 963. [Google Scholar] [CrossRef]
- Bi, G.; Zhou, Z.; Wang, W.; Li, L.; Rao, S.; Wu, Y.; Zhang, X.; Menke, F.L.H.; Chen, S.; Zhou, J.M. Receptor-Like Cytoplasmic Kinases Directly Link Diverse Pattern Recognition Receptors to the Activation of Mitogen-Activated Protein Kinase Cascades in Arabidopsis. Plant Cell 2018, 30, 1543–1561. [Google Scholar] [CrossRef]
- Jaiswal, N.; Liao, C.J.; Mengesha, B.; Han, H.; Lee, S.; Sharon, A.; Zhou, Y.; Mengiste, T. Regulation of plant immunity and growth by tomato receptor-like cytoplasmic kinase TRK1. New Phytol. 2022, 233, 458–478. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Y.; Huang, L.; Ouyang, Z.; Hong, Y.; Zhang, H.; Li, D.; Song, F. Tomato SlMKK2 and SlMKK4 contribute to disease resistance against Botrytis cinerea. BMC Plant Biol. 2014, 14, 166. [Google Scholar] [CrossRef] [PubMed]
- Matilla, M.A. Chapter 10—Metabolic Responses of Plants Upon Different Plant–Pathogen Interactions. In Plant Metabolites and Regulation Under Environmental Stress; Ahmad, P., Ahanger, M.A., Singh, V.P., Tripathi, D.K., Alam, P., Alyemeni, M.N., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 195–214. [Google Scholar]
- Skelly, M.J.; Furniss, J.J.; Grey, H.; Wong, K.W.; Spoel, S.H. Dynamic ubiquitination determines transcriptional activity of the plant immune coactivator NPR1. eLife 2019, 8, e47005. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Wang, F.; Niran, J.; Li, N.; Yin, Y.; Yu, C.; Jiao, C.; Yao, M. Transcriptome analysis reveals defense-related genes and pathways against Xanthomonas campestris pv. vesicatoria in pepper (Capsicum annuum L.). PLoS ONE 2021, 16, e0240279. [Google Scholar] [CrossRef]
- Troncoso-Rojas, R.; Sánchez-Estrada, A.; Carvallo, T.; González-León, A.; Ojeda-Contreras, J.; Aguilar-Valenzuela, A.; Tiznado-Hernández, M.-E. A fungal elicitor enhances the resistance of tomato fruit to Fusarium oxysporum infection by activating the phenylpropanoid metabolic pathway. Phytoparasitica 2013, 41, 133–142. [Google Scholar] [CrossRef]
- Ray, S.; Mondal, S.; Chowdhury, S.; Kundu, S. Differential responses of resistant and susceptible tomato varieties to inoculation with Alternaria solani. Physiol. Mol. Plant Pathol. 2015, 90, 78–88. [Google Scholar] [CrossRef]
- Pandey, A.K.; Dinesh, K.; Sam Nirmala, N.; Kumar, A.; Chakraborti, D.; Bhattacharyya, A. Insight into tomato plant immunity to necrotrophic fungi. Curr. Res. Biotechnol. 2023, 6, 100144. [Google Scholar] [CrossRef]
- McCombe, C.L.; Greenwood, J.R.; Solomon, P.S.; Williams, S.J. Molecular plant immunity against biotrophic, hemibiotrophic, and necrotrophic fungi. Essays Biochem. 2022, 66, 581–593. [Google Scholar] [CrossRef]
- Liao, C.J.; Hailemariam, S.; Sharon, A.; Mengiste, T. Pathogenic strategies and immune mechanisms to necrotrophs: Differences and similarities to biotrophs and hemibiotrophs. Curr. Opin. Plant Biol. 2022, 69, 102291. [Google Scholar] [CrossRef]
- Bano, A.; Gupta, A.; Prusty, M.R.; Kumar, M. Elicitation of Fruit Fungi Infection and Its Protective Response to Improve the Postharvest Quality of Fruits. Stresses 2023, 3, 231–255. [Google Scholar] [CrossRef]
- Alós, E.; Rodrigo, M.J.; Zacarias, L. Chapter 7—Ripening and Senescence. In Postharvest Physiology and Biochemistry of Fruits and Vegetables; Yahia, E.M., Ed.; Woodhead Publishing: Sawston, UK, 2019; pp. 131–155. [Google Scholar]
- Ziv, C.; Zhao, Z.; Gao, Y.G.; Xia, Y. Multifunctional Roles of Plant Cuticle During Plant-Pathogen Interactions. Front. Plant Sci. 2018, 9, 1088. [Google Scholar] [CrossRef] [PubMed]
- Thole, V.; Vain, P.; Yang, R.Y.; Almeida Barros da Silva, J.; Enfissi, E.M.A.; Nogueira, M.; Price, E.J.; Alseekh, S.; Fernie, A.R.; Fraser, P.D.; et al. Analysis of Tomato Post-Harvest Properties: Fruit Color, Shelf Life, and Fungal Susceptibility. Curr. Protoc. Plant Biol. 2020, 5, e20108. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, Y.; Li, B.; Zhang, Z.; Qin, G.; Chen, T.; Tian, S. Molecular mechanisms underlying multi-level defense responses of horticultural crops to fungal pathogens. Hortic. Res. 2022, 9, uhac066. [Google Scholar] [CrossRef] [PubMed]
- Alkan, N.; Friedlander, G.; Ment, D.; Prusky, D.; Fluhr, R. Simultaneous transcriptome analysis of Colletotrichum gloeosporioides and tomato fruit pathosystem reveals novel fungal pathogenicity and fruit defense strategies. New Phytol. 2015, 205, 801–815. [Google Scholar] [CrossRef]
- Prusky, D.; Alkan, N.; Mengiste, T.; Fluhr, R. Quiescent and necrotrophic lifestyle choice during postharvest disease development. Annu. Rev. Phytopathol. 2013, 51, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Lima, S.L.; Colombo, A.L.; de Almeida Junior, J.N. Fungal Cell Wall: Emerging Antifungals and Drug Resistance. Front. Microbiol. 2019, 10, 2573. [Google Scholar] [CrossRef] [PubMed]
- Pusztahelyi, T. Chitin and chitin-related compounds in plant-fungal interactions. Mycology 2018, 9, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, M.; Ortiz, D.; Henningsen, E.C. Tactics of host manipulation by intracellular effectors from plant pathogenic fungi. Curr. Opin. Plant Biol. 2021, 62, 102054. [Google Scholar] [CrossRef]
- Henry García, Y.; Troncoso-Rojas, R.; Tiznado-Hernández, M.E.; Báez-Flores, M.E.; Carvajal-Millan, E.; Rascón-Chu, A.; Lizardi-Mendoza, J.; Martínez-Robinson, K.G. Enzymatic treatments as alternative to produce chitin fragments of low molecular weight from Alternaria alternata. J. Appl. Polym. Sci. 2019, 136, 47339. [Google Scholar] [CrossRef]
- FDA. Fact Sheet for Chitin; Poly-N-acetyl-D-glucosamine; US EPA, Office of Pesticide Programs: Washington, DC, USA, 2011; Volume 2.
- Zhang, H.; Yang, Q.; Ge, L.; Zhang, G.; Zhang, X.; Zhang, X. Chitin enhances biocontrol of Rhodotorula mucilaginosa to postharvest decay of peaches. Int. J. Biol. Macromol. 2016, 88, 465–475. [Google Scholar] [CrossRef]
- Fu, D.; Xiang, H.; Yu, C.; Zheng, X.; Yu, T. Colloidal chitin reduces disease incidence of wounded pear fruit inoculated by Penicillium expansum. Postharvest Biol. Technol. 2016, 111, 1–5. [Google Scholar] [CrossRef]
- Sun, C.; Fu, D.; Jin, L.; Chen, M.; Zheng, X.; Yu, T. Chitin isolated from yeast cell wall induces the resistance of tomato fruit to Botrytis cinerea. Carbohydr. Polym. 2018, 199, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Valle-Sotelo, E.; Troncoso-Rojas, R.; Tiznado-Hernández, M.; Carvajal-Millan, E.; Estrada, A.; García, Y. Bioefficacy of fungal chitin oligomers in the control of postharvest decay in tomato fruit. Int. Food Res. J. 2022, 29, 1131–1142. [Google Scholar] [CrossRef]
- Zhou, Q.; Liu, J.; Wang, J.; Chen, S.; Chen, L.; Wang, J.; Wang, H.B.; Liu, B. The juxtamembrane domains of Arabidopsis CERK1, BAK1, and FLS2 play a conserved role in chitin-induced signaling. J. Integr. Plant Biol. 2020, 62, 556–562. [Google Scholar] [CrossRef]
- Macho, A.P.; Zipfel, C. Plant PRRs and the activation of innate immune signaling. Mol. Cell 2014, 54, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Buendia, L.; Girardin, A.; Wang, T.; Cottret, L.; Lefebvre, B. LysM Receptor-Like Kinase and LysM Receptor-Like Protein Families: An Update on Phylogeny and Functional Characterization. Front. Plant Sci. 2018, 9, 1531. [Google Scholar] [CrossRef] [PubMed]
- Petutschnig, E.K.; Jones, A.M.; Serazetdinova, L.; Lipka, U.; Lipka, V. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 2010, 285, 28902–28911. [Google Scholar] [CrossRef] [PubMed]
- Willmann, R.; Lajunen, H.M.; Erbs, G.; Newman, M.A.; Kolb, D.; Tsuda, K.; Katagiri, F.; Fliegmann, J.; Bono, J.J.; Cullimore, J.V.; et al. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. USA 2011, 108, 19824–19829. [Google Scholar] [CrossRef]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef]
- Wan, J.; Zhang, X.C.; Neece, D.; Ramonell, K.M.; Clough, S.; Kim, S.Y.; Stacey, M.G.; Stacey, G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 2008, 20, 471–481. [Google Scholar] [CrossRef]
- Cao, Y.; Liang, Y.; Tanaka, K.; Nguyen, C.; Jedrzejczak, R.; Joachimiak, A.; Stacey, G. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 2014, 3, e03766. [Google Scholar] [CrossRef] [PubMed]
- Erwig, J.; Ghareeb, H.; Kopischke, M.; Hacke, R.; Matei, A.; Petutschnig, E.; Lipka, V. Chitin-induced and CHITIN ELICITOR RECEPTOR KINASE1 (CERK1) phosphorylation-dependent endocytosis of Arabidopsis thaliana LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE5 (LYK5). New Phytol. 2017, 215, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wang, E.; Liu, J. CERK1, more than a co-receptor in plant-microbe interactions. New Phytol. 2022, 234, 1606–1613. [Google Scholar] [CrossRef] [PubMed]
- Kaku, H.; Nishizawa, Y.; Ishii-Minami, N.; Akimoto-Tomiyama, C.; Dohmae, N.; Takio, K.; Minami, E.; Shibuya, N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 11086–11091. [Google Scholar] [CrossRef] [PubMed]
- Hayafune, M.; Berisio, R.; Marchetti, R.; Silipo, A.; Kayama, M.; Desaki, Y.; Arima, S.; Squeglia, F.; Ruggiero, A.; Tokuyasu, K.; et al. Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad. Sci. USA 2014, 111, E404–E413. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Tian, Y.; Cong, P.; Zhu, Y. Functional characterization of an apple (Malus x domestica) LysM domain receptor encoding gene for its role in defense response. Plant Sci. 2018, 269, 56–65. [Google Scholar] [CrossRef]
- Chen, Q.; Dong, C.; Sun, X.; Zhang, Y.; Dai, H.; Bai, S. Overexpression of an apple LysM-containing protein gene, MdCERK1-2, confers improved resistance to the pathogenic fungus, Alternaria alternata, in Nicotiana benthamiana. BMC Plant Biol. 2020, 20, 146. [Google Scholar] [CrossRef]
- Zhang, L.; Yuan, L.; Staehelin, C.; Li, Y.; Ruan, J.; Liang, Z.; Xie, Z.; Wang, W.; Xie, J.; Huang, S. The LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE 1 protein of banana is required for perception of pathogenic and symbiotic signals. New Phytol. 2019, 223, 1530–1546. [Google Scholar] [CrossRef]
- Brulé, D.; Villano, C.; Davies, L.J.; Trdá, L.; Claverie, J.; Héloir, M.C.; Chiltz, A.; Adrian, M.; Darblade, B.; Tornero, P.; et al. The grapevine (Vitis vinifera) LysM receptor kinases VvLYK1-1 and VvLYK1-2 mediate chitooligosaccharide-triggered immunity. Plant Biotechnol. J. 2019, 17, 812–825. [Google Scholar] [CrossRef]
- Roudaire, T.; Marzari, T.; Landry, D.; Löffelhardt, B.; Gust, A.A.; Jermakow, A.; Dry, I.; Winckler, P.; Héloir, M.C.; Poinssot, B. The grapevine LysM receptor-like kinase VvLYK5-1 recognizes chitin oligomers through its association with VvLYK1-1. Front. Plant Sci. 2023, 14, 1130782. [Google Scholar] [CrossRef]
- Zeng, L.; Velasquez, A.C.; Munkvold, K.R.; Zhang, J.; Martin, G.B. A tomato LysM receptor-like kinase promotes immunity and its kinase activity is inhibited by AvrPtoB. Plant J. 2012, 69, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Liao, D.; Sun, X.; Wang, N.; Song, F.; Liang, Y. Tomato LysM Receptor-Like Kinase SlLYK12 Is Involved in Arbuscular Mycorrhizal Symbiosis. Front. Plant Sci. 2018, 9, 1004. [Google Scholar] [CrossRef] [PubMed]
- Ai, Y.; Li, Q.; Li, C.; Wang, R.; Sun, X.; Chen, S.; Cai, X.Z.; Qi, X.; Liang, Y. Tomato LysM receptor kinase 4 mediates chitin-elicited fungal resistance in both leaves and fruit. Hortic. Res. 2023, 10, uhad082. [Google Scholar] [CrossRef]
- García, Y.H.; Troncoso-Rojas, R.; Báez-Flores, M.E.; Hernández-Oñate, M.Á.; Tiznado-Hernández, M.E. RNA-Seq of Tomato Fruit-Alternaria Chitin Oligomer Interaction Reveals Genes Encoding Chitin Membrane Receptors and the Activation of the Defense Response. Horticulturae 2023, 9, 1064. [Google Scholar] [CrossRef]
- Liang, X.; Zhou, J.M. Receptor-Like Cytoplasmic Kinases: Central Players in Plant Receptor Kinase-Mediated Signaling. Annu. Rev. Plant Biol. 2018, 69, 267–299. [Google Scholar] [CrossRef] [PubMed]
- Kanda, Y.; Shinya, T.; Maeda, S.; Mujiono, K.; Hojo, Y.; Tomita, K.; Okada, K.; Kamakura, T.; Galis, I.; Mori, M. BSR1, a Rice Receptor-like Cytoplasmic Kinase, Positively Regulates Defense Responses to Herbivory. Int. J. Mol. Sci. 2023, 24, 10395. [Google Scholar] [CrossRef]
- Liu, J.; Li, W.; Wu, G.; Ali, K. An update on evolutionary, structural, and functional studies of receptor-like kinases in plants. Front. Plant Sci. 2024, 15, 1305599. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, J. Regulatory role of receptor-like cytoplasmic kinases in early immune signaling events in plants. FEMS Microbiol. Rev. 2020, 44, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Veronese, P.; Nakagami, H.; Bluhm, B.; Abuqamar, S.; Chen, X.; Salmeron, J.; Dietrich, R.A.; Hirt, H.; Mengiste, T. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 2006, 18, 257–273. [Google Scholar] [CrossRef]
- Wang, N.; Liu, Y.; Dong, C.; Zhang, Y.; Bai, S. MdMAPKKK1 Regulates Apple Resistance to Botryosphaeria dothidea by Interacting with MdBSK1. Int. J. Mol. Sci. 2022, 23, 4415. [Google Scholar] [CrossRef]
- Li, W.; Liao, C.-J.; Bluhm, B.H.; Mengiste, T.; Woloshuk, C.P. A Maize (Zea mays L.) BIK1-Like Receptor-Like Cytoplasmic Kinase Contributes to Disease Resistance. Plant Mol. Biol. Rep. 2022, 40, 28–42. [Google Scholar] [CrossRef]
- Kim, D.S.; Kim, N.H.; Hwang, B.K. The Capsicum annuum class IV chitinase ChitIV interacts with receptor-like cytoplasmic protein kinase PIK1 to accelerate PIK1-triggered cell death and defence responses. J. Exp. Bot. 2015, 66, 1987–1999. [Google Scholar] [CrossRef] [PubMed]
- Vij, S.; Giri, J.; Dansana, P.K.; Kapoor, S.; Tyagi, A.K. The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: Organization, phylogenetic relationship, and expression during development and stress. Mol. Plant 2008, 1, 732–750. [Google Scholar] [CrossRef] [PubMed]
- Abuqamar, S.; Chai, M.F.; Luo, H.; Song, F.; Mengiste, T. Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell 2008, 20, 1964–1983. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, S.; Chen, L.; Zhou, Q.; Wang, M.; Feng, D.; Li, J.F.; Wang, J.; Wang, H.B.; Liu, B. BIK1 cooperates with BAK1 to regulate constitutive immunity and cell death in Arabidopsis. J. Integr. Plant Biol. 2017, 59, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Wan, W.L.; Zhang, L.; Pruitt, R.; Zaidem, M.; Brugman, R.; Ma, X.; Krol, E.; Perraki, A.; Kilian, J.; Grossmann, G.; et al. Comparing Arabidopsis receptor kinase and receptor protein-mediated immune signaling reveals BIK1-dependent differences. New Phytol. 2019, 221, 2080–2095. [Google Scholar] [CrossRef] [PubMed]
- Shinya, T.; Yamaguchi, K.; Desaki, Y.; Yamada, K.; Narisawa, T.; Kobayashi, Y.; Maeda, K.; Suzuki, M.; Tanimoto, T.; Takeda, J.; et al. Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J. 2014, 79, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Yamaguchi, K.; Shirakawa, T.; Nakagami, H.; Mine, A.; Ishikawa, K.; Fujiwara, M.; Narusaka, M.; Narusaka, Y.; Ichimura, K.; et al. The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J. 2016, 35, 2468–2483. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Maierhofer, T.; Rybak, K.; Sklenar, J.; Breakspear, A.; Johnston, M.G.; Fliegmann, J.; Huang, S.; Roelfsema, M.R.G.; Felix, G.; et al. Anion channel SLAH3 is a regulatory target of chitin receptor-associated kinase PBL27 in microbial stomatal closure. eLife 2019, 8, e44474. [Google Scholar] [CrossRef]
- Gong, B.Q.; Wang, F.Z.; Li, J.F. Hide-and-Seek: Chitin-Triggered Plant Immunity and Fungal Counterstrategies. Trends Plant Sci. 2020, 25, 805–816. [Google Scholar] [CrossRef]
- Lal, N.K.; Nagalakshmi, U.; Hurlburt, N.K.; Flores, R.; Bak, A.; Sone, P.; Ma, X.; Song, G.; Walley, J.; Shan, L.; et al. The Receptor-like Cytoplasmic Kinase BIK1 Localizes to the Nucleus and Regulates Defense Hormone Expression during Plant Innate Immunity. Cell Host Microbe 2018, 23, 485–497.e485. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Zhou, Z.; Miao, P.; Bi, G.; Hu, M.; Wu, Y.; Feng, F.; Zhang, X.; Zhou, J.M. Roles of Receptor-Like Cytoplasmic Kinase VII Members in Pattern-Triggered Immune Signaling. Plant Physiol. 2018, 177, 1679–1690. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhao, Y.; Shi, H.; Li, J.; Wang, Y.; Tang, D. BRASSINOSTEROID-SIGNALING KINASE1 Phosphorylates MAPKKK5 to Regulate Immunity in Arabidopsis. Plant Physiol. 2018, 176, 2991–3002. [Google Scholar] [CrossRef] [PubMed]
- Bastedo, D.P.; Khan, M.; Martel, A.; Seto, D.; Kireeva, I.; Zhang, J.; Masud, W.; Millar, D.; Lee, J.Y.; Lee, A.H.; et al. Perturbations of the ZED1 pseudokinase activate plant immunity. PLoS Pathog. 2019, 15, e1007900. [Google Scholar] [CrossRef] [PubMed]
- Thor, K.; Jiang, S.; Michard, E.; George, J.; Scherzer, S.; Huang, S.; Dindas, J.; Derbyshire, P.; Leitão, N.; DeFalco, T.A.; et al. The calcium-permeable channel OSCA1.3 regulates plant stomatal immunity. Nature 2020, 585, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Uemura, T.; Hachisu, M.; Desaki, Y.; Ito, A.; Hoshino, R.; Sano, Y.; Nozawa, A.; Mujiono, K.; Galis, I.; Yoshida, A.; et al. Soy and Arabidopsis receptor-like kinases respond to polysaccharide signals from Spodoptera species and mediate herbivore resistance. Commun. Biol. 2020, 3, 224. [Google Scholar] [CrossRef]
- Wang, Z.; Gou, X. The First Line of Defense: Receptor-like Protein Kinase-Mediated Stomatal Immunity. Int. J. Mol. Sci. 2021, 23, 343. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, K.; Tan, M.; Lei, C.; Cao, S. Involvement of a receptor-like kinase complex of FvFLS2 and FvBAK1 in brassinosteroids-induced immunity in postharvest strawberry fruit. Postharvest Biol. Technol. 2023, 198, 112266. [Google Scholar] [CrossRef]
- Maeda, S.; Ackley, W.; Yokotani, N.; Sasaki, K.; Ohtsubo, N.; Oda, K.; Mori, M. Enhanced Resistance to Fungal and Bacterial Diseases Due to Overexpression of BSR1, a Rice RLCK, in Sugarcane, Tomato, and Torenia. Int. J. Mol. Sci. 2023, 24, 3644. [Google Scholar] [CrossRef]
- Xu, S.; Liao, C.J.; Jaiswal, N.; Lee, S.; Yun, D.J.; Lee, S.Y.; Garvey, M.; Kaplan, I.; Mengiste, T. Tomato PEPR1 ORTHOLOG RECEPTOR-LIKE KINASE1 Regulates Responses to Systemin, Necrotrophic Fungi, and Insect Herbivory. Plant Cell 2018, 30, 2214–2229. [Google Scholar] [CrossRef]
- Xiao, X.; Wang, R.; Guo, W.; Khaskhali, S.; Fan, R.; Zhao, R.; Li, C.; He, C.; Niu, X.; Chen, Y. The receptor-like cytoplasmic kinase OsRLCK118 regulates plant development and basal immunity in rice (Oryza sativa L.). Tropical Plants 2022, 1, 4. [Google Scholar] [CrossRef]
- Yamaguchi, K.; Yoshimura, Y.; Nakagawa, S.; Mezaki, H.; Yoshimura, S.; Kawasaki, T. OsDRE2 contributes to chitin-triggered response through its interaction with OsRLCK185. Biosci. Biotechnol. Biochem. 2019, 83, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Hwang, B.K. The pepper receptor-like cytoplasmic protein kinase CaPIK1 is involved in plant signaling of defense and cell-death responses. Plant J. 2011, 66, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Wiweger, M.; Farbos, I.; Ingouff, M.; Lagercrantz, U.; Von Arnold, S. Expression of Chia4-Pa chitinase genes during somatic and zygotic embryo development in Norway spruce (Picea abies): Similarities and differences between gymnosperm and angiosperm class IV chitinases. J. Exp. Bot. 2003, 54, 2691–2699. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Wang, C.; Wang, H.; Li, L.; Wang, C. The Function of MAPK Cascades in Response to Various Stresses in Horticultural Plants. Front. Plant Sci. 2020, 11, 952. [Google Scholar] [CrossRef] [PubMed]
- Hamel, L.P.; Nicole, M.C.; Duplessis, S.; Ellis, B.E. Mitogen-activated protein kinase signaling in plant-interacting fungi: Distinct messages from conserved messengers. Plant Cell 2012, 24, 1327–1351. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Su, J.; Zhang, Y.; Xu, J.; Zhang, S. Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr. Opin. Plant Biol. 2018, 45, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cheval, C.; Samwald, S.; Johnston, M.G.; de Keijzer, J.; Breakspear, A.; Liu, X.; Bellandi, A.; Kadota, Y.; Zipfel, C.; Faulkner, C. Chitin perception in plasmodesmata characterizes submembrane immune-signaling specificity in plants. Proc. Natl. Acad. Sci. USA 2020, 117, 9621–9629. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Yamada, K.; Yoshimura, S.; Yamaguchi, K. Chitin receptor-mediated activation of MAP kinases and ROS production in rice and Arabidopsis. Plant Signal. Behav. 2017, 12, e1361076. [Google Scholar] [CrossRef]
- Yamada, K.; Yamaguchi, K.; Yoshimura, S.; Terauchi, A.; Kawasaki, T. Conservation of Chitin-Induced MAPK Signaling Pathways in Rice and Arabidopsis. Plant Cell Physiol. 2017, 58, 993–1002. [Google Scholar] [CrossRef]
- de Oliveira, M.L.; de Lima Silva, C.C.; Abe, V.Y.; Costa, M.G.; Cernadas, R.A.; Benedetti, C.E. Increased resistance against citrus canker mediated by a citrus mitogen-activated protein kinase. Mol. Plant-Microbe Interact. MPMI 2013, 26, 1190–1199. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Lovato, A.; Polverari, A.; Wang, M.; Liang, Y.H.; Ma, Y.C.; Cheng, Z.M. Genome-wide identification and analysis of mitogen activated protein kinase kinase kinase gene family in grapevine (Vitis vinifera). BMC Plant Biol. 2014, 14, 219. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Wang, J.; Cheng, L.; Liu, S.; Wu, J.; Peng, Z.; Lu, G. Genome-wide analysis of the mitogen-activated protein kinase gene family in Solanum lycopersicum. Gene 2012, 499, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.D.; Félix, M.D.R.; Patanita, M.; Materatski, P.; Varanda, C. High throughput sequencing unravels tomato-pathogen interactions towards a sustainable plant breeding. Hortic. Res. 2021, 8, 171. [Google Scholar] [CrossRef] [PubMed]

| RLCK a | Associated RLK Receptors | Species | Elicitor | RLCK Function | References |
|---|---|---|---|---|---|
| PBL27 | CERK1 | Arabidopsis | Chitin oligomers | Pattern-triggered immunity | [71,72] |
| BIK1 | EFR/BAK1 | Arabidopsis | Pseudomonas syringae | Regulation of JA and SA levels (Interacts with WRKY in the nucleus) | [75] |
| PBL27 | CERK1-LYK5 | Arabidopsis | Fungal chitin | Stomatal-mediated immunity | [73] |
| PORK1-2 | CERK1 | Arabidopsis | P. syringae | Increased ROS production | [76] |
| BSK1 | FLS2 | Arabidopsis | P. syringae | Activation of MAP kinases in the presence of flg22 | [77] |
| ZED1/PBL | ZAR1 | Arabidopsis | P. syringae | They act by forming a complex as part of the defense response against P. syringae | [78] |
| BIK1 | OSCA1.3 | Arabidopsis | Synthetic flg22 | Enables stomatal closure during the immune response to bacterial flagellin | [79] |
| PBL27 | HAK1 | Arabidopsis/Glycine max | Spodoptera litura | Defense response against Spodoptera litura | [80] |
| BIK1 | FLS2/BAK1 | Arabidopsis | P. syringae | Stomatal-mediated immunity | [81] |
| FvBIK1 | FvBAK1 | Fragaria vesca | Brassinosteroid | Provides immunity to the necrotrophic fungus B. cinerea | [82] |
| BSR1 | CERK1 | Saccharum officinarum | Sporisorium scitamineum | Transgenics that overexpress BSR1 are resistant to Sporisorium scitamineum | [83] |
| TPK1b | PORK1 | Solanum lycopersicum | B. cinerea | Regulates the production of jasmonic acid in the systemin-mediated defense response. | [84] |
| TRK1 | SlLYK1 | S. lycopersicum | Commercial chitin | Induces fungal resistance and ROS production in the presence of chitin | [12] |
| BSR1 | Unknown | S. lycopersicum | Rhizoctonia solani | Transgenics that overexpress BSR1 are resistant to Rhizoctonia solani | [83] |
| OsRLCK118 | Unknown - | O. sativa | Xanthomonas oryzae | Silencing of OsRLCK118 increases susceptibility to Xanthomonas oryzae | [85] |
| BSR1 | PEPR1/PEPR2 | O. sativa | Mythimna loreyi | BSR1 mediates the activation of defense responses against chewing herbivores in monocot rice | [60] |
| BIK1 | Unknown | Zea mays | Clavibacter michiganensis subsp. nebraskensis | Defense response against pathogenic bacteria | [65] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reyes Zamora, O.; Troncoso-Rojas, R.; Báez-Flores, M.E.; Tiznado-Hernández, M.E.; Rascón-Chu, A. Signaling of Plant Defense Mediated by Receptor-like Kinases, Receptor-like Cytoplasmic Protein Kinases and MAPKs Triggered by Fungal Chitin in Horticultural Crops. Horticulturae 2024, 10, 361. https://doi.org/10.3390/horticulturae10040361
Reyes Zamora O, Troncoso-Rojas R, Báez-Flores ME, Tiznado-Hernández ME, Rascón-Chu A. Signaling of Plant Defense Mediated by Receptor-like Kinases, Receptor-like Cytoplasmic Protein Kinases and MAPKs Triggered by Fungal Chitin in Horticultural Crops. Horticulturae. 2024; 10(4):361. https://doi.org/10.3390/horticulturae10040361
Chicago/Turabian StyleReyes Zamora, Orlando, Rosalba Troncoso-Rojas, María Elena Báez-Flores, Martín Ernesto Tiznado-Hernández, and Agustín Rascón-Chu. 2024. "Signaling of Plant Defense Mediated by Receptor-like Kinases, Receptor-like Cytoplasmic Protein Kinases and MAPKs Triggered by Fungal Chitin in Horticultural Crops" Horticulturae 10, no. 4: 361. https://doi.org/10.3390/horticulturae10040361
APA StyleReyes Zamora, O., Troncoso-Rojas, R., Báez-Flores, M. E., Tiznado-Hernández, M. E., & Rascón-Chu, A. (2024). Signaling of Plant Defense Mediated by Receptor-like Kinases, Receptor-like Cytoplasmic Protein Kinases and MAPKs Triggered by Fungal Chitin in Horticultural Crops. Horticulturae, 10(4), 361. https://doi.org/10.3390/horticulturae10040361