Genomic Insight into a Potential Biological Control Agent for Fusarium-Related Diseases in Potatoes: Bacillus cabrialesii Subsp. cabrialesii Strain PE1
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria Isolation and Culture Conditions
2.2. Genomic Analysis
2.3. Genome Annotation and Genome Mining
2.4. Evaluation of the Antagonistic Activity of Strain PE1 against Fusarium languescens CE2 through Extracellular Metabolites
3. Results
3.1. Genomic Analysis
3.2. Genome Annotation
3.3. Genome Mining
3.4. Antagonistic Activity of Bacillus cabrialesii subsp. cabrialesii PE1 against Fusarium Languescens CE2, through Extracellular Metabolites
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Calıskan, M.E.; Bakhsh, A.; Jabran, K. (Eds.) Potato Production Worldwide; Academic Press: Cambridge, MA, USA, 2022. [Google Scholar]
- Waheed, A.; Chuang, L.; Murad, M.; Mushtaq, A.; Khalid, A.K.; Hamed, A.; Ghramh Zhongwei, W.; Daoyuan, Z. Sustainable Potato Growth under Straw Mulching Practices. Sustainability 2023, 15, 10442. [Google Scholar] [CrossRef]
- Khedher, S.B.; Mejdoub-Trabelsi, B.; Tounsi, S. Biological potential of Bacillus subtilis V26 for the control of Fusarium wilt and tuber dry rot on potato caused by Fusarium species and the promotion of plant growth. Biol. Control. 2021, 152, 104444. [Google Scholar] [CrossRef]
- García-Ávila, C.J.; Valenzuela-Tirado, G.A.; Florencio Anastasio, J.G.; Ruiz-Galván, I.; Moreno-Velázquez, M.; Hernández-Macías, B.; López-Buenfil, J.A.; Bravo-Pérez, D.; Pineda-Ríos, J.M.; Quezada-Salinas, A.; et al. Organisms associated with damage to post-harvest potato tubers. Rev. Mex. Fitopatol. 2018, 36, 308–320. [Google Scholar]
- Lahlali, R.; Ezrari, S.; Radouane, N.; Kenfaoui, J.; Esmaeel, Q.; El Hamss, H.; Belabess, Z.; Barka, E.A. Biological Control of Plant Pathogens: A Global Perspective. Microorganisms 2022, 10, 596. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, R.K.; Kumar, R.; Sharma, S.; Sagar, V.; Aggarwal, R.; Naga, K.C.; Lal, M.C.; Chourasia, K.N.; Kumar, D.; Kumar, M. Potato dry rot disease: Current status, pathogenomics and management. 3 Biotech 2020, 10, 503. [Google Scholar] [CrossRef] [PubMed]
- Ferrigo, D.; Raiola, A.; Causin, R. Fusarium toxins in cereals: Occurrence, legislation, factors promoting the appearance and their management. Molecules 2016, 21, 627. [Google Scholar] [CrossRef] [PubMed]
- Pfordt, A.; Ramos Romero, L.; Schiwek, S.; Karlovsky, P.; von Tiedemann, A. Impact of environmental conditions and agronomic practices on the prevalence of Fusarium species associated with ear-and stalk rot in maize. Pathogens 2020, 9, 236. [Google Scholar] [CrossRef]
- Montoya-Martínez, A.C.; Figueroa-Brambila, K.M.; Escalante-Beltrán, A.; López-Montoya, N.D.; Valenzuela-Ruíz, V.; Parra-Cota, F.I.; Estrada Alvarado, M.I.; de los Santos-Villalobos, S. Biological Control Mechanisms of Bacillus cabrialesii subsp. tritici TSO2T against Fusarium languescens, the Causal Agent of Wilt in Jalapeño Peppers. Horticulturae 2023, 9, 964. [Google Scholar] [CrossRef]
- Cota-Barreras, C.I.; García-Estrada, R.S.; León-Félix, J.; Valenzuela-Herrera, V.; Mora-Romero, G.A.; Leyva-Madrigal, K.Y.; Tovar-Pedraza, J.M. Phylogeny, distribution, and pathogenicity of fusarioid fungi associated with chickpea wilt in Sinaloa and Sonora, Mexico. Available online: https://doi.org/10.21203/rs.3.rs-2960826/v1 (accessed on 7 July 2023).
- de Chaves, M.A.; Reginatto, P.; da Costa, B.S.; de Paschoal, R.I.; Teixeira, M.L.; Fuentefria, A.M. Fungicide resistance in Fusarium graminearum species complex. Curr. Microbiol. 2022, 79, 62. [Google Scholar] [CrossRef]
- Mao, Y.; Zhang, Z.; Shen, J.; Yin, X.; Wang, T.; Zheng, X.; Duan, Y. The intrinsic resistance of Fusarium solani to the Fusarium-specific fungicide phenamacril is attributed to the natural variation of both T218S and K376M in myosin5. Pestic. Biochem. Physiol. 2023, 196, 105595. [Google Scholar] [CrossRef]
- Zhang, N.; Xu, Y.; Zhang, Q.; Zhao, L.; Zhu, Y.; Wu, Y.; Li, Z.; Yang, W. Detection of fungicide resistance to fludioxonil and tebuconazole in Fusarium pseudograminearum, the causal agent of Fusarium crown rot in wheat. PeerJ 2023, 11, e14705. [Google Scholar] [CrossRef] [PubMed]
- Mir, S.A.; Padhiary, A.; Ekka, N.J.; Baitharu, I.; Nayak, B. Environmental impacts of synthetic and biofungicides. In Current Developments in Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2023; pp. 487–504. [Google Scholar] [CrossRef]
- Sulaiman, M.A.; Bello, S.K. Biological control of soil-borne pathogens in arid lands: A review. J. Plant Dis. Prot. 2023, 131, 293–313. [Google Scholar] [CrossRef]
- Ramlawi, S.; Chiu, J.O.; Cloutier, A.; Avis, T.J. Suppression of Fusarium dry rot of potato using beneficial bacterial treatments. J. Plant Pathol. 2021, 103, 269–281. [Google Scholar] [CrossRef]
- Mnif, I.; Hammami, I.; Triki, M.A.; Azabou, M.C.; Ellouze-Chaabouni, S.; Ghribi, D. Antifungal efficiency of a lipopeptide biosurfactant derived from Bacillus subtilis SPB1 versus the phytopathogenic fungus, Fusarium solani. Environ. Sci. Pollut. Res. Int. 2015, 22, 18137–18147. [Google Scholar] [CrossRef] [PubMed]
- Etesami, H.; Jeong, B.R.; Glick, B.R. Biocontrol of plant diseases by Bacillus spp. Physiol. Mol. Plant Pathol. 2023, 126, 102048. [Google Scholar] [CrossRef]
- Chen, K.; Tian, Z.; He, H.; Long, C.A.; Jiang, F. Bacillus species as potential biocontrol agents against citrus diseases. Biol. Control. 2020, 151, 104419. [Google Scholar] [CrossRef]
- Miljaković, D.; Marinković, J.; Balešević-Tubić, S. The significance of Bacillus spp. in disease suppression and growth promotion of field and vegetable crops. Microorganisms 2020, 8, 1037. [Google Scholar] [CrossRef] [PubMed]
- Shimbori, E.M.; Querino, R.B.; Costa, V.A.; Zucchi, R.A. Taxonomy and biological control: New challenges in an old relationship. Neotrop. Entomol. 2023, 52, 351–372. [Google Scholar] [CrossRef]
- Bertê, R.; Teixeira, G.M.; de Oliveira, J.P.; Nicoletto, M.L.A.; da Silva, D.V.; de Godoy, G.G.; Sanches, D.S.; de Resende, J.T.V.; Pereira, U.d.P.; da Rocha, U.N.; et al. Genome mining reveals high biosynthetic potential of biocontrol agent Bacillus velezensis B. BV10. Genes 2022, 13, 1984. [Google Scholar] [CrossRef]
- Paterson, J.; Jahanshah, G.; Li, Y.; Wang, Q.; Mehnaz, S.; Gross, H. The contribution of genome mining strategies to the understanding of active principles of PGPR strains. FEMS Microbiol. Ecol. 2017, 93, fiw249. [Google Scholar] [CrossRef]
- Montoya-Martínez, A.C.; Valenzuela-Ruíz, V.; Ortega-Urquieta, M.E.; Parra-Cota, F.I.; de los Santos-Villalobos, S. Genomic mining for the identification of promising mechanisms of bioactivity in biological control agents. In Biocontrol Agents for Improved Agriculture; Academic Press: Cambridge, MA, USA, 2024; pp. 143–163. [Google Scholar]
- Panth, M.; Hassler, S.C.; Baysal-Gurel, F. Methods for management of soilborne diseases in crop production. Agriculture 2020, 10, 16. [Google Scholar] [CrossRef]
- Ortega-Urquieta, M.E.; Valenzuela-Ruíz, V.; Mitra, D.; Hyder, S.; Elsheery, N.I.; Kumar Das Mohapatra, D.; Parra-Cota, F.I.; de los Santos-Villalobos, S. Draft Genome Sequence of Priestia sp. Strain TSO9, a Plant Growth-Promoting Bacterium Associated with Wheat (Triticum turgidum subsp. durum) in the Yaqui Valley, Mexico. Plants 2022, 11, 2231. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- de los Santos-Villalobos, S.; Díaz-Rodríguez, A.M.; Ávila-Mascareño, M.F.; Martínez-Vidales, A.D.; Parra-Cota, F.I. COLMENA: A culture collection of native microorganisms for harnessing the agro-biotechnological potential in soils and contributing to food security. Diversity 2021, 13, 337. [Google Scholar] [CrossRef]
- Baard, V.; Bakare, O.O.; Daniel, A.I.; Nkomo, M.; Gokul, A.; Keyster, M.; Klein, A. Biocontrol potential of Bacillus subtilis and Bacillus tequilensis against four fusarium species. Pathogens 2023, 12, 254. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
- R Core Team. R version 4.3.0 (2023–04–21 ucrt) “Already Tomorrow” Copyright (C) 2023 The R Foundation for Statistical Computing Platform: x86_64–w64–mingw32/x64 (64–bit). 2023. Available online: https://www.r-project.org/ (accessed on 25 February 2024).
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef]
- Farris, J.S. Estimating phylogenetic trees from distance matrices. Am. Nat. 1972, 106, 645–667. [Google Scholar] [CrossRef]
- Yi, H.S.; Ahn, Y.R.; Ryu, C.M. Impact of a bacterial volatile 2, 3-butanediol on Bacillus subtilis rhizosphere robustness. Front. Microbiol. 2016, 7, 180263. [Google Scholar] [CrossRef] [PubMed]
- Ayilara, M.S.; Adeleke, B.S.; Babalola, O.O. Bioprospecting and challenges of plant microbiome research for sustainable agriculture, a review on soybean endophytic bacteria. Microb. Ecol. 2023, 85, 1113–1135. [Google Scholar] [CrossRef] [PubMed]
- Collinge, D.B.; Jensen, D.F.; Rabiey, M.; Sarrocco, S.; Shaw, M.W.; Shaw, R.H. Biological control of plant diseases–What has been achieved and what is the direction? Plant Pathol. 2022, 71, 1024–1047. [Google Scholar] [CrossRef]
- Rampersad, S.N. Pathogenomics and management of Fusarium diseases in plants. Pathogens 2020, 9, 340. [Google Scholar] [CrossRef] [PubMed]
- Todorović, I.; Moënne-Loccoz, Y.; Raičević, V.; Jovičić-Petrović, J.; Muller, D. Microbial diversity in soils suppressive to Fusarium diseases. Front. Plant Sci. 2023, 14, 1228749. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Selvaraj, J.; Xing, F.; Zhou, L.; Wang, Y.; Song, H.; Tan, X.; Sun, L.; Sangare, L.; Elodie-Folly, Y.M.; et al. Antagonistic action of Bacillus subtilis strain SG6 on Fusarium graminearum. PLoS ONE 2014, 9, e92486. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, V.V.; Santoyo, G.; Godínez, L.J.G.; Chávez, L.A.C.; Cota, F.I.P.; Villalobos, S.d.L.S. Complete genome sequencing of Bacillus cabrialesii TE3T: A plant growth-promoting and biological control agent isolated from wheat (Triticum turgidum subsp. durum) in the Yaqui Valley. Curr. Res. Microb. Sci. 2023, 4, 100193. [Google Scholar]
- Chen, L.; Heng, J.; Qin, S.; Bian, K. A comprehensive understanding of the biocontrol potential of Bacillus velezensis LM2303 against Fusarium head blight. PLoS ONE 2018, 13, e0198560. [Google Scholar] [CrossRef]
- Villa-Rodriguez, E.; Moreno-Ulloa, A.; Castro-Longoria, E.; Parra-Cota, F.I.; de Los Santos-Villalobos, S. Integrated omics approaches for deciphering antifungal metabolites produced by a novel Bacillus species, B. cabrialesii TE3T, against the spot blotch disease of wheat (Triticum turgidum L. subsp. durum). Microbiol. Res. 2021, 251, 126826. [Google Scholar] [CrossRef]
- Gong, A.D.; Li, H.P.; Yuan, Q.S.; Song, X.S.; Yao, W.; He, W.J.; Zhang, J.B.; Liao, Y.C. Antagonistic mechanism of iturin A and plipastatin A from Bacillus amyloliquefaciens S76-3 from wheat spikes against Fusarium graminearum. PLoS ONE 2015, 10, e0116871. [Google Scholar] [CrossRef] [PubMed]
- Boulahouat, S.; Cherif-Silini, H.; Silini, A.; Bouket, A.C.; Luptakova, L.; Alenezi, F.N.; Belbahri, L. Biocontrol Efficiency of Rhizospheric Bacillus against the Plant Pathogen Fusarium oxysporum: A Promising Approach for Sustainable Agriculture. Microbiol. Res. 2023, 14, 892–908. [Google Scholar] [CrossRef]
- Dimopoulou, A.; Theologidis, I.; Benaki, D.; Koukounia, M.; Zervakou, A.; Tzima, A.; Diallinas, G.; Hatzinikolaou, D.G.; Skandalis, N. Direct antibiotic activity of bacillibactin broadens the biocontrol range of Bacillus amyloliquefaciens MBI600. Msphere 2021, 6, e00376-21. [Google Scholar] [CrossRef]
- Ghosh, S.K.; Bera, T.; Chakrabarty, A.M. Microbial siderophore—A boon to agricultural sciences. Biol. Control. 2020, 144, 104214. [Google Scholar] [CrossRef]
- Palmieri, D.; Ianiri, G.; Del Grosso, C.; Barone, G.; De Curtis, F.; Castoria, R.; Lima, G. Advances and perspectives in the use of biocontrol agents against fungal plant diseases. Horticulturae 2022, 8, 577. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, V.; Dhanjal, D.S.; Singh, J. Biological control agents: Diversity, ecological significances, and biotechnological applications. In Natural Bioactive Products in Sustainable Agriculture; Springer: Singapore, 2020; pp. 31–44. [Google Scholar]
- Bonaterra, A.; Badosa, E.; Daranas, N.; Francés, J.; Roselló, G.; Montesinos, E. Bacteria as biological control agents of plant diseases. Microorganisms 2022, 10, 1759. [Google Scholar] [CrossRef]
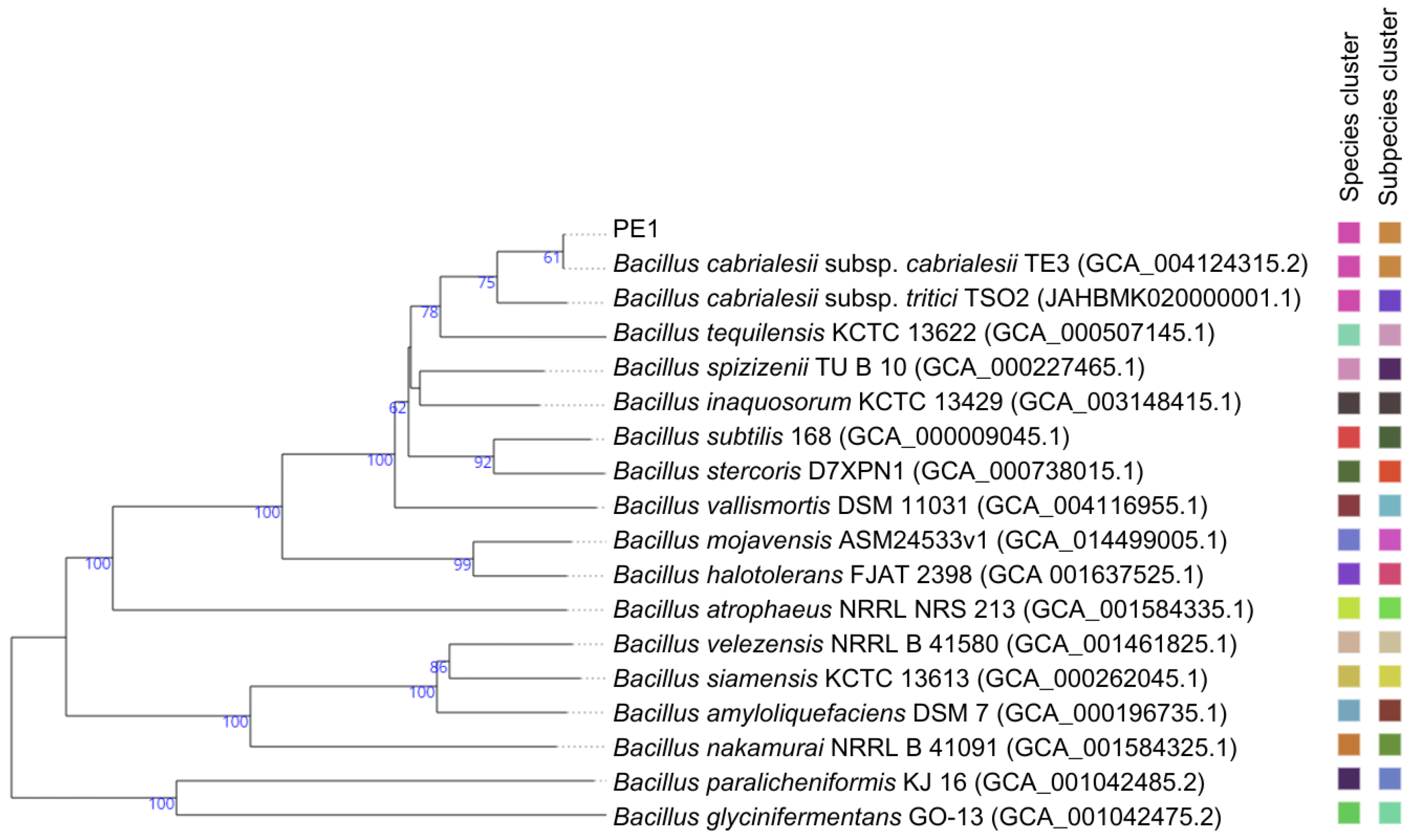


| Taxon Name | Strain | GenBank Accession Number | Similarity (%) | OrthoANI | GGDC (Formula 2) |
|---|---|---|---|---|---|
| Bacillus cabrialesii subsp. cabrialesii | TE3 T | MK462260 | 100 | 100 | 100 |
| Bacillus inaquosorum | KCTC 13429 T | AMXN01000021 | 100 | 93.95 | 54.5 |
| Bacillus tequilensis | KCTC 13622 T | AYTO01000043 | 99.86 | 93.58 | 52.7 |
| Bacillus stercoris | JCM 30051 T | MN536904 | 99.86 | 92.19 | 46.4 |
| Bacillus spizizenii | NRRL B-23049 T | CP002905 | 99.86 | 93.76 | 53.4 |
| Bacillus subtilis | NCIB 3610 T | ABQL01000001 | 99.8 | 92.42 | 47.8 |
| Bacillus halotolerans | ATCC 25096 T | LPVF01000003 | 99.73 | 87.46 | 33.4 |
| Bacillus mojavensis | RO-H-1 T | JH600280 | 99.66 | 87.57 | 33.2 |
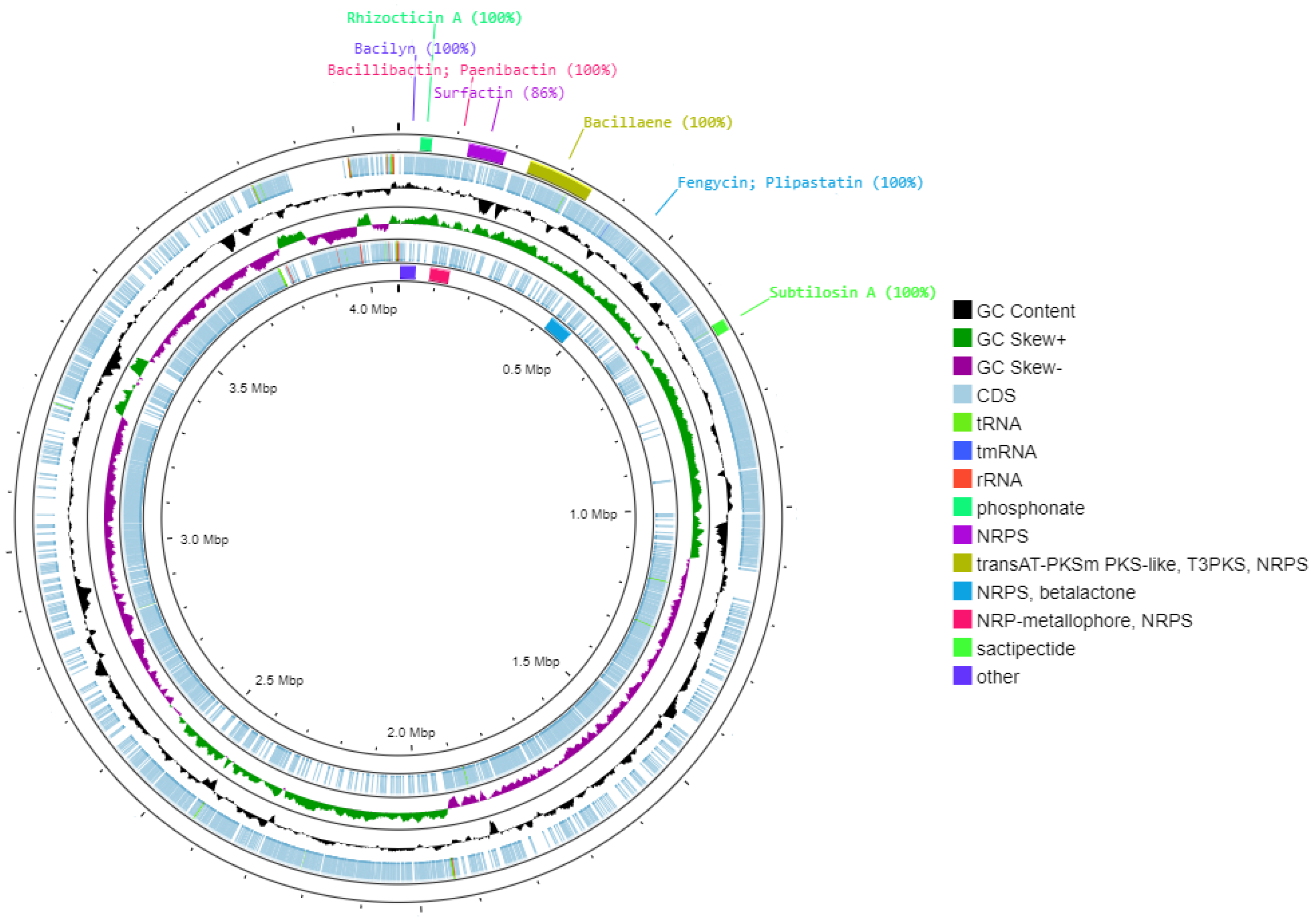
| Region | From | To | BGCs Type | Most Similar Known Cluster | Similarity (%) |
|---|---|---|---|---|---|
| 5.1 | 37,565 | 57,940 | Phosphonate | Rhizocticin A | 100 |
| 6.1 | 121,022 | 186,413 | NRPS | Surfactin | 86 |
| 14.1 | 230,573 | 345,237 | TransAT-PKSm PKS-like, T3PKS, NRPS | Bacillaene | 100 |
| 14.2 | 424,703 | 487,830 | NRPS, betalactone | Fengycin/plipastatin | 100 |
| 20.1 | 82,439 | 134,222 | NRP-metallophore, NRPS | Bacillibactin; paenibactin | 100 |
| 20.2 | 662,012 | 682,579 | Sactipectide | Subtilosin A | 100 |
| 21.1 | 3451 | 44,869 | Other | Bacilyn | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valenzuela-Aragon, B.; Montoya-Martínez, A.C.; Parra-Cota, F.I.; de los Santos-Villalobos, S. Genomic Insight into a Potential Biological Control Agent for Fusarium-Related Diseases in Potatoes: Bacillus cabrialesii Subsp. cabrialesii Strain PE1. Horticulturae 2024, 10, 357. https://doi.org/10.3390/horticulturae10040357
Valenzuela-Aragon B, Montoya-Martínez AC, Parra-Cota FI, de los Santos-Villalobos S. Genomic Insight into a Potential Biological Control Agent for Fusarium-Related Diseases in Potatoes: Bacillus cabrialesii Subsp. cabrialesii Strain PE1. Horticulturae. 2024; 10(4):357. https://doi.org/10.3390/horticulturae10040357
Chicago/Turabian StyleValenzuela-Aragon, Brenda, Amelia C. Montoya-Martínez, Fannie Isela Parra-Cota, and Sergio de los Santos-Villalobos. 2024. "Genomic Insight into a Potential Biological Control Agent for Fusarium-Related Diseases in Potatoes: Bacillus cabrialesii Subsp. cabrialesii Strain PE1" Horticulturae 10, no. 4: 357. https://doi.org/10.3390/horticulturae10040357
APA StyleValenzuela-Aragon, B., Montoya-Martínez, A. C., Parra-Cota, F. I., & de los Santos-Villalobos, S. (2024). Genomic Insight into a Potential Biological Control Agent for Fusarium-Related Diseases in Potatoes: Bacillus cabrialesii Subsp. cabrialesii Strain PE1. Horticulturae, 10(4), 357. https://doi.org/10.3390/horticulturae10040357






