Molecular Mechanisms of the Effects of Sodium Selenite on the Growth, Nutritional Quality, and Species of Organic Selenium in Dandelions
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material Treatment
2.2. Determination of Growth Indicators
2.3. Determination of Antioxidant Indicators
2.4. Determination of Total Se and Se Species
2.5. Transcriptome Sequencing and Data Analysis
2.6. Statistical Analysis
3. Results
3.1. The Effect of Na2SeO3 Hydroponics on the Height and Biomass of Dandelion
3.2. The Effect of Na2SeO3 on the Total Se and Se Species Content of Dandelion
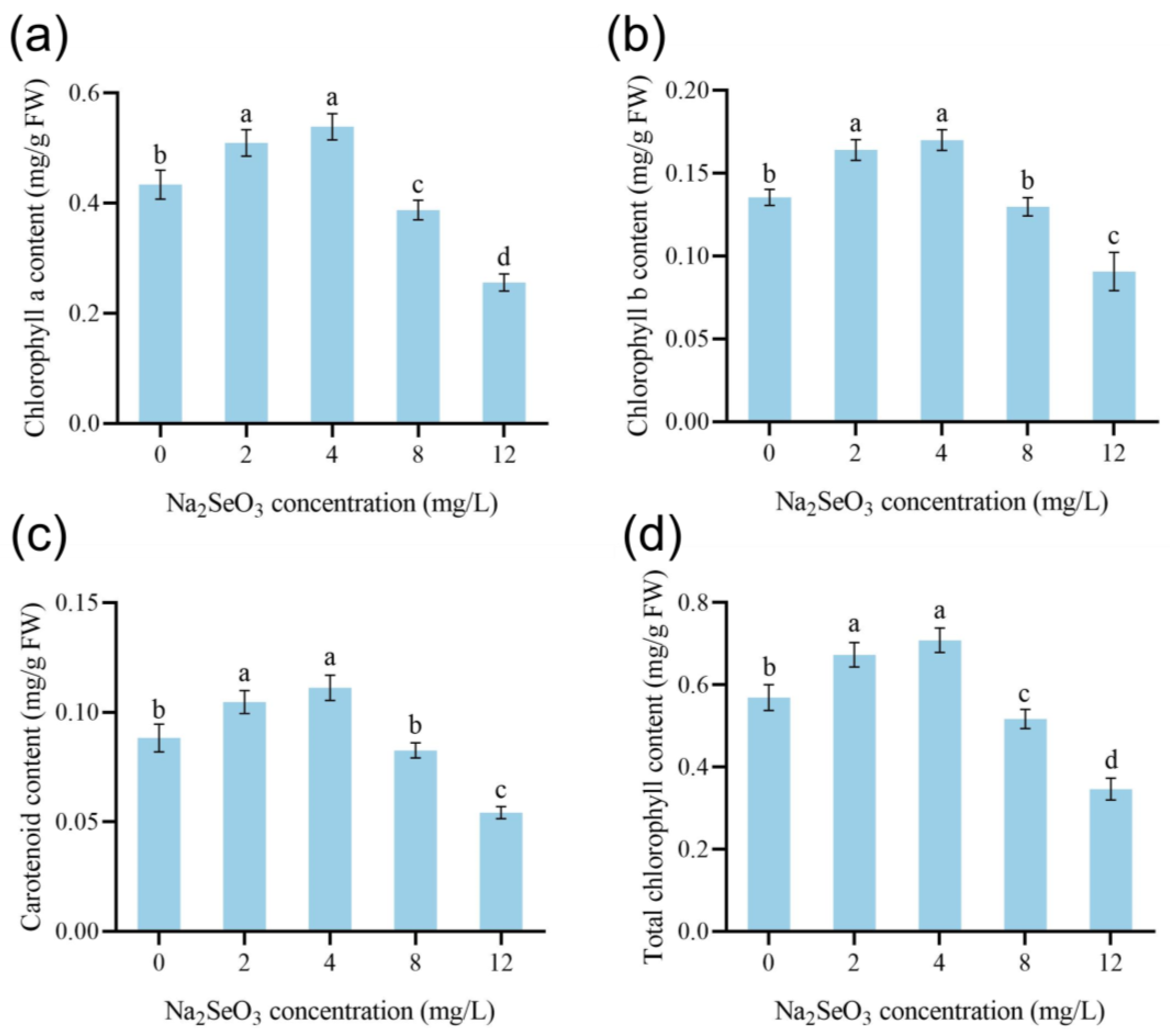
3.3. Effect of Na2SeO3 Treatment on Chlorophyll Content of Dandelion
3.4. The Effect of Na2SeO3 Treatment on the Nutritional Quality of Dandelion
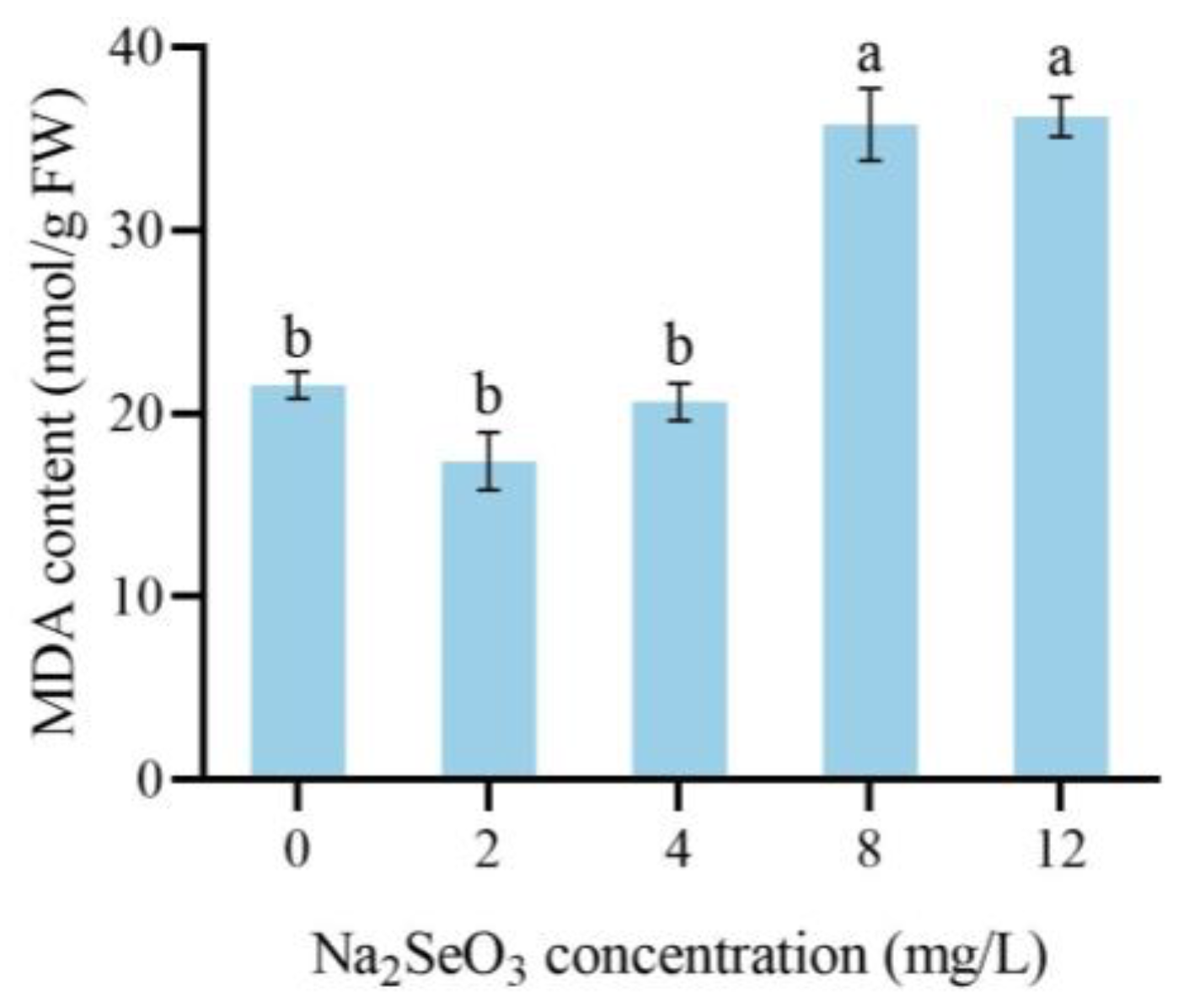
3.5. The Effect of Na2SeO3 Treatment on the MDA Content of Dandelion
3.6. The Effect of Na2SeO3 Treatment on the Stress Resistance of Dandelion
3.7. Transcriptome Data Analysis of Dandelion Treated with Na2SeO3
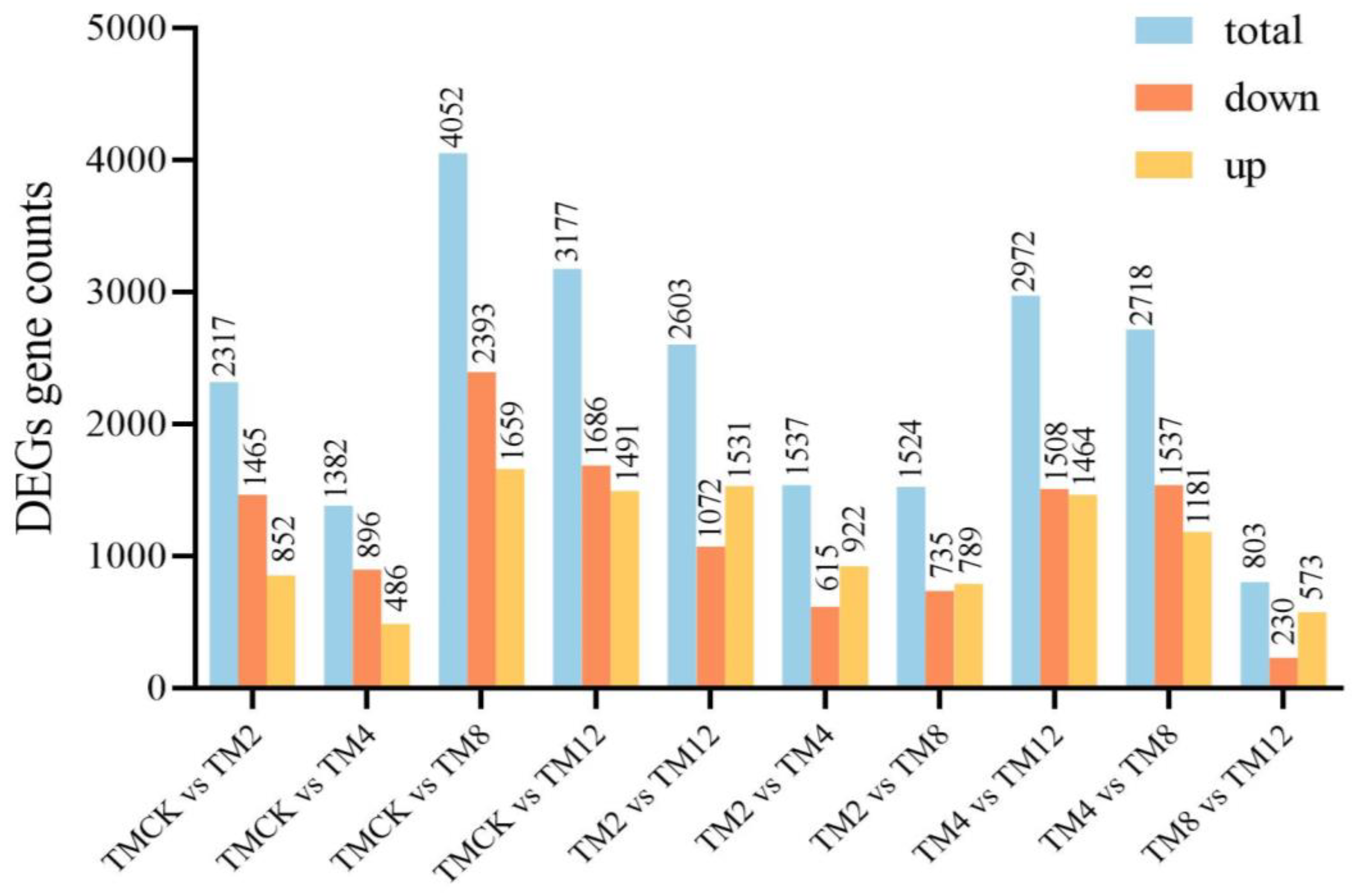
3.8. Analysis of Differentially Expressed Genes (DEGs)
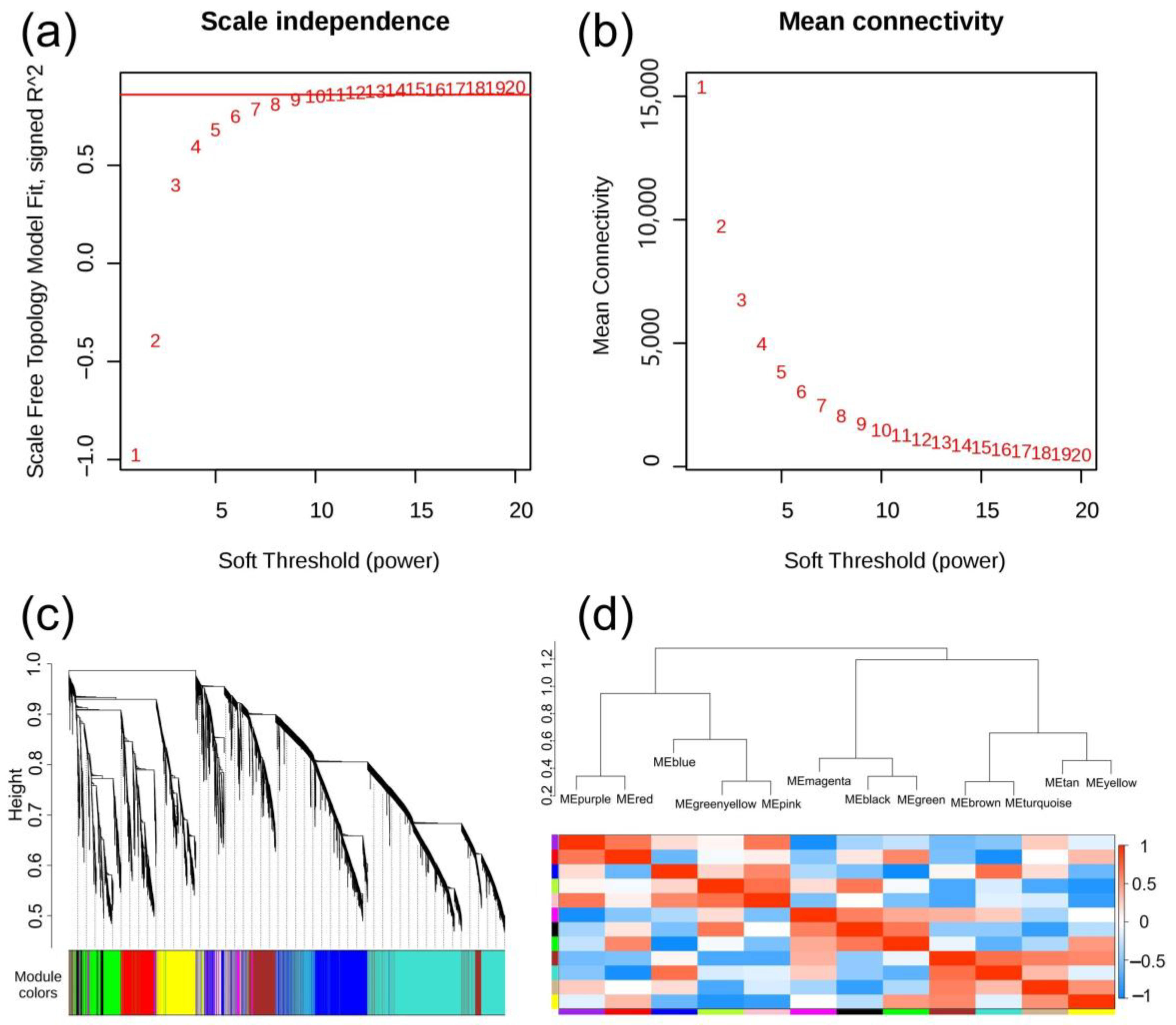
3.9. Weighted Gene Co-Expression Network Analysis (WGCNA) Analysis of DEGs
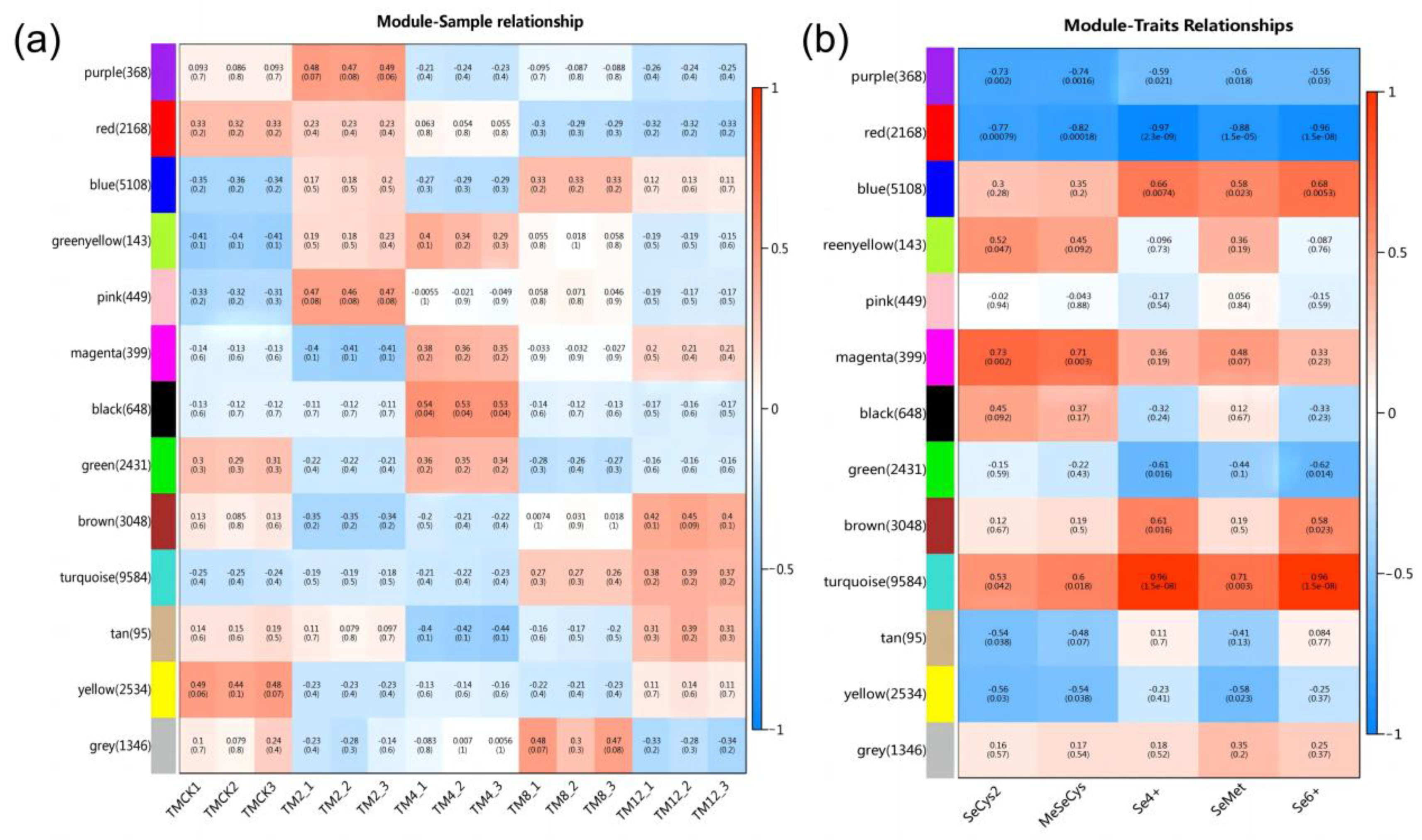
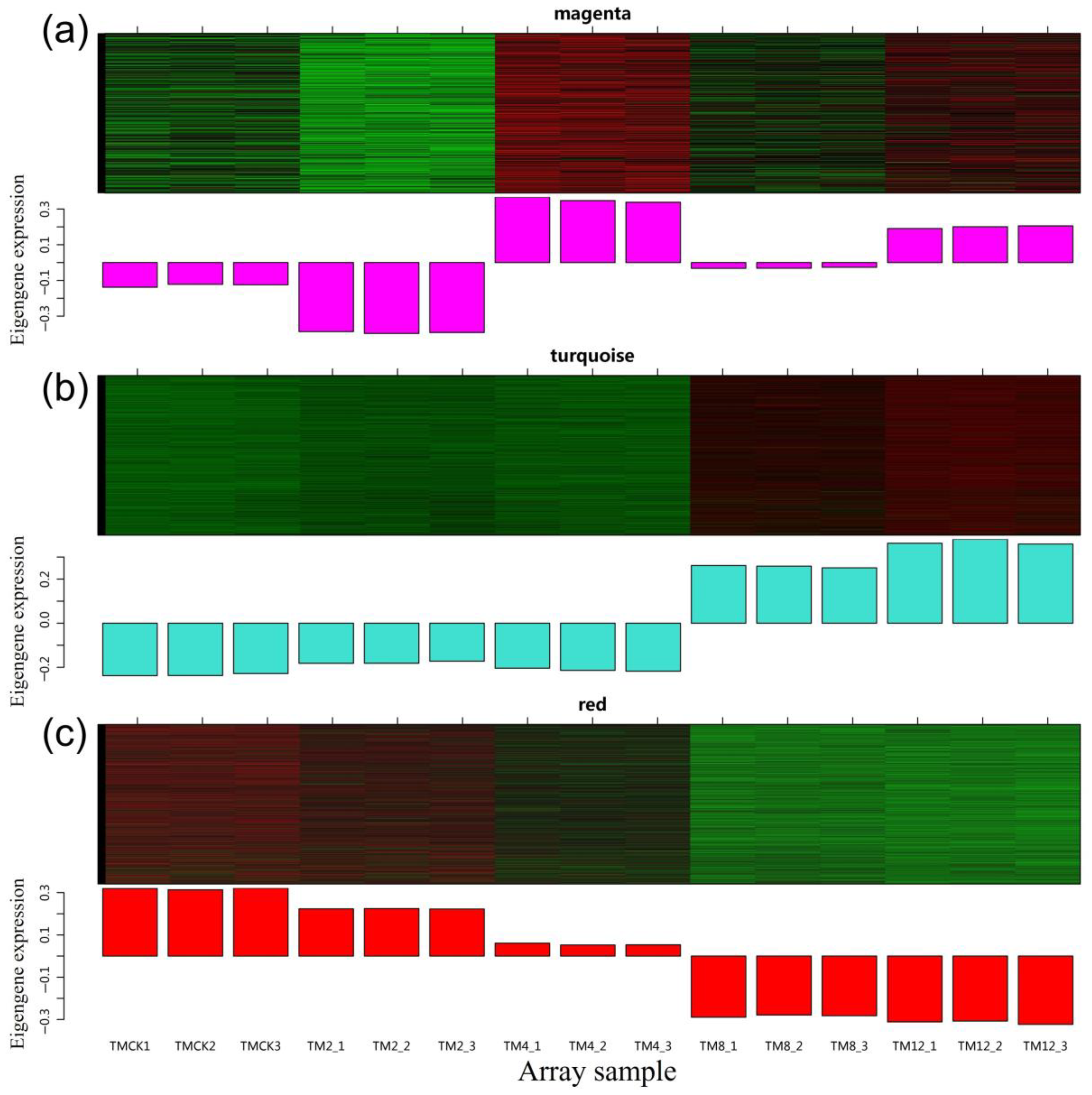
3.10. Characteristic Analysis of Gene Expression Module
3.11. Gene Functional Enrichment Analysis of Key Modules
3.12. Expression Regulation Analysis and Correlation Network of Key Genes
4. Discussion
4.1. The Effect of Se on the Growth of Dandelion
4.2. The Effect of Se on the Nutritional Quality of Dandelion Vegetables
4.3. The Effect of Se on the Antioxidant System of Dandelion
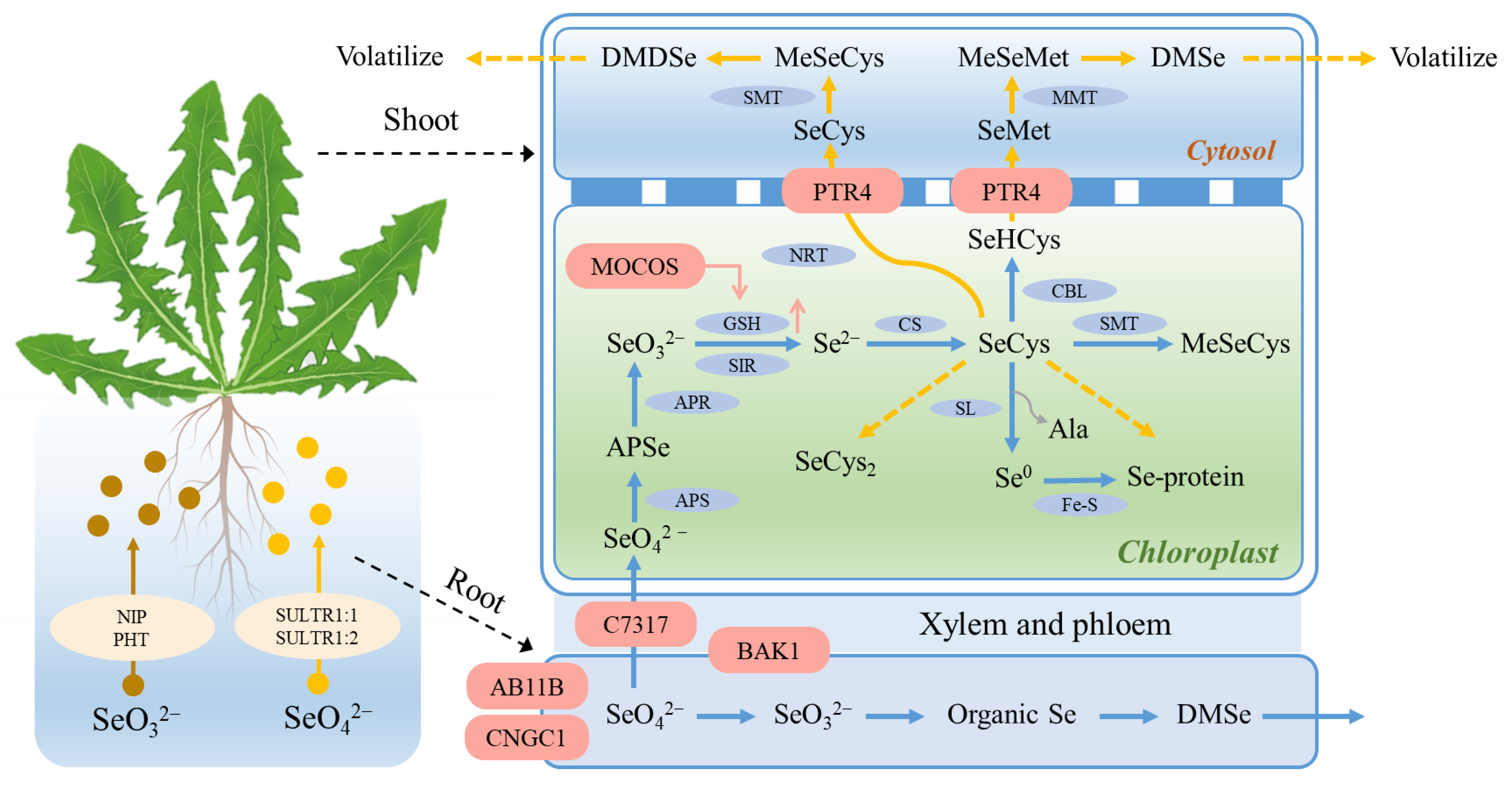
4.4. The Molecular Mechanism of Organic Se Conversion in Dandelion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Schweizer, U.; Fradejas-Villar, N. Why 21? The significance of selenoproteins for human health revealed by inborn errors of metabolism. FASEB J. 2016, 30, 3669–3681. [Google Scholar] [CrossRef]
- Wesselink, E.; Koekkoek, W.A.C.; Grefte, S.; Witkamp, R.F.; van Zanten, A.R.H. Feeding mitochondria: Potential role of nutritional components to improve critical illness convalescence. Clin. Nutr. 2019, 38, 982–995. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Liu, H.; Yang, Z.; Bao, M.; Lin, X.; Han, J.; Qu, C. Progress of selenium deficiency in the pathogenesis of arthropathies and selenium supplement for their treatment. Biol. Trace Elem. Res. 2021, 200, 4238–4249. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.L.U.; Sena-Evangelista, K.C.M.; de Azevedo, E.P.; Pinheiro, F.I.; Cobucci, R.N.; Pedrosa, L.F.C. Selenium in human health and gut microflora: Bioavailability of selenocompounds and relationship with diseases. Front. Nutr. 2021, 8, 685317. [Google Scholar] [CrossRef] [PubMed]
- Radomska, D.; Czarnomysy, R.; Radomski, D.; Bielawska, A.; Bielawski, K. Selenium as a bioactive micronutrient in the human diet and its cancer chemopreventive activity. Nutrients 2021, 13, 1649. [Google Scholar] [CrossRef] [PubMed]
- Danso, O.P.; Asante-Badu, B.; Zhang, Z.; Song, J.; Wang, Z.; Yin, X.; Zhu, R. Selenium biofortification: Strategies, progress and challenges. Agriculture 2023, 13, 416. [Google Scholar] [CrossRef]
- Gong, R.; Ai, C.; Zhang, B.; Cheng, X. Effect of selenite on organic selenium speciation and selenium bioaccessibility in rice grains of two Se-enriched rice cultivars. Food Chem. 2018, 264, 443–448. [Google Scholar] [CrossRef]
- Nothstein, A.K.; Eiche, E.; Riemann, M.; Nick, P.; Winkel, L.H.E.; Göttlicher, J.; Steininger, R.; Brendel, R.; Von Brasch, M.; Konrad, G. Tracking se assimilation and speciation through the rice plant–nutrient competition, toxicity and distribution. PLoS ONE 2016, 11, e0152081. [Google Scholar] [CrossRef]
- Schiavon, M.; Pilon-Smits, E.A.H. Selenium biofortification and phytoremediation phytotechnologies: A review. J. Environ. Qual. 2017, 46, 10–19. [Google Scholar] [CrossRef]
- El Mehdawi, A.F.; Jiang, Y.; Guignardi, Z.S.; Esmat, A.; Pilon, M.; Pilon-Smits, E.A.H.; Schiavon, M. Influence of sulfate supply on selenium uptake dynamics and expression of sulfate/selenate transporters in selenium hyperaccumulator and nonhyperaccumulator Brassicaceae. New Phytol. 2017, 217, 194–205. [Google Scholar] [CrossRef]
- Bai, B.; Zhang, S.; Suo, X.; Chen, W.; Shen, Y. Selenite uptake by Medicago sativa L. roots. Grassl. Sci. 2022, 68, 328–335. [Google Scholar] [CrossRef]
- Li, L.; Wu, S.; Wang, S.; Shi, X.; Cheng, S.; Cheng, H. Molecular mechanism of exogenous selenium affecting the nutritional quality, species and content of organic selenium in mustard. Agronomy 2023, 13, 1425. [Google Scholar] [CrossRef]
- Hossain, A.; Skalicky, M.; Brestic, M.; Maitra, S.; Sarkar, S.; Ahmad, Z.; Vemuri, H.; Garai, S.; Mondal, M.; Bhatt, R.; et al. Selenium biofortification: Roles, mechanisms, responses and prospects. Molecules 2021, 26, 881. [Google Scholar] [CrossRef]
- Gao, D. Analysis of nutritional components of Taraxacum mongolicum and its antibacterial activity. Pharmacogn. J. 2010, 2, 502–505. [Google Scholar] [CrossRef]
- Zhang, S.; Li, C.; Gu, W.; Qiu, R.; Chao, J.; Pei, L.; Ma, L.; Guo, Y.; Tian, R. Metabolomics analysis of dandelions from different geographical regions in China. Phytochem. Anal. 2021, 32, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Zhang, C.; Zhao, Y.; Chang, Y.; Guo, L. Comparison of bioactive phenolic compounds and antioxidant activities of different parts of Taraxacum mongolicum. Molecules 2020, 25, 3260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Song, Z.; Shi, L.; Zhou, L.; Zhang, J.; Cui, J.; Li, Y.; Jin, D.Q.; Ohizumi, Y.; Xu, J.; et al. A dandelion polysaccharide and its selenium nanoparticles: Structure features and evaluation of anti-tumor activity in zebrafish models. Carbohydr. Polym. 2021, 270, 118365. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.Z.; Park, B.J.; Kang, H.M.; Lee, Y.T. Influence of selenium biofortification on the bioactive compounds and antioxidant activity of wheat microgreen extract. Food Chem. 2019, 309, 125763. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983, 603, 591–592. [Google Scholar] [CrossRef]
- Matos Reyes, M.N.; Cervera, M.L.; de la Guardia, M. Determination of total Sb, Se, Te, and Bi and evaluation of their inorganic species in garlic by hydride generation atomic fluorescence spectrometry. Anal. Bioanal. Chem. 2009, 394, 1557–1562. [Google Scholar] [CrossRef]
- Goenaga Infante, H.; Arias Borrego, A.; Peachey, E.; Hearn, R.; O’Connor, G.; García Barrera, T.; Gómez Ariza, J.L. Study of the effect of sample preparation and cooking on the selenium speciation of selenized potatoes by HPLC with ICP-MS and electrospray ionization MS/MS. J. Agric. Food Chem. 2008, 57, 38–45. [Google Scholar] [CrossRef]
- Shumate, A.; Wong, B.; Pertea, G.; Pertea, M. Improved transcriptome assembly using a hybrid of long and short reads with StringTie. Biorxiv Bioinform. 2021, 18, e1009730. [Google Scholar] [CrossRef]
- Saleem, M.; Fariduddin, Q. Novel mechanistic insights of selenium induced microscopic, histochemical and physio-biochemical changes in tomato (Solanum lycopersicum L.) plant. An account of beneficiality or toxicity. J. Hazard. Mater. 2022, 434, 128830. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Tu, S.; Dai, Z.; Huang, W.; Jia, W.; Xu, Z.; Shao, H. Comparison of selenite and selenate in alleviation of drought stress in Nicotiana tabacum L. Chemosphere 2022, 287, 132136. [Google Scholar] [CrossRef] [PubMed]
- Bai, B.; Wang, Z.; Gao, L.; Chen, W.; Shen, Y. Effects of selenite on the growth of alfalfa (Medicago sativa L. cv. Sadie 7) and related physiological mechanisms. Acta Physiol. Plant. 2019, 41, 78. [Google Scholar] [CrossRef]
- Yin, H.; Qi, Z.; Li, M.; Ahammed, G.J.; Chu, X.; Zhou, J. Selenium forms and methods of application differentially modulate plant growth, photosynthesis, stress tolerance, selenium content and speciation in Oryza sativa L. Ecotoxicol. Environ. Saf. 2019, 169, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Gui, J.-Y.; Rao, S.; Gou, Y.; Xu, F.; Cheng, S. Comparative study of the effects of selenium yeast and sodium selenite on selenium content and nutrient quality in broccoli florets (Brassica oleracea L. var. italica). J. Sci. Food Agric. 2022, 102, 1707–1718. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Gao, M.; Shi, R.; Song, S.; Zhang, Y.; Su, W.; Liu, H. The combination of selenium and LED light quality affects growth and nutritional properties of broccoli sprouts. Molecules 2020, 25, 4788. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wang, J.; Wu, H.; Yuan, Q.; Wang, J.; Cui, J.; Lin, A. Effects of selenium fertilizer application and tomato varieties on tomato fruit quality: A meta-analysis. Sci. Hortic. 2022, 304, 111242. [Google Scholar] [CrossRef]
- Wang, K.; Yuan, Y.; Luo, X.; Shen, Z.; Huang, Y.; Zhou, H.; Gao, X. Effects of exogenous selenium application on nutritional quality and metabolomic characteristics of mung bean (Vigna radiata L.). Front. Plant Sci. 2022, 13, 961447. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, Y.; Liu, J.; Chen, Y.; Zhang, X. Exploring the effects of selenium treatment on the nutritional quality of tomato fruit. Food Chem. 2018, 252, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhou, C.; Zhang, J.; An, Q.; Wu, Y.; Li, J.; Pan, C. Nanoselenium foliar applications enhance the nutrient quality of pepper by activating the capsaicinoid synthetic pathway. J. Agric. Food Chem. 2020, 68, 9888–9895. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, J.; Zhang, K.; Wen, Q.; Ming, K.; Xiong, H.; Ning, F. Peanut selenium distribution, concentration, speciation, and effects on proteins after exogenous selenium biofortification. Food Chem. 2021, 354, 129515. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, S.; Wu, S.; Rao, S.; Li, L.; Cheng, S.; Cheng, H. Morphological and physiological indicators and transcriptome analyses reveal the mechanism of selenium multilevel mitigation of cadmium damage in Brassica juncea. Plants 2023, 12, 1583. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Parvin, K.; Bardhan, K.; Nahar, K.; Anee, T.I.; Masud, A.A.C.; Fotopoulos, V. Biostimulants for the regulation of reactive oxygen species metabolism in plants under abiotic stress. Cells 2021, 10, 2537. [Google Scholar] [CrossRef] [PubMed]
- Santos, E.F.; Kondo Santini, J.M.; Paixão, A.P.; Júnior, E.F.; Lavres, J.; Campos, M.; Reis, A.R.d. Physiological highlights of manganese toxicity symptoms in soybean plants: Mn toxicity responses. Plant Physiol. Biochem. 2017, 113, 6–19. [Google Scholar] [CrossRef]
- Michalak, M. Plant derived antioxidants: Significance in skin health and the ageing process. Int. J. Mol. Sci. 2022, 23, 585. [Google Scholar] [CrossRef]
- Wang, X.; Lu, W.; Zhao, Z.; Hao, W.; Du, R.; Li, Z.; Wang, Z.; Lv, X.; Wang, J.; Liang, D.; et al. Abscisic acid promotes selenium absorption, metabolism and toxicity via stress-related phytohormones regulation in Cyphomandra betacea Sendt. (Solanum betaceum Cav.). J. Hazard. Mater. 2023, 461, 132642. [Google Scholar] [CrossRef]
- Khanna, K.; Kumar, P.; Ohri, P.; Bhardwaj, R. Harnessing the role of selenium in soil–plant-microbe ecosystem: Ecophysiological mechanisms and future prospects. Plant Growth Regul. 2022, 100, 197–217. [Google Scholar] [CrossRef]
- Mroczek-Zdyrska, M.; Strubińska, J.; Hanaka, A. Selenium improves physiological parameters and alleviates oxidative stress in shoots of lead-exposed Vicia faba L. minor plants grown under phosphorus-deficient conditions. J. Plant Growth Regul. 2016, 36, 186–199. [Google Scholar] [CrossRef]
- Silva, V.M.; Rimoldi Tavanti, R.F.; Gratão, P.L.; Alcock, T.D.; Reis, A.R.D. Selenate and selenite affect photosynthetic pigments and ROS scavenging through distinct mechanisms in cowpea (Vigna unguiculata (L.) walp) plants. Ecotoxicol. Environ. Saf. 2020, 201, 110777. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Q.; Zou, D.; Yang, Y. Effects of selenite on growth, photosynthesis and antioxidant system in seaweeds, Ulva fasciata (Chlorophyta) and Gracilaria lemaneiformis (Rhodophyta). Algal Res. 2018, 36, 115–124. [Google Scholar] [CrossRef]
- Hosseinzadeh Rostam Kalaei, M.; Abdossi, V.; Danaee, E. Evaluation of foliar application of selenium and flowering stages on selected properties of Iranian Borage as a medicinal plant. Sci. Rep. 2022, 12, 12568. [Google Scholar] [CrossRef]
- Qu, L.; Xu, J.; Dai, Z.; Elyamine, A.M.; Huang, W.; Han, D.; Dang, B.; Xu, Z.; Jia, W. Selenium in soil-plant system: Transport, detoxification and bioremediation. J. Hazard. Mater. 2023, 452, 131272. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, T.; Hayashi, N.; Yamaya, T.; Takahashi, H. Root-to-shoot transport of sulfate in arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol. 2004, 136, 4198–4204. [Google Scholar] [CrossRef] [PubMed]
- Trippe, R.C.; Pilon-Smits, E.A.H. Selenium transport and metabolism in plants: Phytoremediation and biofortification implications. J. Hazard. Mater. 2020, 404, 124178. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Gupta, S. An overview of selenium uptake, metabolism, and toxicity in plants. Front. Plant Sci. 2017, 7, 2074. [Google Scholar] [CrossRef]
- Cheng, H.; Li, L.; Dong, J.; Wang, S.; Wu, S.; Rao, S.; Li, L.; Cheng, S.; Li, L. Transcriptome and physiological determination reveal the effects of selenite on the growth and selenium metabolism in mung bean sprouts. Food Res. Int. 2023, 169, 112880. [Google Scholar] [CrossRef] [PubMed]
- Dou, L.; Tian, Z.; Zhao, Q.; Xu, M.; Zhu, Y.; Luo, X.; Qiao, X.; Ren, R.; Zhang, X.; Li, H. Transcriptomic characterization of the effects of selenium on maize seedling growth. Front. Plant Sci. 2021, 12, 737029. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Yu, T.; Cong, X.; Lai, X.; Xiang, J.; Cao, J.; Liao, X.; Gou, Y.; Chao, W.; Xue, H.; et al. Transcriptome, proteome, and metabolome reveal the mechanism of tolerance to selenate toxicity in Cardamine violifolia. J. Hazard. Mater. 2021, 406, 124283. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Aller, S.G.; Beis, K.; Carpenter, E.P.; Chang, G.; Chen, L.; Dassa, E.; Dean, M.; Duong Van Hoa, F.; Ekiert, D.; et al. Structural and functional diversity calls for a new classification of ABC transporters. FEBS Lett. 2020, 594, 3767–3775. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xiong, Y.; Wang, Y.; Wu, S.; Xiao, C.; Wang, S.; Cheng, S.; Cheng, H. Effect of nano-selenium on nutritional quality of cowpea and response of ABCC transporter family. Molecules 2023, 28, 1398. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yu, L.; Chao, W.; Xiang, J.; Yang, X.; Ye, J.; Liao, X.; Zhou, X.; Rao, S.; Cheng, S.; et al. Comparative physiological and transcriptome analysis reveals the potential mechanism of selenium accumulation and tolerance to selenate toxicity of Broussonetia papyrifera. Tree Physiol. 2022, 42, 2578–2595. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.; Hu, H.; Hu, J.; Xiang, M.; Yang, Q. Comparative proteomics analysis of selenium responses in selenium-enriched alfalfa (Medicago sativa L.) leaves. Plant Physiol. Biochem. 2021, 165, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Guo, L.; Huang, J.; Hao, X.; Li, X.; Li, N.; Wang, Y.; Zhang, K.; Wang, X.; Wang, L.; et al. Comparative transcriptomics provides novel insights into the mechanisms of selenium accumulation and transportation in tea cultivars (Camellia sinensis (L.) O. Kuntze). Front. Plant Sci. 2023, 14, 1268537. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.; Cho, H. The function of ABCB transporters in auxin transport. Plant Signal. Behav. 2013, 8, e22990. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, F.; Wang, Y.; Zhang, S.; Wang, F.; Ma, Y. The cytochrome P450 gene, MdCYP716B1, is involved in regulating plant growth and anthracnose resistance in apple. Plant Sci. 2023, 335, 111832. [Google Scholar] [CrossRef]
- Zheng, X.; Li, P.; Lu, X. Research advances in cytochrome P450-catalysed pharmaceutical terpenoid biosynthesis in plants. J. Exp. Bot. 2019, 70, 4619–4630. [Google Scholar] [CrossRef]
- Singh, A.; Panwar, R.; Mittal, P.; Hassan, M.I.; Singh, I.K. Plant cytochrome P450s: Role in stress tolerance and potential applications for human welfare. Int. J. Biol. Macromol. 2021, 184, 874–886. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, W.; Xu, Y.; Li, G.; Liao, Y.; Fu, F. Differential expression of candidate genes for lignin biosynthesis under drought stress in maize leaves. J. Appl. Genet. 2009, 50, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Corratgé-Faillie, C.; Lacombe, B. Substrate (un)specificity of arabidopsis NRT1/PTR FAMILY (NPF) proteins. J. Exp. Bot. 2017, 68, 3107–3113. [Google Scholar] [CrossRef]
- Zhang, L.; Hu, B.; Deng, K.; Gao, X.; Sun, G.; Zhang, Z.; Li, P.; Wang, W.; Li, H.; Zhang, Z.; et al. NRT1.1B improves selenium concentrations in rice grains by facilitating selenomethinone translocation. Plant Biotechnol. J. 2019, 17, 1058–1068. [Google Scholar] [CrossRef] [PubMed]
- Weichert, A.; Brinkmann, C.; Komarova, N.Y.; Dietrich, D.; Thor, K.; Meier, S.; Suter Grotemeyer, M.; Rentsch, D. AtPTR4 and AtPTR6 are differentially expressed, tonoplast-localized members of the peptide transporter/nitrate transporter 1 (PTR/NRT1) family. Planta 2011, 235, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Chen, J.; Wang, S. Tissue specific regulation of rice molybdenum cofactor sulfurase gene in response to salt stress and ABA. Acta Physiol. Plant. 2009, 31, 545–551. [Google Scholar] [CrossRef]
- Contour-Ansel, D.; Torres-Franklin, M.L.; Cruz De Carvalho, M.H.; D’Arcy-Lameta, A.; Zuily-Fodil, Y. Glutathione reductase in leaves of cowpea: Cloning of two cDNAs, expression and enzymatic activity under progressive drought stress, desiccation and abscisic acid treatment. Ann. Bot. 2006, 98, 1279–1287. [Google Scholar] [CrossRef] [PubMed]
- Schiavon, M.; Pilon-Smits, E.A.H. The fascinating facets of plant selenium accumulation-biochemistry, physiology, evolution and ecology. New Phytol. 2016, 213, 1582–1596. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Yong, R.; Zhang, J.; Cai, G.; Wang, R.; Li, J.; Wang, Y.; Zhang, H.; Gao, X.; Huang, J. OsBAK2/OsSERK2 expression is repressed by OsBZR1 to modulate brassinosteroid response and grain length in rice. J. Exp. Bot. 2023, 74, 4978–4993. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, S.; Chen, L.; Zhou, Q.; Wang, M.; Feng, D.; Li, J.; Wang, J.; Wang, H.; Liu, B. BIK1 cooperates with BAK1 to regulate constitutive immunity and cell death in Arabidopsis. J. Integr. Plant Biol. 2017, 59, 234–239. [Google Scholar] [CrossRef]
- Kim, M.H.; Kim, Y.; Kim, J.W.; Lee, H.S.; Lee, W.S.; Kim, S.K.; Wang, Z.; Kim, S.H. Identification of arabidopsis BAK1-associating receptor-like kinase 1 (BARK1) and characterization of its gene expression and brassinosteroid-regulated root phenotypes. Plant Cell Physiol. 2013, 54, 1620–1634. [Google Scholar] [CrossRef]
- Marchesi, A.; Gao, X.; Adaixo, R.; Rheinberger, J.; Stahlberg, H.; Nimigean, C.; Scheuring, S. An iris diaphragm mechanism to gate a cyclic nucleotide-gated ion channel. Nat. Commun. 2018, 9, 3978. [Google Scholar] [CrossRef]
- Chakraborty, S.; Toyota, M.; Moeder, W.; Chin, K.; Fortuna, A.; Champigny, M.; Vanneste, S.; Gilroy, S.; Beeckman, T.; Nambara, E.; et al. Cyclic Nucleotide-Gated Ion Channel 2 modulates auxin homeostasis and signaling. Plant Physiol. 2021, 187, 1690–1703. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Chai, X.; Gao, Q.; Zhou, L.; Zhang, S.; Li, L.; Luan, S. Dynamic interactions of plant CNGC subunits and calmodulins drive oscillatory Ca2+ channel activities. Dev. Cell 2019, 48, 710–725. [Google Scholar] [CrossRef] [PubMed]

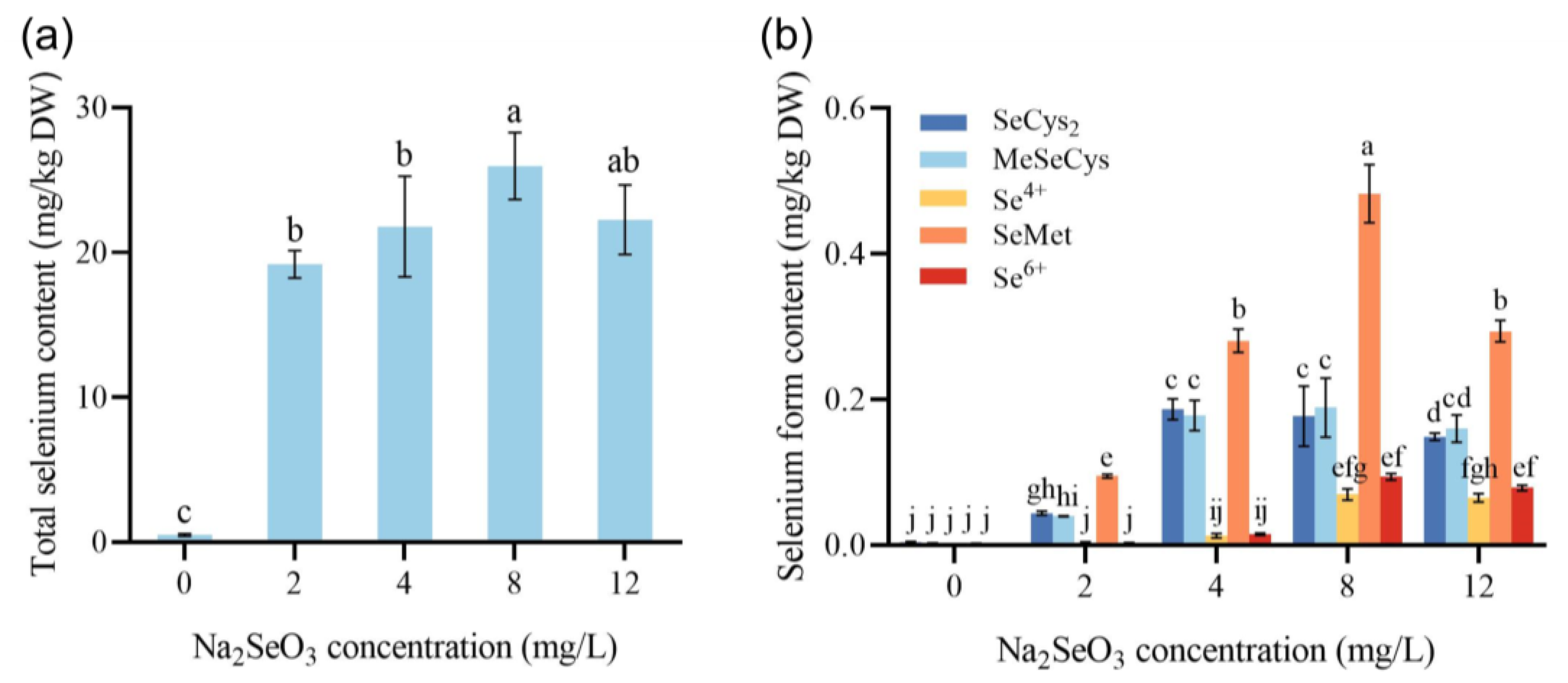



| Na2SeO3 Concentration (mg/L) | Content of Different Se Species (mg/kg DW) | ||||
|---|---|---|---|---|---|
| SeCys2 | MeSeCys | Se4+ | SeMet | Se6+ | |
| 0 | 0.0039 ± 0.0008 d | 0.0018 ± 0.0009 b | 0.0005 ± 0.0002 c | 0.0019 ± 0.0005 d | 0.0008 ± 0.0003 d |
| 2 | 0.044 ± 0.0028 c | 0.0398 ± 0.0004 b | 0.0024 ± 0.002 c | 0.0947 ± 0.0024 c | 0.0028 ± 0.0002 d |
| 4 | 0.1866 ± 0.0144 a | 0.178 ± 0.0207 a | 0.0126 ± 0.0031 b | 0.2808 ± 0.0159 b | 0.0146 ± 0.0015 c |
| 8 | 0.1771 ± 0.0413 ab | 0.1888 ± 0.0404 a | 0.0695 ± 0.0079 a | 0.4827 ± 0.0399 a | 0.0937 ± 0.0046 a |
| 12 | 0.1488 ± 0.0049 b | 0.16 ± 0.0187 a | 0.0651 ± 0.0057 a | 0.2939 ± 0.0148 b | 0.0786 ± 0.0038 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, H.; Chang, S.; Shi, X.; Chen, Y.; Cong, X.; Cheng, S.; Li, L. Molecular Mechanisms of the Effects of Sodium Selenite on the Growth, Nutritional Quality, and Species of Organic Selenium in Dandelions. Horticulturae 2024, 10, 209. https://doi.org/10.3390/horticulturae10030209
Cheng H, Chang S, Shi X, Chen Y, Cong X, Cheng S, Li L. Molecular Mechanisms of the Effects of Sodium Selenite on the Growth, Nutritional Quality, and Species of Organic Selenium in Dandelions. Horticulturae. 2024; 10(3):209. https://doi.org/10.3390/horticulturae10030209
Chicago/Turabian StyleCheng, Hua, Siyuan Chang, Xinyu Shi, Yuanfei Chen, Xin Cong, Shuiyuan Cheng, and Linling Li. 2024. "Molecular Mechanisms of the Effects of Sodium Selenite on the Growth, Nutritional Quality, and Species of Organic Selenium in Dandelions" Horticulturae 10, no. 3: 209. https://doi.org/10.3390/horticulturae10030209
APA StyleCheng, H., Chang, S., Shi, X., Chen, Y., Cong, X., Cheng, S., & Li, L. (2024). Molecular Mechanisms of the Effects of Sodium Selenite on the Growth, Nutritional Quality, and Species of Organic Selenium in Dandelions. Horticulturae, 10(3), 209. https://doi.org/10.3390/horticulturae10030209





