Identification and Characterization of Relict Olive Varieties (Olea europaea L.) in the Northwest of the Iberian Peninsula
Abstract
1. Introduction
2. Materials and Methods
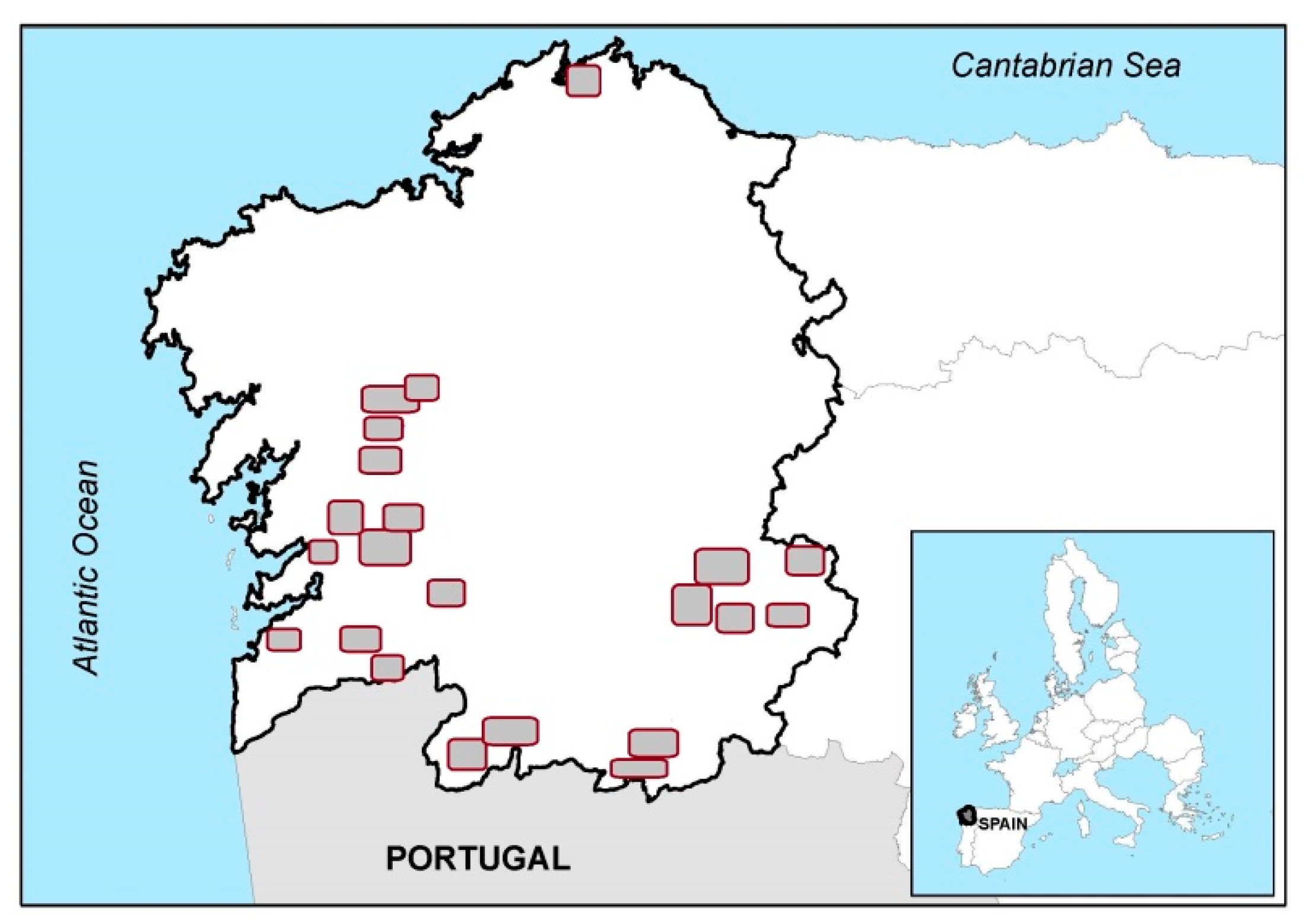
2.1. Plant Material
2.2. DNA Extraction and Microsatellite Analysis
2.3. Botanical Characterization
2.4. Physicochemical Characterization of the Drupes and of the Oil Obtained from Them
2.5. Plant Health
- Fungi: Verticillium dahliae (a regulated, nonquarantinable disease (RNQD))
- Bacteria: Xylella fastidiosa (a priority quarantinable disease (QD)) and Pseudomonas savastanoi pv. Savastanoi (RNQD)
- Viruses: Arabis mosaic virus (ArMVoo), cherry leaf roll virus (CLRVoo), strawberry latent ring spot virus (SLRSVo), and cucumber mosaic virus (CMVoo).
3. Results
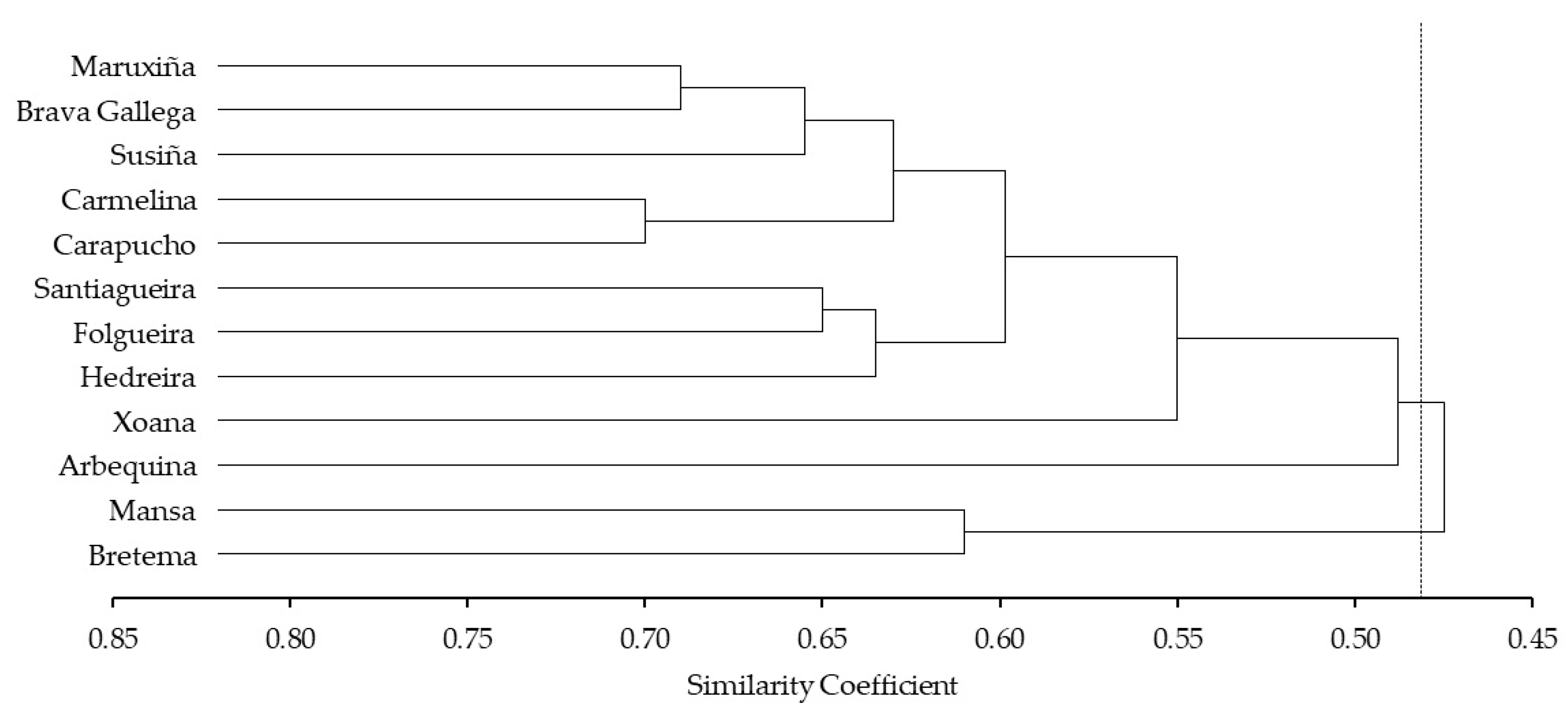
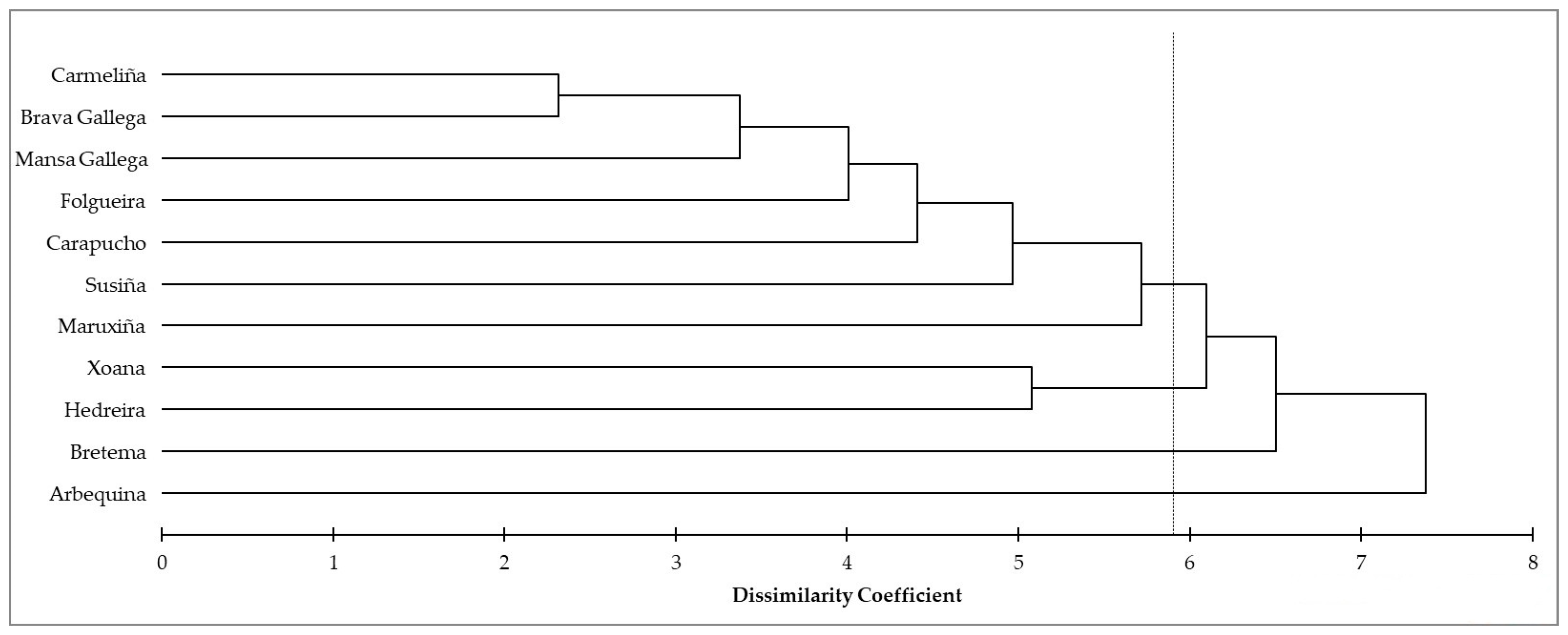
3.1. SSR Analyses
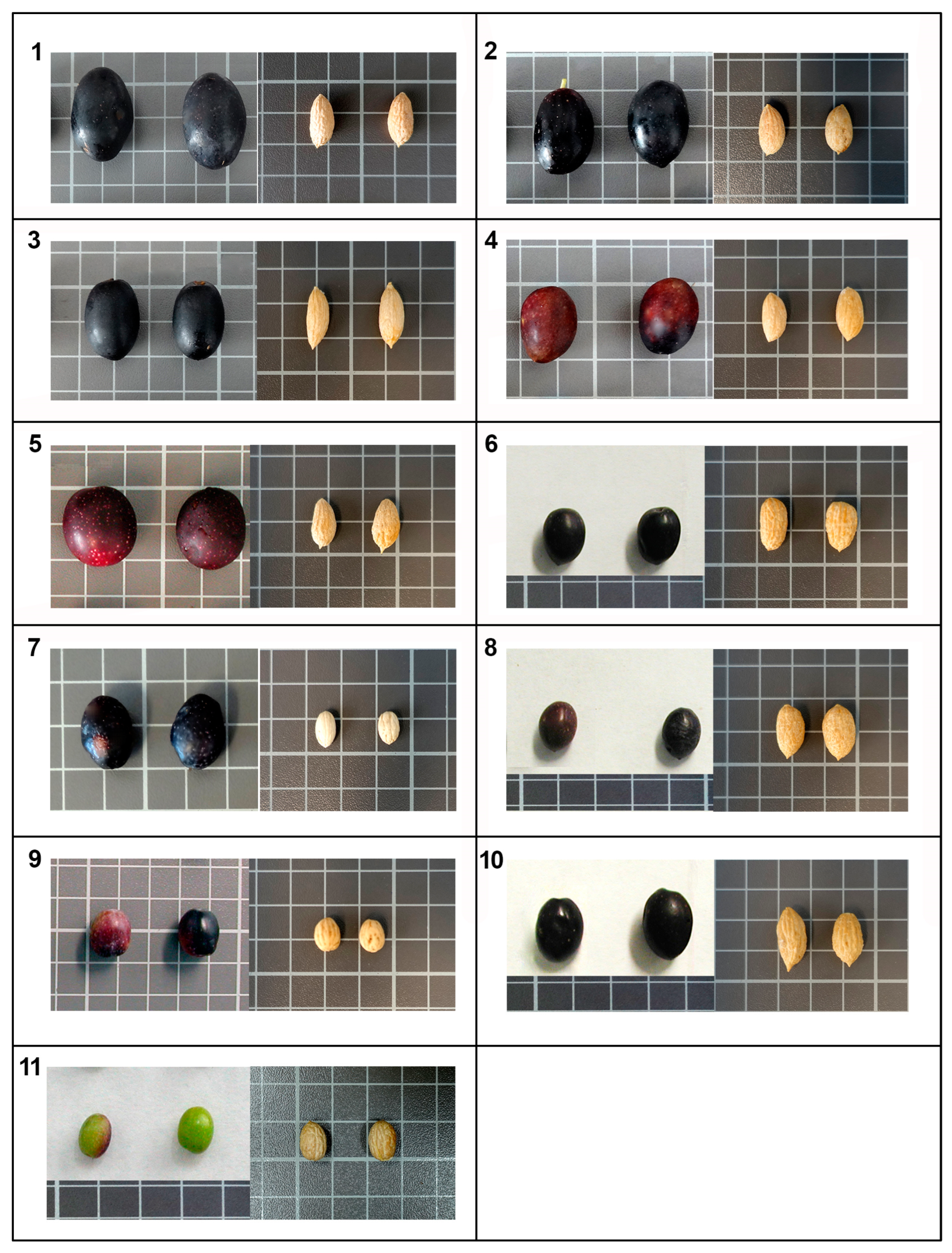
3.2. Botanical Characterization Results
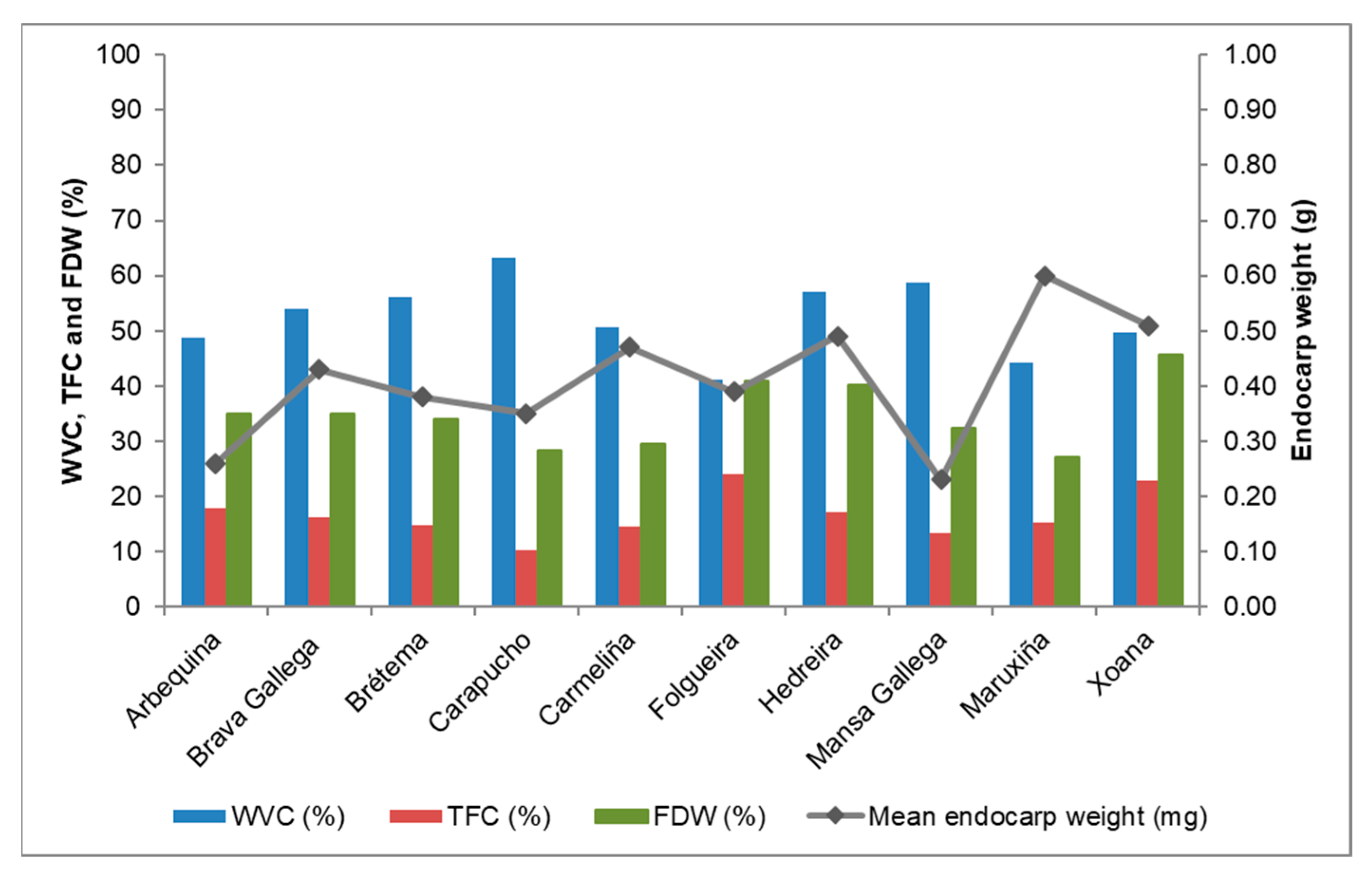
3.3. ABENCOR® Variables
3.4. Health Status of the Examined Trees
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ninot, A.; Howad, W.; Aranzana, M.J.; Senar, R.; Romero, A.; Mariotti, R.; Baldoni, L.; Belaj, A. Survey of over 4,500 Monumental Olive Trees Preserved on-Farm in the Northeast Iberian Peninsula, Their Genotyping and Characterization. Sci. Hortic. 2018, 231, 253–264. [Google Scholar] [CrossRef]
- Trujillo, I.; Ojeda, M.A.; Urdiroz, N.M.; Potter, D.; Barranco, D.; Rallo, L.; Diez, C.M. Identification of the Worldwide Olive Germplasm Bank of Córdoba (Spain) Using SSR and Morphological Markers. Tree Genet. Genomes 2014, 10, 141–155. [Google Scholar] [CrossRef]
- Fernández de la Cigoña, E.; Martínez Tamuxe, X. O aceite en Galicia: Guía das Lagaretas Castrexo-Romanas, Medievais e Modernas; Martínez Tamuxe, X., Ed.; Asociación galega para a cultura e a ecoloxía: Pontevedra, Spain, 2003. [Google Scholar]
- Teira-Brión, A. Understanding the Plant Economy of the Westernmost Territory of the Roman State through Waste: The Wet Site of O Areal (Vigo, Spain). Veg. Hist. Archaeobot. 2022, 31, 595–610. [Google Scholar] [CrossRef]
- Sarmiento, M. Obra de 660 Pliegos: De Historia Natural y de Todo Género de Erudición; Monteagudo, H., Ed.; Obras de Martín Sarmiento; Consello da Cultura Galega Consejo Superior de Investigaciones Científicas: Santiago de Compostela, Spain, 1762; ISBN 978-84-96530-34-8. [Google Scholar]
- Meijide-Pardo, A. Apuntes Históricos Sobre Oleicultura Gallega. Rev. Econ. De Galicia 1964, 37–38, 93–102. [Google Scholar]
- Ramos-Cabrer, A.M.; Dìaz-Hernàndez, M.B.; Pereira-Lorenzo, S. Morphology and Microsatellites in Spanish Apple Collections. J. Hortic. Sci. Biotechnol. 2007, 82, 257–265. [Google Scholar] [CrossRef]
- Fernández-Cruz, J.; Míguez-Soto, B.; Fernández-López, J. Origin of Traditional Sweet Chestnut (Castanea Sativa Mill.) Varieties from the Northwest of the Iberian Peninsula. Tree Genet. Genomes 2022, 18, 34. [Google Scholar] [CrossRef]
- Rodríguez, V.M.; Cartea, M.E.; Padilla, G.; Velasco, P.; Ordás, A. The Nabicol: A Horticultural Crop in Northwestern Spain. Euphytica 2005, 142, 237–246. [Google Scholar] [CrossRef]
- Martínez, S.; Losada, P.; Franco, I.; Carballo, J. Protein, Amino Acid, Ash and Mineral Contents in Brassica Spp. Grown in Northwest Spain. Int. J. Food Sci. Technol. 2011, 46, 146–153. [Google Scholar] [CrossRef]
- Moreno-Larrazabal, A.; Teira-Brión, A.; Sopelana-Salcedo, I.; Arranz-Otaegui, A.; Zapata, L. Ethnobotany of Millet Cultivation in the North of the Iberian Peninsula. Veg. Hist. Archaeobot. 2015, 24, 541–554. [Google Scholar] [CrossRef]
- Sandalio de Arias, A. Agricultura General de Gabriel Alonso de Herrera Corregida y Adicionada Por La Real Sociedad Económica Matritense (Apartado Del Olivo); Imprenta Real: Madrid, Spain, 1818. [Google Scholar]
- Sánchez Rodríguez, A.M. La Agricultura Gallega En La Crisis Del Antiguo Régimen: Tentativas Modernizadoras. Obradoiro Hist. Mod. 2003, 12, 223–246. [Google Scholar] [CrossRef]
- Marchese, A.; Bonanno, F.; Marra, F.P.; Trippa, D.A.; Zelasco, S.; Rizzo, S.; Giovino, A.; Imperiale, V.; Ioppolo, A.; Sala, G.; et al. Recovery and Genotyping Ancient Sicilian Monumental Olive Trees. Front. Conserv. Sci. 2023, 4, 1206832. [Google Scholar] [CrossRef]
- Sarmiento, M. Viaje a Galicia (1745); Pensado, J.L., Ed.; Salamanca Universidad: Salamanca, Spain, 1975. [Google Scholar]
- Labrada, J.L. Descripción Económca Del Reyno de Galicia; Imprenta de Don Lorenzo José Riesgo y Montero: Ferrol, España, 1804. [Google Scholar]
- Gago, P.; Santiago, J.L.; Boso, S.; Martínez, M.C. The Forgotten, Ancient Olive Trees of the Spanish Northwest: A First Molecular and Botanical Analysis. Span. J. Agric. Res. 2019, 17, e0702. [Google Scholar] [CrossRef]
- Reboredo-Rodríguez, P.; González-Barreiro, C.; Cancho-Grande, B.; Simal-Gándara, J.; Trujillo, I. Genotypic and Phenotypic Identification of Olive Cultivars from North-Western Spain and Characterization of Their Extra Virgin Olive Oils in Terms of Fatty Acid Composition and Minor Compounds. Sci. Hortic. 2018, 232, 269–279. [Google Scholar] [CrossRef]
- Carvalho, J.; Yadav, S.; Garrido-Maestu, A.; Azinheiro, S.; Trujillo, I.; Barros-Velázquez, J.; Prado, M. Evaluation of Simple Sequence Repeats (SSR) and Single Nucleotide Polymorphism (SNP)-Based Methods in Olive Varieties from the Northwest of Spain and Potential for Miniaturization. Food Chem. Mol. Sci. 2021, 3, 100038. [Google Scholar] [CrossRef] [PubMed]
- Martínez, M.C.; Santiago, J.L.; Boso, S.; Gago, P. Bases Científicas Para La Creación de Una DOP o IGP “Aceites de Galicia. ” Almazaras 2019, 17, 36–44. [Google Scholar]
- Haddad, B.; Gristina, A.S.; Mercati, F.; Saadi, A.E.; Aiter, N.; Martorana, A.; Sharaf, A.; Carimi, F. Molecular Analysis of the Official Algerian Olive Collection Highlighted a Hotspot of Biodiversity in the Central Mediterranean Basin. Genes 2020, 11, 303. [Google Scholar] [CrossRef] [PubMed]
- Atrouz, K.; Bousba, R.; Marra, F.P.; Marchese, A.; Conforti, F.L.; Perrone, B.; Harkat, H.; Salimonti, A.; Zelasco, S. Algerian Olive Germplasm and Its Relationships with the Central-western Mediterranean Varieties Contributes to Clarify Cultivated Olive Diversification. Plants 2021, 10, 678. [Google Scholar] [CrossRef]
- Lazović, B.; Adakalić, M.; Pucci, C.; Perović, T.; Bandelj, D.; Belaj, A.; Mariotti, R.; Baldoni, L. Characterizing Ancient and Local Olive Germplasm from Montenegro. Sci. Hortic. 2016, 209, 117–123. [Google Scholar] [CrossRef]
- Baldoni, L.; Cultrera, N.G.; Mariotti, R.; Ricciolini, C.; Arcioni, S.; Vendramin, G.G.; Buonamici, A.; Porceddu, A.; Sarri, V.; Ojeda, M.A.; et al. A Consensus List of Microsatellite Markers for Olive Genotyping. Mol. Breed. 2009, 24, 213–231. [Google Scholar] [CrossRef]
- Marra, F.P.; Caruso, T.; Costa, F.; Di Vaio, C.; Mafrica, R.; Marchese, A. Genetic Relationships, Structure and Parentage Simulation among the Olive Tree (Olea europaea L. Subsp. Europaea) Cultivated in Southern Italy Revealed by SSR Markers. Tree Genet. Genomes 2013, 9, 961–973. [Google Scholar] [CrossRef]
- Rotondi, A.; Cultrera, N.G.M.; Mariotti, R.; Baldoni, L. Genotyping and Evaluation of Local Olive Varieties of a Climatically Disfavoured Region through Molecular, Morphological and Oil Quality Parameters. Sci. Hortic. 2011, 130, 562–569. [Google Scholar] [CrossRef]
- Linos, A.; Nikoloudakis, N.; Katsiotis, A.; Hagidimitriou, M. Genetic Structure of the Greek Olive Germplasm Revealed by RAPD, ISSR and SSR Markers. Sci. Hortic. 2014, 175, 33–43. [Google Scholar] [CrossRef]
- Bombarely, A.; Doulis, A.G.; Lambrou, K.K.; Zioutis, C.; Margaritis, E.; Koubouris, G. Elucidation of the Origin of the Monumental Olive Tree of Vouves in Crete, Greece. Plants 2021, 10, 2374. [Google Scholar] [CrossRef] [PubMed]
- Unver, H.; Sakar, E.; Ulas, M.; Ercisli, S.; Ak, B.E. Molecular Characterization of Indigenous Olive Genotypes Based on SSR Analysis. Genetika 2016, 48, 1017–1025. [Google Scholar] [CrossRef]
- Sakar, E.; Unver, H.; Ercisli, S. Genetic Diversity Among Historical Olive (Olea europaea L.) Genotypes from Southern Anatolia Based on SSR Markers. Biochem. Genet. 2016, 54, 842–853. [Google Scholar] [CrossRef]
- Valeri, M.C.; Mifsud, D.; Sammut, C.; Pandolfi, S.; Lilli, E.; Bufacchi, M.; Stanzione, V.; Passeri, V.; Baldoni, L.; Mariotti, R.; et al. Exploring Olive Genetic Diversity in the Maltese Islands. Sustainability 2022, 14, 684. [Google Scholar] [CrossRef]
- Haouane, H.; El Bakkali, A.; Moukhli, A.; Tollon, C.; Santoni, S.; Oukabli, A.; El Modafar, C.; Khadari, B. Genetic Structure and Core Collection of the World Olive Germplasm Bank of Marrakech: Towards the Optimised Management and Use of Mediterranean Olive Genetic Resources. Genetica 2011, 139, 1083–1094. [Google Scholar] [CrossRef]
- El Bakkali, A.; Essalouh, L.; Tollon, C.; Rivallan, R.; Mournet, P.; Moukhli, A.; Zaher, H.; Mekkaoui, A.; Hadidou, A.; Sikaoui, L.; et al. Characterization of Worldwide Olive Germplasm Banks of Marrakech (Morocco) and Córdoba (Spain): Towards Management and Use of Olive Germplasm in Breeding Programs. PLoS One 2019, 14, e0223716. [Google Scholar] [CrossRef]
- Yadav, S.; Carvalho, J.; Trujillo, I.; Prado, M. Microsatellite Markers in Olives (Olea europaea L.): Utility in the Cataloging of Germplasm, Food Authenticity and Traceability Studies. Foods 2021, 1, 1907. [Google Scholar] [CrossRef]
- Sion, S.; Savoia, M.A.; Gadaleta, S.; Piarulli, L.; Mascio, I.; Fanelli, V.; Montemurro, C.; Miazzi, M.M. How to Choose a Good Marker to Analyze the Olive Germplasm (Olea europaea L.) and Derived Products. Genes 2021, 12, 1474. [Google Scholar] [CrossRef]
- Sion, S.; Taranto, F.; Montemurro, C.; Mangini, G.; Camposeo, S.; Falco, V.; Gallo, A.; Mita, G.; Debbabi, O.S.; Amar, F.B.; et al. Genetic Characterization of Apulian Olive Germplasm as Potential Source in New Breeding Programs. Plants 2019, 8, 268. [Google Scholar] [CrossRef]
- International Organisation of Vine and Wine. OIV Descriptor List for Grape Varieties and Vitis Species, 2nd ed.; International Organisation of Vine and Wine: Dijon, France, 2009. [Google Scholar]
- Community Plant Variety Office-CPVO Protocol for Distinctness, Uniformity and Stability Tests, Olea europaea L. Available online: https://cpvo.europa.eu/sites/default/files/documents/olea_europaea_1.pdf (accessed on 25 March 2022).
- Miazzi, M.M.; di Rienzo, V.; Mascio, I.; Montemurro, C.; Sion, S.; Sabetta, W.; Vivaldi, G.A.; Camposeo, S.; Caponio, F.; Squeo, G.; et al. Ger.O.P.: An Integrated Project for the Recovery of Ancient and Rare Olive Germplasm. Front. Plant Sci. 2020, 11, 73. [Google Scholar] [CrossRef]
- Fontana, A.; Piscopo, A.; De Bruno, A.; Tiberini, A.; Muzzalupo, I.; Albanese, G. Impact of Olive Leaf Yellowing Associated Virus on Olive (Olea europaea L.) Oil. Eur. J. Lipid Sci. Technol. 2019, 121, 180047. [Google Scholar] [CrossRef]
- De La Rosa, R.; James, C.M.; Tobutt, K.R. Isolation and Characterization of Polymorphic Microsatellites in Olive (Olea europaea L.) and Their Transferability to Other Genera in the Oleaceae. Mol. Ecol. Notes 2002, 2, 265–267. [Google Scholar] [CrossRef]
- Sefc, K.M.; Lopes, M.S.; Mendonça, D.; Santos, M.R.D.; Machado, M.L.D.C.; Machado, A.D.C. Identification of Microsatellite Loci in Olive (Olea Europaea) and Their Characterization in Italian and Iberian Olive Trees. Mol. Ecol. 2000, 9, 1171–1173. [Google Scholar] [CrossRef] [PubMed]
- Carriero, F.; Fontanazza, G.; Cellini, F.; Giorio, G. Identification of Simple Sequence Repeats (SSRs) in Olive (Olea europaea L.). Theor. Appl. Genet. 2002, 104, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Cipriani, G.; Marrazzo, M.T.; Marconi, R.; Cimato, A.; Testolin, R. Microsatellite Markers Isolated in Olive (Olea europaea L.) Are Suitable for Individual Fingerprinting and Reveal Polymorphism within Ancient Cultivars. Theor. Appl. Genet. 2002, 104, 223–228. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Barranco, D.; Trujillo, I.; Rallo, L. Variedades de Olivo En España; Mundi-Prensa: Madrid, Spain, 2000. [Google Scholar]
- Rallo, L.; Barranco, D.; Caballero, J.M.; del Río, C.; Martín, A.; Tous, J.; Trujillo, I. Variedades de Olivo En España; Rallo, L., Barranco, D., Caballero, J.M., Del Río, C., Martín, A., Tous, J., Trujillo, I., Eds.; Mundi-Prensa: Madrid, Spain, 2005. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Martínez-Suárez, J.M.; Muñoz-Aranda, E.; Alba-Mendoza, J.; Lanzón-Rey, A. Informe Sobre Utilización Del Analizador de Rendimientos “Abencor. ” Grasas Y Aceites 1975, 26, 379–385. [Google Scholar]
- Belaj, A.; Dominguez-García, M.d.C.; Atienza, S.G.; Martín Urdíroz, N.; de la Rosa, R.; Satovic, Z.; Martín, A.; Kilian, A.; Trujillo, I.; Valpuesta, V.; et al. Developing a Core Collection of Olive (Olea europaea L.) Based on Molecular Markers (DArTs, SSRs, SNPs) and Agronomic Traits. Tree Genet. Genomes 2012, 8, 365–378. [Google Scholar] [CrossRef]
- Laaribi, I.; Gouta, H.; Mezghani Ayachi, M.; Labidi, F.; Mars, M. Combination of Morphological and Molecular Markers for the Characterization of Ancient Native Olive Accessions in Central-Eastern Tunisia. C R Biol. 2017, 340, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Muzzalupo, I.; Stefanizzi, F.; Perri, E. Evaluation of Olives Cultivated in Southern Italy by Simple Sequence Repeat Markers. HortScience 2009, 44, 582–588. [Google Scholar] [CrossRef]
- Zellama, M.S.; Varanda, C.M.R.; Materatski, P.; Nabi, N.; Hafsa, A.B.; Saamali, B.M.; Chaouachi, M.; Félix, M.R. An Integrated Approach for Understanding the High Infection Rates of Olive Viruses in Tunisia. Eur. J. Plant Pathol. 2019, 153, 1043–1054. [Google Scholar] [CrossRef]
- Luigi, M.; Godena, S.; Dermić, E.; Barba, M.; Faggioli, F. Detection of Viruses in Olive Trees in Croatian Istria. Phytopathol. Mediterr. 2011, 50, 150–153. [Google Scholar]
- Xylogianni, E.; Margaria, P.; Knierim, D.; Sareli, K.; Winter, S.; Chatzivassiliou, E.K.; Ali, A. Virus Surveys in Olive Orchards in Greece Identify Olive Virus T, a Novel Member of the Genus Tepovirus. Pathogens 2021, 10, 574. [Google Scholar] [CrossRef]
| SSRs LOCI | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name Given | N | ssrOeUA-DCA03 | ssrOeUA-DCA05 | ssrOeUA-DCA09 | ssrOeUA-DCA11 | ssrOeUA-DCA14 | ssrOeUA-DCA15 | ssrOeUA-DCA18 | |||||||
| Brava Gallega | 53 | 237–251 | 207–207 | 184–194 | 140–179 | 190–190 | 243–254 | 171–181 | |||||||
| Brétema | 28 | 228–251 | 201–207 | 172–184 | 130–161 | 173–180 | 243–254 | 171–181 | |||||||
| Carapucho | 3 | 237–243 | 207–207 | 182–206 | 140–140 | 190–190 | 254–254 | 171–187 | |||||||
| Carmeliña | 2 | 243–247 | 207–207 | 162–184 | 140–179 | 190–190 | 254–263 | 173–177 | |||||||
| Folgueira | 11 | 243–247 | 207–207 | 162–206 | 161–179 | 180–190 | 263–263 | 173–181 | |||||||
| Hedreira | 1 | 237–251 | 207–207 | 162–208 | 161–179 | 180–190 | 243–263 | 173–177 | |||||||
| Mansa Gallega | 13 | 228–243 | 201–207 | 182–184 | 130–140 | 173–190 | 254–254 | 171–187 | |||||||
| Maruxiña | 1 | 237–251 | 207–207 | 162–184 | 179–179 | 190–190 | 243–254 | 173–181 | |||||||
| Susiña | 1 | 237–247 | 207–207 | 162–184 | 140–179 | 190–190 | 254–263 | 179–181 | |||||||
| Xoana | 3 | 241–247 | 195–207 | 172–194 | 146–161 | 178–190 | 243–263 | 173–181 | |||||||
| Santiagueira | 1 | 243–251 | 207–207 | 184–194 | 179–179 | 190–190 | 243–263 | 173–181 | |||||||
| Arbequina (Control) | 3 | 230–241 | 203–207 | 184–206 | 140–179 | 190–190 | 243–263 | 169–179 | |||||||
| SSRs LOCI | |||||||||||||||
| Name Given | N | GAPU-59 | GAPU-71B | GAPU-101 | GAPU-103-A | UDO99-011 | UDO99-019 | UDO99-024 | UDO99-043 | ||||||
| Brava Gallega | 53 | 212–222 | 127–141 | 192–218 | 138–138 | 114–127 | 130–130 | 166–186 | 174–206 | ||||||
| Brétema | 28 | 212–222 | 124–141 | 190–192 | 165–165 | 110–112 | 130–130 | 178–186 | 172–214 | ||||||
| Carapucho | 3 | 212–222 | 124–141 | 190–218 | 138–165 | 112–114 | 100–130 | 178–186 | 172–218 | ||||||
| Carmeliña | 2 | 212–222 | 127–141 | 198–218 | 138–153 | 114–127 | 130–130 | 166–186 | 210–214 | ||||||
| Folgueira | 11 | 212–222 | 127–141 | 192–218 | 189–189 | 122–127 | 130–130 | 186–186 | 174–218 | ||||||
| Hedreira | 1 | 212–212 | 121–141 | 198–218 | 189–189 | 112–114 | 130–130 | 186–186 | 174–218 | ||||||
| Mansa Gallega | 13 | 222–222 | 124–127 | 190–192 | 165–165 | 112–127 | 100–130 | 166–178 | 172–216 | ||||||
| Maruxiña | 1 | 212–222 | 127–141 | 198–218 | 138–153 | 112–114 | 130–130 | 186–186 | 174–204 | ||||||
| Susiña | 1 | 222–222 | 141–141 | 192–192 | 138–138 | 122–127 | 130–130 | 166–186 | 174–206 | ||||||
| Xoana | 3 | 212–212 | 127–141 | 198–218 | 177–189 | 120–122 | 130–130 | 186–186 | 174–176 | ||||||
| Santiagueira | 1 | 212–222 | 127–141 | 198–200 | 189–189 | 114–127 | 130–130 | 186–186 | 210–218 | ||||||
| Arbequina (Control) | 3 | 222–222 | 121–141 | 184–206 | 153–162 | 112–124 | 130–155 | 202–202 | 176–176 | ||||||
| SSR Locus | Size Range | Na | Ne | I | Ho | He |
|---|---|---|---|---|---|---|
| ssrOeUA-DCA03 | 228–251 | 7 | 5.633 | 1.809 | 1.000 | 0.822 |
| ssrOeUA-DCA05 | 195–207 | 4 | 1.380 | 0.589 | 0.308 | 0.275 |
| ssrOeUA-DCA09 | 162–208 | 8 | 5.045 | 1.828 | 1.000 | 0.802 |
| ssrOeUA-DCA11 | 130–179 | 6 | 3.634 | 1.466 | 0.769 | 0.725 |
| ssrOeUA-DCA14 | 173–190 | 4 | 1.633 | 0.774 | 0.385 | 0.388 |
| ssrOeUA-DCA15 | 243–263 | 3 | 2.965 | 1.093 | 0.769 | 0.663 |
| ssrOeUA-DCA18 | 169–187 | 7 | 5.281 | 1.778 | 1.000 | 0.811 |
| GAPU-59 | 212–222 | 2 | 1.988 | 0.690 | 0.615 | 0.497 |
| GAPU-71B | 118–141 | 5 | 3.045 | 1.291 | 0.923 | 0.672 |
| GAPU-101 | 184–218 | 7 | 4.507 | 1.658 | 0.923 | 0.778 |
| GAPU-103-A | 138–189 | 6 | 4.072 | 1.537 | 0.385 | 0.754 |
| UDO99-011 | 110–127 | 7 | 4.630 | 1.670 | 1.000 | 0.784 |
| UDO99-019 | 100–155 | 3 | 1.266 | 0.431 | 0.231 | 0.210 |
| UDO99-024 | 166–202 | 4 | 2.126 | 1.012 | 0.462 | 0.530 |
| UDO99-043 | 172–218 | 9 | 7.191 | 2.071 | 0.923 | 0.861 |
| All loci | 100–263 | 65 | 5.633 | 1.809 | 1.000 | 0.822 |
| Mean | 5.467 | 3.626 | 1.313 | 0.713 | 0.638 |
| Leaf Descriptors | ||||
|---|---|---|---|---|
| UPOV 5 Length | UPOV 6 Width | UPOV 7 Ratio Length/Width | UPOV 9 Curvature of Longitudinal Axis | |
| Brava Gallega | 5 | 3 | 7 | 2 |
| Medium | Narrow | Very elongated | Straight | |
| Brétema | 5 | 5 | 3–5 | 2 |
| Medium | Medium | Slightly-Moderately elongated | Straight | |
| Carapucho | 5 | 3 | 5 | 2 |
| Medium | Narrow | Moderately elongated | Straight | |
| Carmeliña | 5 | 3 | 7 | 2 |
| Medium | Narrow | Very elongated | Straight | |
| Folgueira | 5 | 3 | 7 | 2 |
| Medium | Narrow | Very elongated | Straight | |
| Hedreira | 5 | 5 | 5 | 2 |
| Medium | Medium | Moderately elongated | Straight | |
| Mansa Gallega | 5 | 5 | 5 | 2 |
| Medium | Medium | Moderately elongated | Straight | |
| Maruxiña | 5 | 3 | 7 | 2 |
| Medium | Narrow | Very elongated | Straight | |
| Susiña | 5 | 3 | 5 | 2 |
| Medium | Narrow | Moderately elongated | Straight | |
| Santiagueira | 5 | 3 | 7 | 2 |
| Medium | Narrow | Very elongated | Straight | |
| Xoana | 5 | 3 | 7 | 2 |
| Medium | Narrow | Very elongated | Straight | |
| Arbequina | 3 | 5 | 3 | 3 |
| Short | Medium | Slightly elongated | Recurved | |
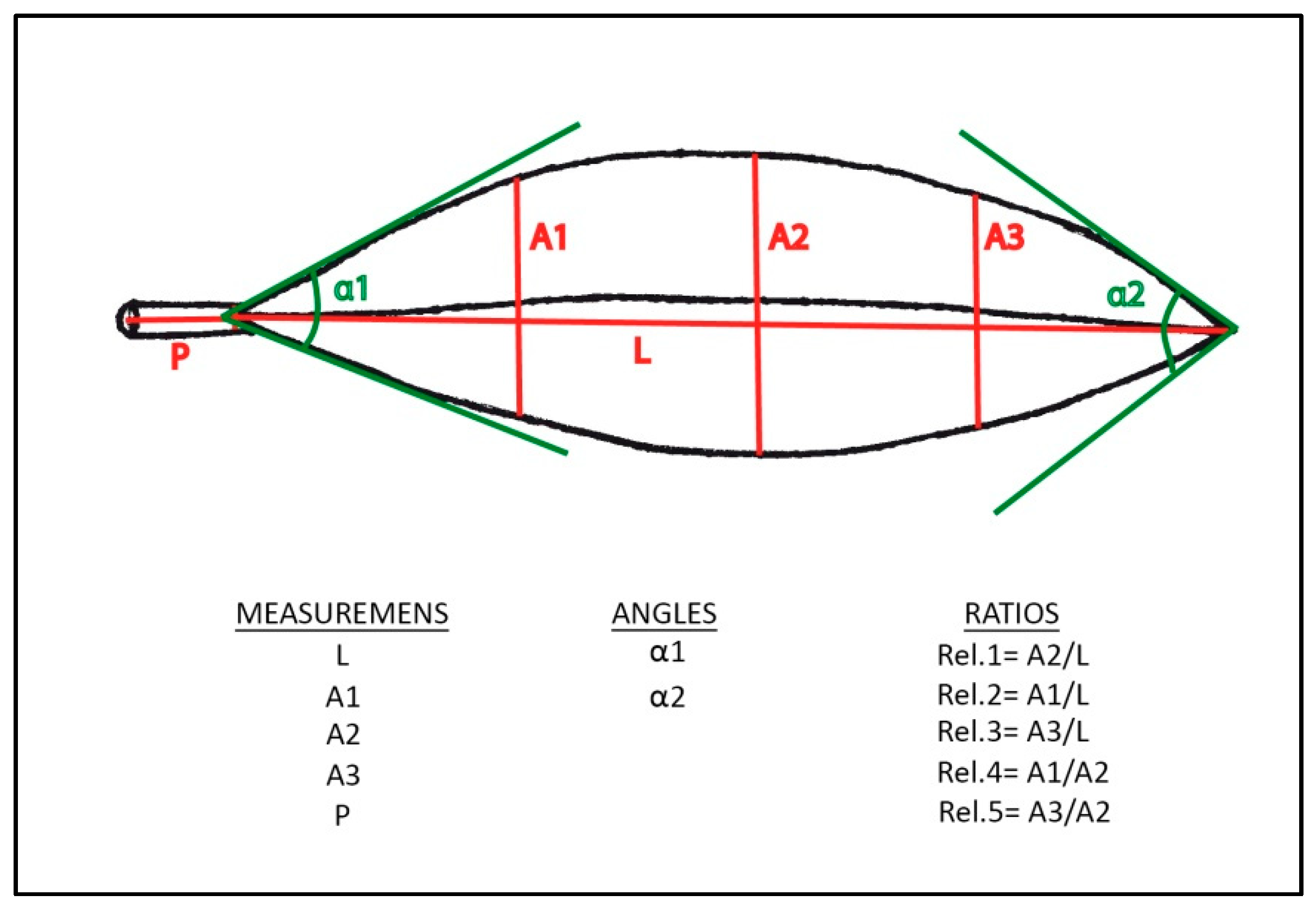
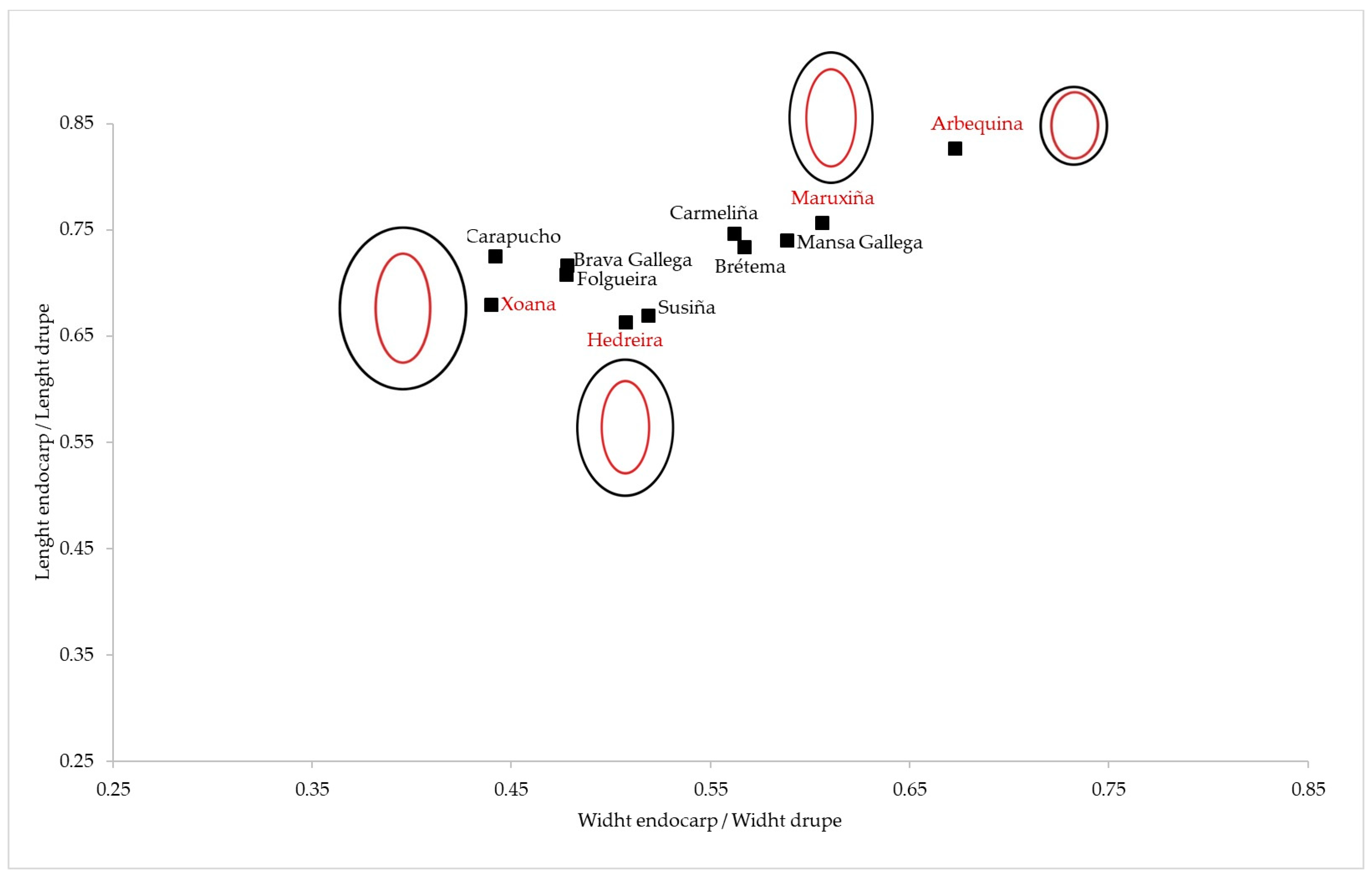
| REL.1 = A2/L | REL.2 = A1/L | REL.3 = A3/L | REL.4 = A1/A2 | REL.5 = A3/A2 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | SD | CV | M | SD | CV | M | SD | CV | M | SD | CV | M | SD | CV | |
| Brava Gallega | 0.18 | 0.02 | 0.12 | 0.12 | 0.02 | 0.14 | 0.14 | 0.02 | 0.11 | 1.54 | 0.18 | 0.12 | 1.23 | 0.15 | 0.12 |
| Brétema | 0.23 | 0.03 | 0.15 | 0.24 | 0.09 | 0.38 | 0.23 | 0.05 | 0.21 | 1.12 | 0.44 | 0.39 | 1.04 | 0.26 | 0.25 |
| Carapucho | 0.15 | 0.03 | 0.17 | 0.19 | 0.04 | 0.19 | 0.16 | 0.03 | 0.19 | 0.81 | 0.06 | 0.08 | 0.85 | 0.06 | 0.08 |
| Carmeliña | 0.18 | 0.02 | 0.09 | 0.12 | 0.01 | 0.12 | 0.15 | 0.01 | 0.09 | 1.50 | 0.14 | 0.09 | 1.24 | 0.17 | 0.14 |
| Folgueira | 0.16 | 0.02 | 0.15 | 0.10 | 0.02 | 0.16 | 0.13 | 0.02 | 0.16 | 1.52 | 0.18 | 0.12 | 1.27 | 0.19 | 0.15 |
| Hedreira | 0.15 | 0.03 | 0.17 | 0.20 | 0.03 | 0.17 | 0.16 | 0.03 | 0.21 | 0.77 | 0.07 | 0.09 | 0.83 | 0.10 | 0.12 |
| Mansa Gallega | 0.14 | 0.03 | 0.25 | 0.20 | 0.04 | 0.19 | 0.17 | 0.04 | 0.23 | 0.70 | 0.09 | 0.13 | 0.84 | 0.08 | 0.10 |
| Maruxiña | 0.17 | 0.02 | 0.12 | 0.13 | 0.02 | 0.15 | 0.14 | 0.02 | 0.15 | 1.32 | 0.12 | 0.09 | 1.09 | 0.15 | 0.14 |
| Santiagueira | 0.23 | 0.02 | 0.11 | 0.17 | 0.02 | 0.12 | 0.17 | 0.02 | 0.12 | 1.33 | 0.13 | 0.10 | 1.01 | 0.13 | 0.13 |
| Susiña | 0.18 | 0.02 | 0.14 | 0.13 | 0.02 | 0.16 | 0.14 | 0.02 | 0.15 | 1.43 | 0.12 | 0.08 | 1.14 | 0.14 | 0.12 |
| Xoana | 0.16 | 0.03 | 0.16 | 0.12 | 0.02 | 0.17 | 0.13 | 0.02 | 0.19 | 1.26 | 0.13 | 0.10 | 1.06 | 0.18 | 0.17 |
| Arbequina | 0.17 | 0.04 | 0.22 | 0.23 | 0.04 | 0.17 | 0.18 | 0.03 | 0.18 | 0.14 | 0.18 | 0.12 | 1.08 | 0.18 | 0.16 |
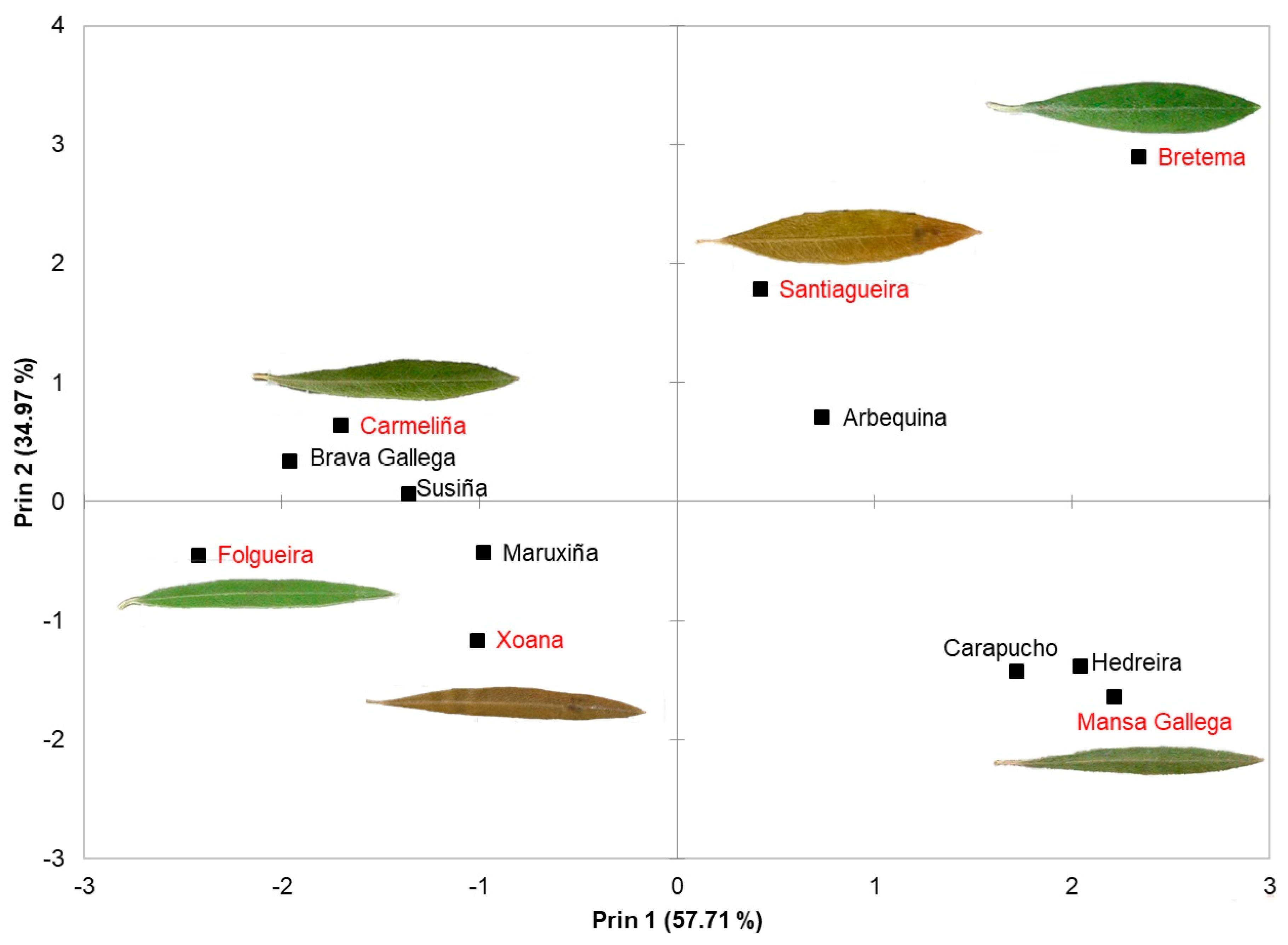
| PCA Variable | |||
|---|---|---|---|
| Component | Autovalue | Proportion | Acc. Var. |
| 1 | 2.89 | 0.5771 | 0.5771 |
| 2 | 1.75 | 0.3497 | 0.9268 |
| 3 | 0.28 | 0.0559 | 0.9826 |
| 4 | 0.09 | 0.0173 | 0.9999 |
| 5 | 0.00 | 0.0001 | 1.0000 |
| Drupe Descriptors * | ||||||||
|---|---|---|---|---|---|---|---|---|
| UPOV16 | UPOV18 | UPOV22 | UPOV23 | DiamMaxDrup | UPOV24 | UPOV25 | UPOV26 | |
| Brava Gallega | 5 | 5 | 3 | 2 | 2 | 3 | 1 | 3 |
| Medium | Moderately elongated | Black | Weakly asymmetric | Center | Rounded | Absent | Truncate | |
| Brétema | 5 | 5 | 3 | 2 | 2 | 3 | 1 | 3 |
| Medium | Moderately elongated | Black | Weakly asymmetric | Center | Rounded | Absent | Truncate | |
| Carapucho | 5 | 7 | 3 | 2 | 2 | 3 | 1 | 3 |
| Medium | Very elongated | Black | Weakly asymmetric | Center | Rounded | Absent | Truncate | |
| Carmeliña | 5 | 5 | 2 | 2 | 2 | 3 | 1 | 3 |
| Medium | Moderately elongated | Dark violet | Weakly asymmetric | Center | Rounded | Absent | Truncate | |
| Folgueira | 5 | 5 | 3 | 2 | 2 | 3 | 1 | 1 |
| Medium | Moderately elongated | Black | Weakly asymmetric | Center | Rounded | Absent | Rounded | |
| Hedreira | 5 | 5 | 2 | 3 | 1 | 2 | 2 | 3 |
| Medium | Moderately elongated | Dark violet | Strongly asymmetric | Toward the base | Obtuse | Moderate | Truncate | |
| Mansa Gallega | 3 | 5 | 3 | 2 | 2 | 3 | 1 | 3 |
| Low | Moderately elongated | Black | Weakly asymmetric | Center | Rounded | Absent | Truncate | |
| Maruxiña | 5 | 5 | 2 | 2 | 2 | 3 | 3 | 1 |
| Medium | Moderately elongated | Dark violet | Weakly asymmetric | Center | Rounded | Strong | Rounded | |
| Susiña | 3 | 3 | 1 | 1 | 2 | 3 | 1 | 3 |
| Low | Slightly elongated | Medium violet | Symmetric | Center | Rounded | Absent | Truncate | |
| Xoana | 7 | 5 | 1 | 3 | 2 | 2 | 2 | 3 |
| High | Moderately elongated | Medium violet | Strongly asymmetric | Center | Obtuse | Moderate | Truncate | |
| Arbequina | 3 | 3 | 3 | 1 | 1 | 3 | 1 | 3 |
| Low | Slightly elongated | Black | Symmetric | Toward the base | Rounded | Absent | Truncate | |
| Endocarp Descriptors * | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| UPOV31 | UPOV32 | UPOV33 | UPOV34 | UPOV35 | UPOV36 | UPOV37 | UPOV38 | UPOV39 | UPOV40 | DMax Endo | |
| Brava Gallega | 2 | 5 | 2 | 1 | 2 | 1 | 3 | 9 | 2 | 2 | 2 |
| Moderately elongated | Medium | Weakly asymmetric | Symmetric | Between 7 and 10 | Evenly distributed | Rounded | Present | Rounded | Medium | Centered | |
| Brétema | 2 | 5 | 3 | 1 | 1 | 3 | 3 | 9 | 2 | 2 | 2 |
| Moderately elongated | Medium | Strongly asymmetric | Symmetric | Less than 7 | Strongly grouped | Rounded | Present | Rounded | Medium | Centered | |
| Carapucho | 3 | 5 | 2 | 1 | 2 | 1 | 1 | 9 | 1 | 2 | 2 |
| Very elongated | Medium | Weakly asymmetric | Symmetric | Between 7 and 10 | Evenly distributed | Acute | Present | Acute | Medium | Centered | |
| Carmeliña | 2 | 7 | 2 | 1 | 2 | 1 | 3 | 9 | 1 | 2 | 2 |
| Moderately elongated | High | Weakly asymmetric | Symmetric | Between 7 and 10 | Evenly distributed | Rounded | Present | Acute | Medium | Centered | |
| Folgueira | 2 | 5 | 2 | 1 | 2 | 1 | 3 | 9 | 1 | 2 | 3 |
| Moderately elongated | Medium | Weakly asymmetric | Symmetric | Between 7 and 10 | Evenly distributed | Rounded | Present | Acute | Medium | Toward the apex | |
| Hedreira | 2 | 7 | 2 | 1 | 2 | 1 | 3 | 9 | 2 | 2 | 1 |
| Moderately elongated | High | Weakly asymmetric | Symmetric | Between 7 and 10 | Evenly distributed | Rounded | Present | Rounded | Medium | Toward the base | |
| Mansa Gallega | 2 | 3 | 2 | 1 | 2 | 1 | 3 | 9 | 2 | 1 | 2 |
| Moderately elongated | Low | Weakly asymmetric | Symmetric | Between 7 and 10 | Evenly distributed | Rounded | Present | Rounded | Weak | Centered | |
| Maruxiña | 2 | 7 | 2 | 1 | 2 | 1 | 1 | 9 | 3 | 2 | 2 |
| Moderately elongated | High | Weakly asymmetric | Symmetric | Between 7 and 10 | Evenly distributed | Acute | Present | Truncate | Medium | Centered | |
| Susiña | u | 3 | 2 | 1 | 2 | 1 | 3 | 9 | 2 | 1 | 2 |
| Moderately elongated | Low | Weakly asymmetric | Symmetric | Between 7 and 10 | Evenly distributed | Rounded | Present | Rounded | Weak | Centered | |
| Xoana | 2 | 7 | 2 | 1 | 2 | 1 | 1 | 9 | 1 | 3 | 2 |
| Moderately elongated | High | Weakly asymmetric | Symmetric | Between 7 and 10 | Evenly distributed | Acute | Present | Acute | Strong | Centered | |
| Arbequina | 1 | 3 | 1 | 1 | 2 | 1 | 3 | 1 | 2 | 2 | 2 |
| Slightly elongated | Low | Symmetric | Symmetric | Between 7 and 10 | Evenly distributed | Rounded | Absent | Rounded | medium | Centered | |
| Drupes | Endocarps | ||||||
|---|---|---|---|---|---|---|---|
| Variable | Variety | Mean | SD | CV (%) | Mean | SD | CV (%) |
| Weight (g) | Brava Gallega | 3.16 | 0.92 | 29.16 | 0.43 | 0.08 | 19.07 |
| Brétema | 2.01 | 0.56 | 27.96 | 0.38 | 0.09 | 23.98 | |
| Carapucho | 2.94 | 0.71 | 24.16 | 0.35 | 0.06 | 18.07 | |
| Carmeliña | 2.17 | 0.48 | 22.25 | 0.47 | 0.06 | 13.6 | |
| Folgueira | 2.84 | 1.00 | 35.36 | 0.39 | 0.08 | 21.67 | |
| Hedreira | 2.70 | 0.31 | 11.51 | 0.49 | 0.06 | 12.97 | |
| Mansa Gallega | 1.03 | 0.20 | 19.8 | 0.23 | 0.04 | 18.34 | |
| Maruxiña | 2.41 | 0.62 | 25.83 | 0.60 | 0.09 | 15.56 | |
| Susiña | 1.66 | 0.32 | 19.28 | 0.26 | 0.06 | 20.92 | |
| Xoana | 4.16 | 0.84 | 20.27 | 0.51 | 0.09 | 16.78 | |
| Arbequina | 0.77 | 0.11 | 14.65 | 0.26 | 0.03 | 11.55 | |
| Length (mm) | Brava Gallega | 21.08 | 2.21 | 10.47 | 14.93 | 1.69 | 11.3 |
| Brétema | 19.18 | 2.02 | 10.51 | 14.09 | 1.50 | 10.64 | |
| Carapucho | 21.62 | 2.02 | 9.34 | 15.69 | 1.82 | 11.58 | |
| Carmeliña | 18.88 | 1.47 | 7.77 | 14.10 | 0.93 | 6.58 | |
| Folgueira | 20.01 | 2.53 | 12.65 | 14.34 | 1.44 | 10.01 | |
| Hedreira | 19.72 | 1.11 | 5.63 | 13.08 | 1.09 | 8.3 | |
| Mansa Gallega | 15.18 | 1.08 | 7.09 | 11.24 | 0.87 | 7.76 | |
| Maruxiña | 19.96 | 1.76 | 8.83 | 15.11 | 0.97 | 6.38 | |
| Susiña | 14.70 | 0.92 | 6.27 | 9.84 | 1.00 | 10.12 | |
| Xoana | 23.95 | 2.12 | 8.85 | 16.27 | 1.75 | 10.76 | |
| Arbequina | 12.00 | 0.60 | 5 | 9.92 | 0.81 | 8.2 | |
| Width (mm) | Brava Gallega | 15.53 | 1.75 | 11.27 | 7.42 | 0.59 | 7.91 |
| Brétema | 13.33 | 1.45 | 10.86 | 7.55 | 0.67 | 8.87 | |
| Carapucho | 14.77 | 1.49 | 10.06 | 6.53 | 0.41 | 6.32 | |
| Carmeliña | 13.70 | 1.20 | 8.73 | 7.70 | 0.52 | 6.8 | |
| Folgueira | 15.04 | 2.09 | 13.89 | 7.19 | 0.61 | 8.5 | |
| Hedreira | 15.42 | 0.69 | 4.47 | 7.82 | 0.27 | 3.5 | |
| Mansa Gallega | 10.53 | 0.74 | 7.06 | 6.19 | 0.49 | 7.83 | |
| Maruxiña | 14.14 | 1.59 | 11.22 | 8.57 | 0.75 | 8.76 | |
| Susiña | 13.31 | 1.00 | 7.47 | 6.91 | 0.48 | 6.97 | |
| Xoana | 17.80 | 1.58 | 8.85 | 7.83 | 0.58 | 7.4 | |
| Arbequina | 9.87 | 0.82 | 8.32 | 6.64 | 0.35 | 5.22 | |
| Width/Length ratio | Brava Gallega | 0.74 | 0.07 | 8.98 | 0.50 | 0.10 | 18.86 |
| Brétema | 0.70 | 0.06 | 8.66 | 0.53 | 0.05 | 9.41 | |
| Carapucho | 0.69 | 0.09 | 12.44 | 0.42 | 0.04 | 10.30 | |
| Carmeliña | 0.73 | 0.05 | 7.08 | 0.55 | 0.04 | 7.97 | |
| Folgueira | 0.75 | 0.05 | 7.12 | 0.50 | 0.04 | 8.43 | |
| Hedreira | 0.78 | 0.05 | 5.96 | 0.60 | 0.06 | 9.29 | |
| Mansa Gallega | 0.70 | 0.05 | 6.64 | 0.55 | 0.04 | 8.04 | |
| Maruxiña | 0.71 | 0.04 | 5.11 | 0.57 | 0.04 | 6.75 | |
| Susiña | 0.91 | 0.06 | 8.05 | 0.71 | 0.08 | 11.26 | |
| Xoana | 0.75 | 0.06 | 7.87 | 0.49 | 0.07 | 14.13 | |
| Arbequina | 0.82 | 0.06 | 7.26 | 0.67 | 0.07 | 10.60 | |
| Free Acidity (% Oleic Acid) | Water and Volatile Compound (% m/m) | Ether-Soluble Impurities (% m/m) | Peroxide Index (meq O2 Peroxidized per kg Oil) | K 270 * | K 232 ** | ∆K | |
|---|---|---|---|---|---|---|---|
| Variety | Rule 2568/91 CEE Annex II | UNE 55 020 | UNE 55 020 | Rule 2568/91 CEE Annex III | Rule 2568/91 CEE Annex IX | Rule 2568/91 CEE Annex IX | Rule 2568/91 CEE Annex IX |
| Brava Gallega | 0.37 | 0.32 | 0.03 | 3.05 | 0.13 | 1.76 | 0.00 |
| Brétema | 0.47 | 0.29 | 0.02 | 6.00 | 0.13 | 1.53 | 0.00 |
| Carapucho | 0.18 | MD | MD | MD | MD | MD | MD |
| Carmeliña | 0.46 | 0.82 | 0.03 | 3.90 | 0.13 | 1.42 | 0.00 |
| Folgueira | 0.20 | 0.32 | 0.03 | 2.50 | 0.12 | 1.57 | 0.00 |
| Hedreira | 0.22 | 0.30 | 0.04 | 4.30 | 0.17 | 1.43 | 0.00 |
| Mansa Gallega | 0.23 | 0.24 | 0.03 | 8.45 | 0.11 | 1.20 | 0.00 |
| Maruxiña | 0.27 | MD | MD | MD | MD | MD | MD |
| Xoana | 0.33 | 0.30 | 0.03 | 3.70 | 0.18 | 1.60 | 0.00 |
| Arbequina | 0.29 | 0.54 | 0.03 | 2.45 | 0.09 | 1.28 | 0.00 |
| EVOO reference $ | ≤0.80 | ≤0.2 | ≤0.1 | ≤20 | ≤0.22 | ≤2.5 | ≤0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gago, P.; Boso, S.; Santiago, J.-L.; Martínez, M.-C. Identification and Characterization of Relict Olive Varieties (Olea europaea L.) in the Northwest of the Iberian Peninsula. Horticulturae 2024, 10, 175. https://doi.org/10.3390/horticulturae10020175
Gago P, Boso S, Santiago J-L, Martínez M-C. Identification and Characterization of Relict Olive Varieties (Olea europaea L.) in the Northwest of the Iberian Peninsula. Horticulturae. 2024; 10(2):175. https://doi.org/10.3390/horticulturae10020175
Chicago/Turabian StyleGago, Pilar, Susana Boso, José-Luis Santiago, and María-Carmen Martínez. 2024. "Identification and Characterization of Relict Olive Varieties (Olea europaea L.) in the Northwest of the Iberian Peninsula" Horticulturae 10, no. 2: 175. https://doi.org/10.3390/horticulturae10020175
APA StyleGago, P., Boso, S., Santiago, J.-L., & Martínez, M.-C. (2024). Identification and Characterization of Relict Olive Varieties (Olea europaea L.) in the Northwest of the Iberian Peninsula. Horticulturae, 10(2), 175. https://doi.org/10.3390/horticulturae10020175








