Volatile Constituents of Cymbopogon citratus (DC.) Stapf Grown in Greenhouse in Serbia: Chemical Analysis and Chemometrics
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growing Conditions
2.2. Harvest and Postharvest Processing
2.3. Analysis of Volatile Compounds
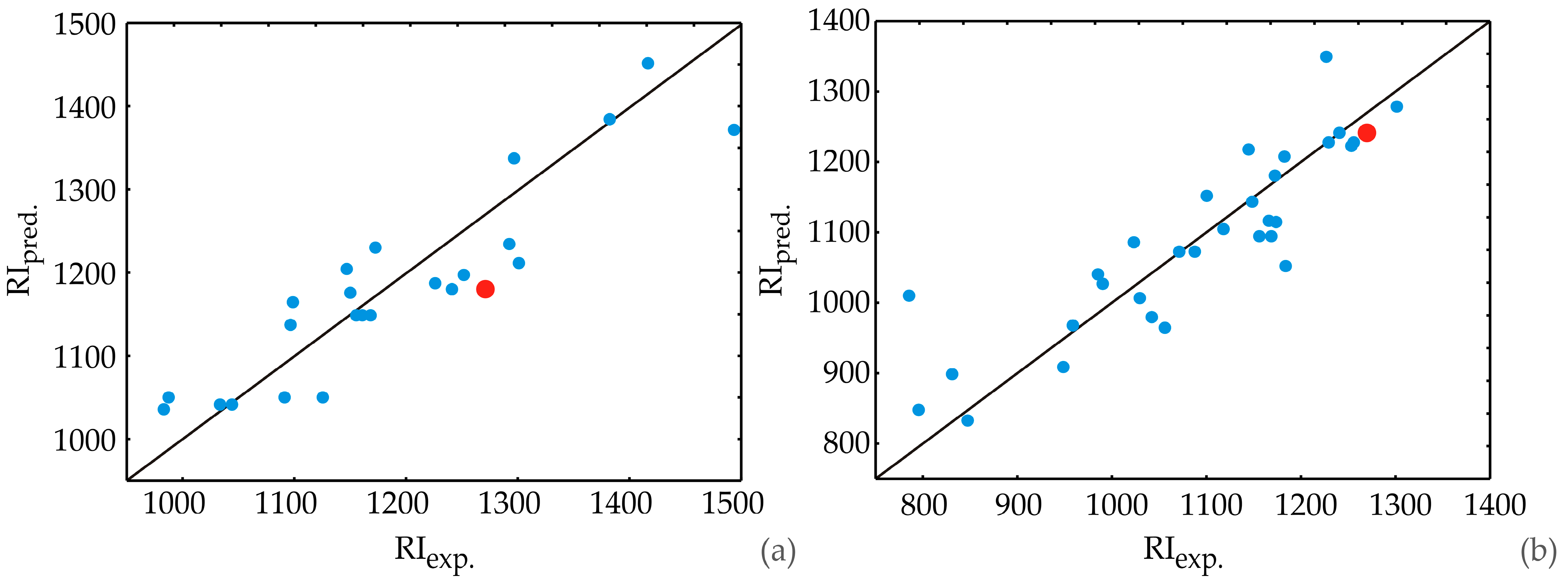
2.4. Quantitative Structure Retention Relationship (QSRR) Analysis
2.5. Artificial Neural Network (ANN)
2.6. Statistical Tests of the Model Fit
2.7. Global Sensitivity Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yeşil, Y.; Akalın, E. Comparative morphological and anatomical characteristics of the species known as lemongrass (limonotu): Melissa officinalis L., Cymbopogon citratus (DC) Stapf. and Aloysia citriodora Palau. J. Fac. Pharm. Istanb. 2015, 45, 29–37. [Google Scholar]
- Bekele, L.D.; Zhang, W.; Liu, Y.; Duns, G.J.; Yu, C.; Jin, L.; Li, X.; Jia, Q.; Chen, J. Preparation and characterization of lemongrass fiber (Cymbopogon species) for reinforcing application in thermoplastic composites. BioResources 2017, 12, 5664–5681. [Google Scholar] [CrossRef]
- Kamaruddin, Z.H.; Jumaidin, R.; Selamat, M.Z.; Ilyas, R.A. Characteristics and properties of lemongrass (Cymbopogan citratus): A comprehensive review. J. Nat. Fibers 2022, 19, 8101–8118. [Google Scholar] [CrossRef]
- Aungtikun, J.; Soonwera, M.; Sittichok, S. Insecticidal synergy of essential oils from Cymbopogon citratus (Stapf.), Myristica fragrans (Houtt.), and Illicium verum Hook. f. and their major active constituents. Ind. Crops Prod. 2021, 164, 113386. [Google Scholar] [CrossRef]
- Manokari, M.; Cokul Raj, M.; Dey, A.; Faisal, M.; Alatar, A.A.; Joshee, N.; Shekhawat, M.S. Structural alterations of Cymbopogon citratus (DC.) Stapf leaves and roots caused by silicon nanoparticles during in vitro propagation. Ind. Crops Prod. 2023, 197, 116648. [Google Scholar] [CrossRef]
- Rodrigues, L.; Coelho, E.; Madeira, R.; Teixeira, P.; Henriques, I.; Coimbra, M.A. Food ingredients derived from lemongrass byproduct hydrodistillation: Essential oil, hydrolate, and decoction. Molecules 2022, 27, 2493. [Google Scholar] [CrossRef]
- Dewi, A.O.T.; Antari, E.D.; Dewi, W.E.R. Antioxidant activity of lemongrass (Cympogon nardus L.) hydrosol with various extraction time. In Proceedings of the 1st International Innovation Technology, Surakarta, Indonesia, 27 July 2023; Volume 1, pp. 130–137. [Google Scholar]
- Aćimović, M.; Čabarkapa, I.; Cvetković, M.; Stanković, J.; Kiprovski, B.; Gvozdenac, S.; Puvača, N. Cymbopogon citratus (DC.) Staph: Chemical composition, antimicrobial and antioxidant activities, use in medicinal and cosmetic purpose. J. Agron. Technol. Eng. Manag. 2019, 2, 344–360. [Google Scholar]
- Wolfender, J.L.; Marti, G.; Thomas, A.; Bertrand, S. Current approaches and challenges for the metabolite profiling of complex natural extracts. J. Chromatogr. A 2015, 1382, 136–164. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- AlSheikh, H.M.A.; Sultan, I.; Kumar, V.; Rather, I.A.; Al-Sheikh, H.; Tasleem Jan, A.; Haq, Q.M.R. Plant-based phytochemicals as possible alternative to antibiotics in combating bacterial drug resistance. Antibiotics 2020, 9, 480. [Google Scholar] [CrossRef]
- Vaou, N.; Stavropoulou, E.; Voidarou, C.; Tsakris, Z.; Rozos, G.; Tsigalou, C.; Bezirtzoglou, E. Interactions between medical plant-derived bioactive compounds: Focus on antimicrobial combination effects. Antibiotics 2022, 11, 1014. [Google Scholar] [CrossRef] [PubMed]
- Dulo, B.; Phan, K.; Githaiga, J.; Raes, K.; De Meester, S. Natural quinone dyes: A review on structure, extraction techniques, analysis and application potential. Waste Biomass Valorization 2021, 12, 6339–6374. [Google Scholar] [CrossRef]
- Kaliszan, R. QSRR: Quantitative structure- (Chromatographic) retention relationships. Chem. Rev. 2007, 107, 3212–3246. [Google Scholar] [CrossRef] [PubMed]
- Blaženović, I.; Kind, T.; Ji, J.; Fiehn, O. Software tools and approaches for compound identification of LC-MS/MS data in metabolomics. Metabolites 2018, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Kind, T.; Fiehn, O. Advances in structure elucidation of small molecules using mass spectrometry. Bioanal. Rev. 2010, 2, 23–60. [Google Scholar] [CrossRef]
- Creek, D.J.; Jankevics, A.; Breitling, R.; Watson, D.G.; Barrett, M.P.; Burgess, K.E. Toward global metabolomics analysis with hydrophilic interaction liquid chromatography–mass spectrometry: Improved metabolite identification by retention time prediction. Anal. Chem. 2011, 83, 8703–8710. [Google Scholar] [CrossRef]
- Martínez-Santiago, O.; Millán-Cabrera, R.; Marrero-Ponce, Y.; Barigye, S.J.; Martínez-López, Y.; Torrens, F.; Pérez-Giménez, F. Discrete derivatives for atom-pairs as a novel graph-theoretical invariant for generating new molecular descriptors: Orthogonality, interpretation and QSARs/QSPRs on Benchmark Databases. Mol. Inform. 2014, 33, 343–368. [Google Scholar] [CrossRef]
- Marrero-Ponce, Y.; Barigye, S.J.; Jorge-Rodríguez, M.E.; Tran-Thi-Thu, T. QSRR prediction of gas chromatography retention indices of essential oil components. Chem. Pap. 2018, 72, 57–69. [Google Scholar] [CrossRef]
- Usman, A.G.; Işik, S.; Abba, S.I. A novel multi-model data-driven ensemble technique for the prediction of retention factor in HPLC method development. Chromatographia 2020, 83, 933–945. [Google Scholar] [CrossRef]
- Matyushin, D.D.; Sholokhova, A.Y.; Buryak, A.K. A deep convolutional neural network for the estimation of gas chromatographic retention indices. J. Chromatogr. A 2019, 1607, 460395. [Google Scholar] [CrossRef]
- Kensert, A.; Bouwmeester, R.; Efthymiadis, K.; Van Broeck, P.; Desmet, G.; Cabooter, D. Graph convolutional networks for improved prediction and interpretability of chromatographic retention data. Anal. Chem. 2021, 93, 15633–15641. [Google Scholar] [CrossRef]
- Wu, L.; Gong, P.; Wu, Y.; Liao, K.; Shen, H.; Qi, Q.; Liu, H.; Wang, G.; Hao, H. An integral strategy toward the rapid identification of analogous nontarget compounds from complex mixtures. J. Chromatogr. A 2013, 1303, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Héberger, K. Quantitative structure–(chromatographic) retention relationships. J. Chromatogr. A 2007, 1158, 273–305. [Google Scholar] [CrossRef] [PubMed]
- Khezeli, T.; Daneshfar, A.; Sahraei, R. A green ultrasonic-assisted liquid–liquid microextraction based on deep eutectic solvent for the HPLC-UV determination of ferulic, caffeic and cinnamic acid from olive, almond, sesame and cinnamon oil. Talanta 2016, 150, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Tropsha, A.; Golbraikh, A. Predictive QSAR modeling workflow, model applicability domains, and virtual screening. Curr. Pharm. Des. 2007, 13, 3494–3504. [Google Scholar] [CrossRef]
- Liebal, U.W.; Phan, A.N.; Sudhakar, M.; Raman, K.; Blank, L.M. Machine learning applications for mass spectrometry-based metabolomics. Metabolites 2020, 10, 243. [Google Scholar] [CrossRef]
- Zisi, C.; Sampsonidis, I.; Fasoula, S.; Papachristos, K.; Witting, M.; Gika, H.G.; Nikitas, P.; Pappa-Louisi, A. QSRR modeling for metabolite standards analyzed by two different chromatographic columns using multiple linear regression. Metabolites 2017, 7, 7. [Google Scholar] [CrossRef]
- Dong, J.; Cao, D.S.; Miao, H.Y.; Liu, S.; Deng, B.C.; Yun, Y.H.; Wang, N.N.; Lu, A.P.; Zeng, W.B.; Chen, A.F. ChemDes: An integrated web-based platform for molecular descriptor and fingerprint computation. J. Cheminform. 2015, 7, 60. [Google Scholar] [CrossRef]
- Yap, C.W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011, 32, 1446–1474. [Google Scholar] [CrossRef]
- Goldberg, D.E. Genetic Algorithms in Search, Optimisation and Machine Learning; Addison-Wesley: Boston, MA, USA, 1989. [Google Scholar]
- Tropsha, A. Best practices for QSAR model development, validation, and exploitation. Mol. Inform. 2010, 29, 476–488. [Google Scholar] [CrossRef]
- Aalizadeh, R.; Thomaidis, N.S.; Bletsou, A.A.; Gago-Ferrero, P. Quantitative structure–retention relationship models to support nontarget high-resolution mass spectrometric screening of emerging contaminants in environmental samples. J. Chem. Inf. Model. 2016, 56, 1384–1398. [Google Scholar] [CrossRef] [PubMed]
- Badem, H.; Basturk, A.; Caliskan, A.; Yuksel, M.E. A new hybrid optimization method combining artificial bee colony and limited-memory BFGS algorithms for efficient numerical optimization. Appl. Soft Comput. 2018, 70, 826–844. [Google Scholar] [CrossRef]
- Amirian, E.; Leung, J.Y.; Zanon, S.; Dzurman, P. Integrated cluster analysis and artificial neural network modeling for steam-assisted gravity drainage performance prediction in heterogeneous reservoirs. Expert Syst. Appl. 2015, 42, 723–740. [Google Scholar] [CrossRef]
- Xu, Q.; Wei, C.; Liu, R.; Gu, S.; Xu, J. Quantitative structure–property relationship study of β-cyclodextrin complexation free energies of organic compounds. Chemometr. Intell. Lab. Syst. 2015, 146, 313–321. [Google Scholar] [CrossRef]
- Kojic, P.; Omorjan, R. Predicting hydrodynamic parameters and volumetric gas–liquid mass transfer coefficient in an external-loop airlift reactor by support vector regression. Chem. Eng. Res. Des. 2018, 125, 398–407. [Google Scholar] [CrossRef]
- Zanin, M.; Papo, D.; Sousa, P.A.; Menasalvas, E.; Nicchi, A.; Kubik, E.; Boccaletti, S. Combining complex networks and data mining: Why and how. Phys. Rep. 2016, 35, 1–44. [Google Scholar] [CrossRef]
- Zhu, C.; Ni, R.; Xu, Z.; Kong, K.; Huang, W.R.; Goldstein, T. Gradinit: Learning to initialize neural networks for stable and efficient training. Adv. Neur. Inf. Process. Syst. 2021, 34, 16410–16422. [Google Scholar] [CrossRef]
- Statistica. Statistica 10 Software (StatSoft, Inc. STATISTICA, Ver. 10, Data Analysis Software System). 2010. Available online: https://www.statsoft.de/en/data-science-applications/tibco-statistica/ (accessed on 15 December 2018).
- Bakshaev, A.; Rudzkis, R. Goodness-of-fit tests based on the empirical characteristic function. Lith. Math. J. 2017, 57, 155–170. [Google Scholar] [CrossRef]
- Maydeu-Olivares, A. Maximum likelihood estimation of structural equation models for continuous data: Standard errors and goodness of fit. Struct. Equ. Model. 2017, 24, 383–394. [Google Scholar] [CrossRef]
- Yoon, Y.; Swales, G.; Margavio, T.M. A comparison of discriminant analysis versus artificial neural networks. J. Oper. Res. Soc. 2017, 44, 51–60. [Google Scholar] [CrossRef]
- Aćimović, M.; Kiprovski, B.; Gvozdenac, S. Application of Cymbopogon citratus in agro-food industry. J. Agron. Technol. Eng. Manag. 2020, 3, 423–436. [Google Scholar]
- Mukarram, M.; Choudhary, S.; Khan, M.A.; Poltronieri, P.; Khan, M.M.A.; Ali, J.; Kurjak, D.; Shahid, M. Lemongrass essential oil components with antimicrobial and anticancer activities. Antioxidants 2021, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- Tazi, A.; Zinedine, A.; Rocha, J.M.; Errachidi, F. Review on the pharmacological properties of lemongrass (Cymbopogon citratus) as a promising source of bioactive compounds. Pharmacol. Res. Nat. Prod. 2024, 3, 100046. [Google Scholar] [CrossRef]
- Du, X.; Zhang, M.; Wang, S.; Li, J.; Zhang, J.; Liu, D. Ethnopharmacology, chemical composition and functions of Cymbopogon citratus. Chin. Herb. Med. 2024, 16, 358–374. [Google Scholar] [CrossRef] [PubMed]
- Poonpaiboonpipat, T.; Pangnakorn, U.; Suvunnamek, U.; Teerarak, M.; Charoenying, P.; Laosinwattan, C. Phytotoxic effects of essential oil from Cymbopogon citratus and its physiological mechanisms on barnyardgrass (Echinochloa crus-galli). Ind. Crops Prod. 2013, 41, 403–407. [Google Scholar] [CrossRef]
- Bassolé, I.H.N.; Lamien-Meda, A.; Bayalaa, B.; Obame, L.C.; Ilboudo, A.J.; Franz, C.; Novak, J.; Nebiéc, R.C.; Dicko, M.H. Chemical composition and antimicrobial activity of Cymbopogon citratus and Cymbopogon giganteus essential oils alone and in combination. Phytomedicine 2011, 18, 1070–1074. [Google Scholar] [CrossRef]
- Mohamed Hanaa, A.R.; Sallam, Y.I.; El-Leithy, A.S.; Aly, S.E. Lemongrass (Cymbopogon citratus) essential oil as affected by drying methods. Ann. Agric. Sci. 2012, 57, 113–116. [Google Scholar] [CrossRef]
- Mwithiga, G.; Maina, S.; Muturi, P.; Gitari, J. Lemongrass (Cymbopogon flexuosus) agronomic traits, oil yield and oil quality under different agro-ecological zones. J. Agric. Food Res. 2022, 10, 100422. [Google Scholar] [CrossRef]
- Ajayi, E.O.; Sadimenko, A.P.; Afolayan, A.J. GC–MS evaluation of Cymbopogon citratus (DC) Stapf oil obtained using modified hydrodistillation and microwave extraction methods. Food Chem. 2016, 209, 262–266. [Google Scholar] [CrossRef]
- Benoudjit, F.; Hamoudi, I.; Aboulouz, A. Extraction and characterization of essential oil and hydrolate obtained from an Algerian lemongrass (Cymbopogon citratus). Alger. J. Environ. Sci. Technol. 2022, 8, 2256–2263. [Google Scholar]
- Morales-Aranibar, L.; Yucra, F.E.Y.; Estrada, N.M.P.; Flores, P.Q.; Zevallos, R.N.M.; Zegarra, J.C.L.; Trujillo, U.P.; Aranibar, C.G.M.; Gonzales, H.H.S.; Aguilera, J.G.; et al. Production of new biopesticides from Cymbopogon citratus for the control of coffee rust (Hemileia vastatrix) under laboratory and field conditions. Plants 2023, 12, 1166. [Google Scholar] [CrossRef] [PubMed]
- Erceg, T.; Šovljanski, O.; Tomić, A.; Aćimović, M.; Stupar, A.; Baloš, S. Comparison of the properties of pullulan-based active edible coatings implemented for improving sliced cheese shelf life. Polymers 2024, 16, 178. [Google Scholar] [CrossRef] [PubMed]
- Noorizadeh, H.; Farmany, A. QSRR models to predict retention indices of cyclic compounds of essential oils. Chromatographia 2010, 72, 563–569. [Google Scholar] [CrossRef]
- Li, X.; Wang, H.; Jiang, M.; Ding, M.; Xu, X.; Xu, B.; Zou, Y.; Yu, Y.; Yang, W. Collision cross section prediction based on machine learning. Molecules 2023, 28, 4050. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Shi, L.M.; Kohn, K.W.; Pommier, Y.; Weinstein, J.N. Quantitative structure-antitumor activity relationships of camptothecin analogues: Cluster analysis and genetic algorithm-based studies. J. Med. Chem. 2001, 44, 3254–3263. [Google Scholar] [CrossRef]
- Leardi, R. Genetic algorithms in chemometrics and chemistry: A review. J. Chemometr. 2001, 15, 559–569. [Google Scholar] [CrossRef]
- Edache, E.I.; Uzairu, A.; Abechi, S.E. Development and estimation of an in silico model for anti-HIV-1 integrase inhibitor using genetic function approximation. J. Adv. Med. 2016, 5, 1–18. [Google Scholar] [CrossRef]
- Moreau, G.; Broto, P. Autocorrelation of molecular structures, application to SAR studies. Nouv. J. Chim. 1980, 4, 757–764. [Google Scholar]
- Hollas, B.; Gutman, I.; Trinajstić, N. On reducing correlation between topological indices. Croat. Chem. Acta 2005, 78, 489–492. [Google Scholar]
- Todeschini, R.; Consonni, V. Handbook of Molecular Descriptors; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Moran, P.A. Notes on continuous stochastic phenomena. Biometrika 1950, 37, 17–23. [Google Scholar] [CrossRef]
- Hall, L.H.; Kier, L.B. Electrotopological state indices for atom types: A novel combination of electronic, topological, and valence state information. J. Chem. Inf. Comp. Sci. 1995, 35, 1039–1045. [Google Scholar] [CrossRef]
- Kier, L.B.; Hall, L.H.; Frazer, J.W. An index of electrotopological state for atoms in molecules. J. Math. Chem. 1991, 7, 229–241. [Google Scholar] [CrossRef]
- García-Domenech, R.; Gálvez, J.; de Julián-Ortiz, J.V.; Pogliani, L. Some new trends in chemical graph theory. Chem. Rev. 2008, 108, 1127–1169. [Google Scholar] [CrossRef] [PubMed]

| pH | CaCO3 *** | Humus **** | Total Nitrogen ***** | P2O5 ****** | K2O ****** | |
|---|---|---|---|---|---|---|
| in KCl * | in H2O ** | |||||
| 7.46 | 8.39 | 0.84% | 4.18% | 0.268% | 30.03 mg/100 g soil | 44.15 mg/100 g soil |
| 1st Season | 2nd Season | 3rd Season | |
|---|---|---|---|
| Dry leaf yield (kg)/greenhouse | 102.00 ± 7.90 | 83.00 ± 1.88 | 58.00 ± 3.83 |
| Essential oil yield (L)/greenhouse | 0.93 ± 0.06 | 0.77 ± 0.05 | 0.50 ± 0.05 |
| Wight of 100 mL essential oil (g) | 86.63 ± 7.63 | 86.90 ± 5.00 | 87.20 ± 8.43 |
| No. | Compound | RIlit | RIexp | RIpred. | RIerror | R.T. (min) | 1st Season | 2nd Season | 3rd Season |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 6-Methyl-5-hepten-2-one | 986 | 983 | 1036.071 | −53.071 | 7.270 | 4.50 ± 0.09 | 2.50 ± 0.00 | 2.00 ± 0.08 |
| 2 | Myrcene | 988 | 988 | 1050.141 | −62.141 | 7.424 | 22.50 ± 2.13 | 15.60 ± 0.91 | 25.50 ± 0.98 |
| 3 | cis-β-Ocimene | 1032 | 1034 | 1041.030 | −7.030 | 8.994 | 0.40 ± 0.00 | 0.20 ± 0.02 | 0.20 ± 0.01 |
| 4 | trans-β-Ocimene | 1044 | 1044 | 1041.030 | 2.970 | 9.379 | 0.30 ± 0.02 | 0.20 ± 0.01 | 0.10 ± 0.00 |
| 5 | 6,7-Epoxymyrcene | 1088 | 1091 | 1050.141 | 40.859 | 11.115 | 0.10 ± 0.01 | 0.10 ± 0.00 | nd |
| 6 | Rosefuran | 1106 | 1097 | 1136.960 | −39.960 | 11.342 | nd | 0.10 ± 0.01 | nd |
| 7 | Linalool | 1095 | 1097 | 1137.469 | −40.469 | 11.366 | 1.10 ± 0.04 | 0.70 ± 0.07 | 0.80 ± 0.08 |
| 8 | Perillene | 1002 | 1099 | 1164.838 | −65.838 | 11.435 | 0.40 ± 0.03 | 0.30 ± 0.00 | 0.20 ± 0.00 |
| 9 | allo-Ocimene | 1128 | 1126 | 1049.858 | 76.142 | 12.597 | 0.10 ± 0.01 | nd | nd |
| 10 | exo-Isocitral | 1140 | 1143 | 1125.798 | 17.202 | 13.270 | 0.10 ± 0.00 | tr | nd |
| 11 | trans-Chrysanthemal | 1153 | 1147 | 1203.735 | −56.735 | 13.472 | 0.10 ± 0.01 | 0.10 ± 0.01 | nd |
| 12 | Citronellal | 1148 | 1150 | 1176.408 | −26.408 | 13.597 | 0.40 ± 0.04 | 0.30 ± 0.00 | 0.30 ± 0.02 |
| 13 | trans-Pinocamphone | 1158 | 1155 | 1149.154 | 5.846 | 13.872 | nd | nd | 0.10 ± 0.00 |
| 14 | cis-Isocitral | 1160 | 1161 | 1149.154 | 11.846 | 14.098 | 0.60 ± 0.04 | 0.80 ± 0.01 | 0.90 ± 0.06 |
| 15 | cis-Pinocamphone | 1172 | 1168 | 1149.154 | 18.846 | 14.448 | nd | nd | 0.50 ± 0.00 |
| 16 | Rosefuran epoxide | 1173 | 1173 | 1229.515 | −56.515 | 14.587 | 0.10 ± 0.01 | 0.10 ± 0.01 | 0.20 ± 0.01 |
| 17 | trans-Isocitral | 1177 | 1180 | 1201.643 | −21.643 | 14.884 | 1.00 ± 0.08 | 1.30 ± 0.02 | 1.40 ± 0.13 |
| 18 | Citronellol | 1223 | 1226 | 1187.474 | 38.526 | 16.913 | 0.50 ± 0.04 | 0.40 ± 0.03 | nd |
| 19 | Neral | 1227 | 1241 | 1180.087 | 60.913 | 17.562 | 27.10 ± 0.75 | 32.30 ± 2.28 | 28.70 ± 0.29 |
| 20 | Geraniol | 1249 | 1252 | 1197.480 | 54.520 | 18.085 | 3.10 ± 0.12 | 2.80 ± 0.05 | 1.80 ± 0.15 |
| 21 | Geranial | 1264 | 1271 | 1180.087 | 90.913 | 18.916 | 34.70 ± 1.40 | 40.70 ± 3.56 | 35.60 ± 0.82 |
| 22 | 2-Undecanone | 1294 | 1292 | 1233.893 | 58.107 | 19.820 | 0.40 ± 0.04 | 0.10 ± 0.00 | tr |
| 23 | Thymol | 1289 | 1297 | 1336.532 | −39.532 | 19.978 | nd | 0.10 ± 0.00 | nd |
| 24 | Carvacrol | 1298 | 1301 | 1212.075 | 88.925 | 20.221 | nd | 0.10 ± 0.01 | nd |
| 25 | Geranyl acetate | 1379 | 1382 | 1383.747 | −1.747 | 23.801 | 0.30 ± 0.00 | 0.40 ± 0.01 | 0.40 ± 0.03 |
| 26 | trans-Caryophyllene | 1408 | 1417 | 1451.016 | −34.016 | 25.326 | 0.20 ± 0.02 | 0.20 ± 0.01 | 0.30 ± 0.00 |
| 27 | trans-α-Bergamotene | 1432 | 1433 | 1403.273 | 29.727 | 26.012 | 0.20 ± 0.00 | 0.20 ± 0.01 | 0.20 ± 0.01 |
| 28 | 2-Tridecanone | 1497 | 1494 | 1371.961 | 122.039 | 28.536 | 0.20 ± 0.01 | 0.10 ± 0.01 | nd |
| 29 | Caryophyllene oxide | 1582 | 1581 | 1536.723 | 44.277 | 32.118 | 0.10 ± 0.00 | 0.10 ± 0.00 | 0.10 ± 0.01 |
| Sum | 98.5 | 99.8 | 99.3 |
| No. | Compound | RIlit | RIexp | RIpred. | RIerror | R.T. (min) | 1st Season | 2nd Season | 3rd Season |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 3-Methyl-2-butenal | 784 | 785 | 1009.350 | −224,350 | 3.109 | 0.30 ± 0.00 | 0.30 ± 0.02 | nd |
| 2 | Hexanal | 801 | 796 | 848.331 | −52,331 | 3.288 | 0.10 ± 0.00 | 0.10 ± 0.00 | nd |
| 3 | 2,2-Dimethyl-3(2H)-furanone | 834 | 831 | 898.210 | −67,210 | 3.875 | 0.40 ± 0.02 | 0.40 ± 0.04 | 0.40 ± 0.04 |
| 4 | 3Z-Hexenol | 850 | 847 | 832.563 | 14,437 | 4.126 | 0.40 ± 0.04 | 0.70 ± 0.00 | tr |
| 5 | 5,5-Dimethyl-2(5H)-furanone | 952 | 949 | 909.646 | 39,354 | 6.364 | 0.10 ± 0.00 | nd | nd |
| 6 | Benzaldehyde | 952 | 959 | 968.564 | −9564 | 6.614 | 0.10 ± 0.01 | nd | nd |
| 7 | 6-Methyl-5-hepten-2-one | 981 | 985 | 1039.808 | −54,808 | 7.368 | 23.70 ± 2.17 | 17.60 ± 1.33 | 20.90 ± 1.19 |
| 8 | Dehydro-1,8-cineole | 995 | 990 | 1026.268 | −36,268 | 7.493 | 3.50 ± 0.21 | 4.20 ± 0.39 | 4.00 ± 0.34 |
| 9 | p-Cymene | 1020 | 1023 | 1085.259 | −62,259 | 8.605 | 0.10 ± 0.01 | nd | 0.10 ± 0.01 |
| 10 | 1,8-Cineole | 1026 | 1030 | 1006.697 | 23,303 | 8.852 | 0.10 ± 0.01 | 0.10 ± 0.01 | tr |
| 11 | cis-β-Ocimene | 1032 | 1033 | 1011.734 | 21,266 | 9.035 | nd | nd | 0.20 ± 0.01 |
| 12 | Benzene acetaldehyde | 1036 | 1042 | 980.087 | 61,913 | 9.300 | 0.20 ± 0.00 | 0.20 ± 0.02 | 0.10 ± 0.01 |
| 13 | 2,5,5-Trimethyl-3-hexyn-2-ol # | / | 1056 | 965.121 | 90,879 | 9.889 | nd | nd | 0.20 ± 0.01 |
| 14 | cis-Linalool oxide (furanoid) | 1067 | 1071 | 1073.100 | −2100 | 10.358 | 0.60 ± 0.03 | 0.60 ± 0.02 | 0.90 ± 0.07 |
| 15 | trans-Linalool oxide (furanoid) | 1084 | 1088 | 1073.100 | 14,900 | 10.973 | 0.60 ± 0.02 | 0.60 ± 0.04 | 0.70 ± 0.06 |
| 16 | Linalool | 1095 | 1100 | 1152.415 | −52,415 | 11.424 | 2.80 ± 0.27 | 2.00 ± 0.03 | 2.80 ± 0.07 |
| 17 | trans-2,8-p-Mentha-dien-1-ol | 1118 | 1118 | 1105.124 | 12,876 | 12.221 | nd | 0.40 ± 0.03 | 0.30 ± 0.00 |
| 18 | cis-p-Mentha-2,8-dien-1-ol | 1133 | 1133 | 1134.732 | −1732 | 12.880 | 0.30 ± 0.03 | 0.30 ± 0.03 | 0.30 ± 0.01 |
| 19 | Ipsdienol | 1140 | 1145 | 1217.318 | −72,318 | 13.377 | nd | 0.10 ± 0.00 | nd |
| 20 | trans-Chrysanthemal | 1153 | 1149 | 1143.801 | 5199 | 13.519 | nd | 0.10 ± 0.01 | 0.10 ± 0.01 |
| 21 | trans-Pinocamphone | 1158 | 1156 | 1095.148 | 60,852 | 13.876 | nd | nd | 0.30 ± 0.03 |
| 22 | p-Mentha-1,5-dien-8-ol | 1166 | 1166 | 1116.561 | 49,439 | 14.259 | nd | 1.10 ± 0.03 | 4.60 ± 0.08 |
| 23 | cis-Pinocamphone | 1172 | 1168 | 1095.148 | 72,852 | 14.471 | nd | nd | 0.80 ± 0.00 |
| 24 | Terpinen-4-ol | 1174 | 1173 | 1114.630 | 58,370 | 14.634 | nd | nd | 0.10 ± 0.00 |
| 25 | Menthol | 1167 | 1172 | 1180.735 | −8735 | 14.552 | nd | 0.10 ± 0.01 | nd |
| 26 | trans-Isocitral | 1177 | 1183 | 1174.781 | 8219 | 14.978 | nd | 0.10 ± 0.00 | 0.30 ± 0.03 |
| 27 | NI-1 * | / | 1183 | 1207.915 | −24,915 | 15.068 | 1.60 ± 0.11 | nd | nd |
| 28 | p-Cymen-8-ol | 1179 | 1184 | 1053.132 | 130,868 | 15.093 | nd | nd | 0.90 ± 0.01 |
| 29 | α-Terpineol | 1186 | 1186 | 1201.723 | −15,723 | 15.121 | nd | 0.10 ± 0.00 | nd |
| 30 | NI-2 ** | / | 1191 | 1210.783 | −19,783 | 15.377 | 4.60 ± 0.27 | nd | nd |
| 31 | Citronellol | 1223 | 1227 | 1350.247 | −123,247 | 16.972 | 0.60 ± 0.02 | nd | nd |
| 32 | Nerol | 1227 | 1229 | 1228.622 | 0378 | 17.023 | nd | 0.40 ± 0.03 | nd |
| 33 | Neral | 1235 | 1241 | 1242.116 | −1116 | 17.609 | 18.40 ± 1.56 | 26.10 ± 0.73 | 20.00 ± 1.31 |
| 34 | Geraniol | 1249 | 1256 | 1228.622 | 27,378 | 18.202 | nd | 5.20 ± 0.24 | 0.10 ± 0.01 |
| 35 | Piperitone | 1249 | 1253 | 1222.984 | 30,016 | 18.164 | 5.00 ± 0.20 | nd | tr |
| 36 | Geranial | 1270 | 1270 | 1242.116 | 27,884 | 18.968 | 26.50 ± 2.19 | 32.60 ± 0.02 | 31.20 ± 2.59 |
| 37 | Geranyl formate | 1300 | 1302 | 1278.547 | 23,453 | 20.278 | 0.10 ± 0.01 | nd | nd |
| Sum | 90.1 | 93.4 | 89.3 |
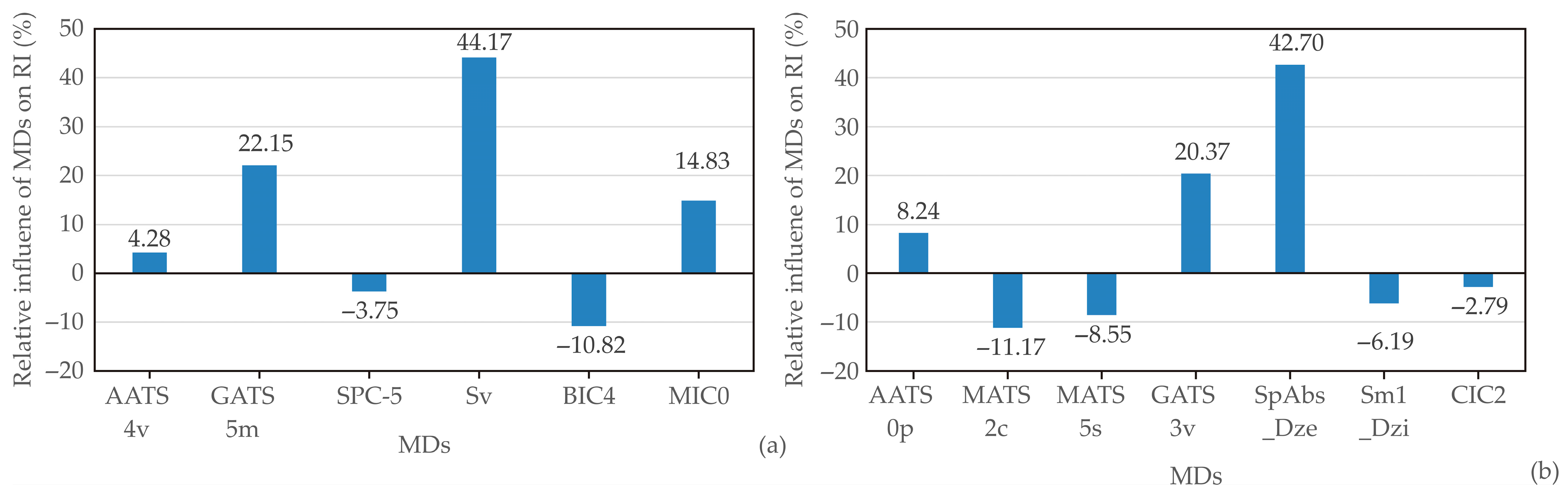
| MD | MD Group | MD Meaning |
|---|---|---|
| AATS4v | Autocorrelation | Average Broto–Moreau autocorrelation—lag 4/weighted by van der Waals volumes |
| GATS5m | Autocorrelation | Geary autocorrelation—lag 5/weighted by mass |
| SPC-5 | Chi path cluster | Simple path cluster, order 5 |
| Sv | Constitutional | Sum of atomic van der Waals volumes (scaled on carbon atom) |
| BIC4 | Information content | Bond information content index (neighborhood symmetry of 4th order) |
| MIC0 | Information content | Modified information content index (neighborhood symmetry of 0th order) |
| GATS5m | SPC-5 | Sv | BIC4 | MIC0 | |
|---|---|---|---|---|---|
| AATS4v | 0.200 p = 0.339 | 0.094 p = 0.655 | 0.172 p = 0.411 | 0.258 p = 0.213 | 0.038 p = 0.856 |
| GATS5m | −0.076 p = 0.720 | 0.167 p = 0.425 | −0.038 p = 0.857 | 0.022 p = 0.918 | |
| SPC-5 | 0.095 p = 0.651 | 0.372 p = 0.067 | 0.189 p = 0.366 | ||
| Sv | −0.066 p = 0.754 | −0.050 p = 0.811 | |||
| BIC4 | 0.176 p = 0.399 |
| MD | MD Group | MD Meaning |
|---|---|---|
| AATS0p | Autocorrelation | Average Broto–Moreau autocorrelation—lag 0/weighted by polarizabilities |
| MATS2c | Autocorrelation | Moran autocorrelation—lag 2/weighted by charges |
| MATS5s | Autocorrelation | Moran autocorrelation—lag 5/weighted by I state |
| GATS3v | Autocorrelation | Geary autocorrelation—lag 3/weighted by van der Waals volumes |
| SpAbs_Dze | Barysz matrix | Graph energy from Barysz matrix/weighted by Sanderson electronegativities |
| SM1_Dzi | Barysz matrix | Spectral moment of order 1 from Barysz matrix/weighted by first ionization potential |
| CIC2 | Information content | Complementary information content index (neighborhood symmetry of 2nd order) |
| MATS2c | MATS5s | GATS3v | SpAbs_Dze | SM1_Dzi | CIC2 | |
|---|---|---|---|---|---|---|
| AATS0p | −0.153 p = 0.404 | −0.230 p = 0.206 | −0.224 p = 0.218 | −0.005 p = 0.978 | 0.309 p = 0.085 | 0.073 p = 0.690 |
| MATS2c | 0.085 p = 0.646 | 0.239 p = 0.188 | 0.056 p = 0.759 | 0.087 p = 0.636 | 0.287 p = 0.111 | |
| MATS5s | 0.019 p = 0.917 | 0.223 p = 0.220 | −0.050 p = 0.786 | 0.187 p = 0.306 | ||
| GATS3v | 0.239 p = 0.187 | −0.098 p = 0.594 | 0.292 p = 0.105 | |||
| SpAbs_Dze | 0.259 p = 0.152 | 0.183 p = 0.315 | ||||
| SM1_Dzi | −0.004 p = 0.985 |
| Network | Performance | Error | Training Algorithm | Error Function | Activation | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Training | Testing | Validation | Training | Testing | Validation | Hidden | Output | |||
| MLP 6-10-1 | 0.819 | 0.241 | 1.000 | 1274.580 | 1427.840 | 2957.379 | BFGS 5 | SOS | Exp. | Exp. |
| MLP 7-11-1 | 0.872 | 0.746 | 1.000 | 1060.173 | 9060.502 | 2018.322 | BFGS 12 | SOS | Log. | Exp. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aćimović, M.; Lončar, B.; Todosijević, M.; Lekić, S.; Erceg, T.; Pezo, M.; Pezo, L. Volatile Constituents of Cymbopogon citratus (DC.) Stapf Grown in Greenhouse in Serbia: Chemical Analysis and Chemometrics. Horticulturae 2024, 10, 1116. https://doi.org/10.3390/horticulturae10101116
Aćimović M, Lončar B, Todosijević M, Lekić S, Erceg T, Pezo M, Pezo L. Volatile Constituents of Cymbopogon citratus (DC.) Stapf Grown in Greenhouse in Serbia: Chemical Analysis and Chemometrics. Horticulturae. 2024; 10(10):1116. https://doi.org/10.3390/horticulturae10101116
Chicago/Turabian StyleAćimović, Milica, Biljana Lončar, Marina Todosijević, Stefan Lekić, Tamara Erceg, Milada Pezo, and Lato Pezo. 2024. "Volatile Constituents of Cymbopogon citratus (DC.) Stapf Grown in Greenhouse in Serbia: Chemical Analysis and Chemometrics" Horticulturae 10, no. 10: 1116. https://doi.org/10.3390/horticulturae10101116
APA StyleAćimović, M., Lončar, B., Todosijević, M., Lekić, S., Erceg, T., Pezo, M., & Pezo, L. (2024). Volatile Constituents of Cymbopogon citratus (DC.) Stapf Grown in Greenhouse in Serbia: Chemical Analysis and Chemometrics. Horticulturae, 10(10), 1116. https://doi.org/10.3390/horticulturae10101116









