Full-Length Transcriptome Analysis and Characterization of DFRs Involved in the Formation of Anthocyanin in Allium wallichii
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Anthocyanins in Flowers of A. wallichii
2.3. RNA Extraction, Library Construction, and Sequencing
2.4. Raw Data Processing and Full-Length Tanscriptome Analysis
2.5. Cloning and Sequence Analysis of AwDFRs
2.6. Relative Expression of AwDFRs in Different Tissues of A. wallichii
2.7. Assay of AwDFRs Activities
2.8. Anthocyanin Analysis of Transformed Arabidopsis and Tobacco
3. Results
3.1. UPLC-MS Analysis of Anthocyanins in A. wallichii Flower
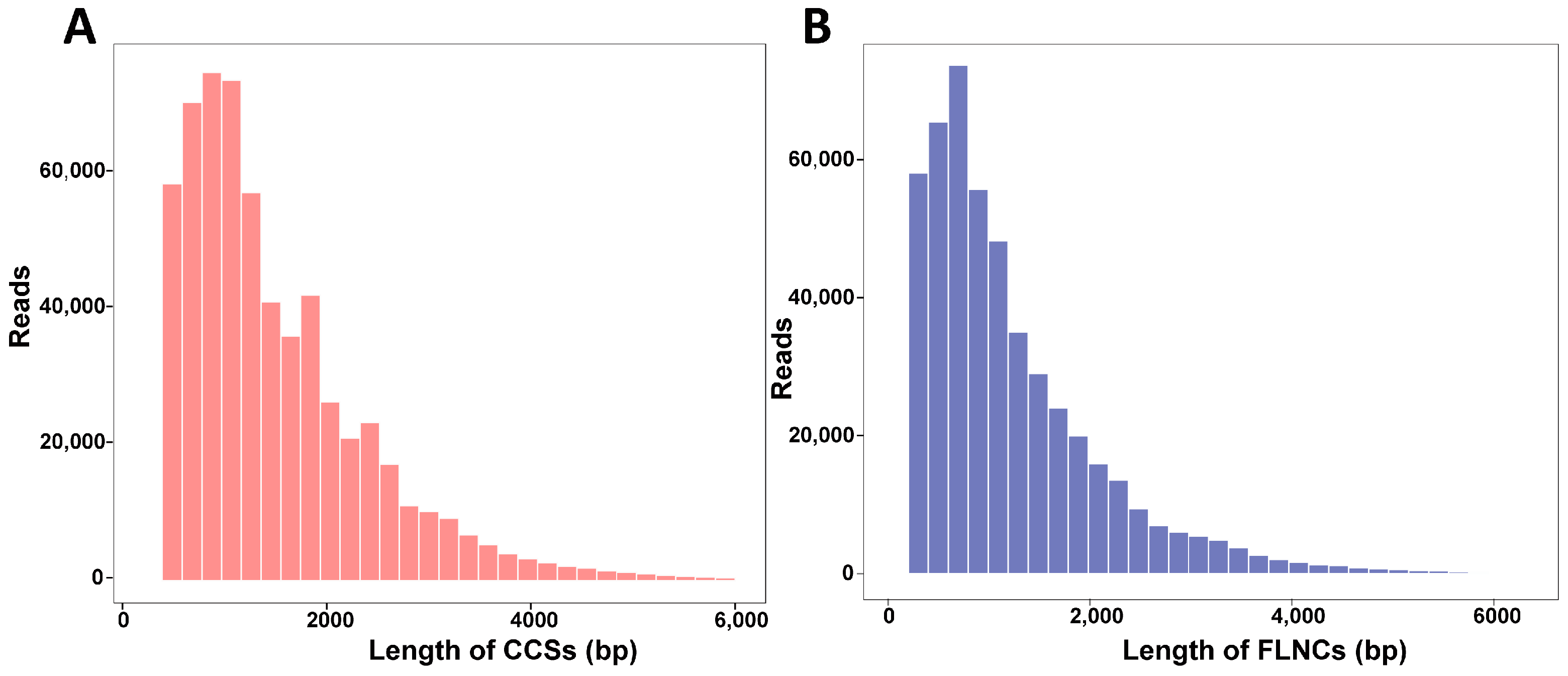
3.2. PacBio SMART Sequencing and Data Analysis
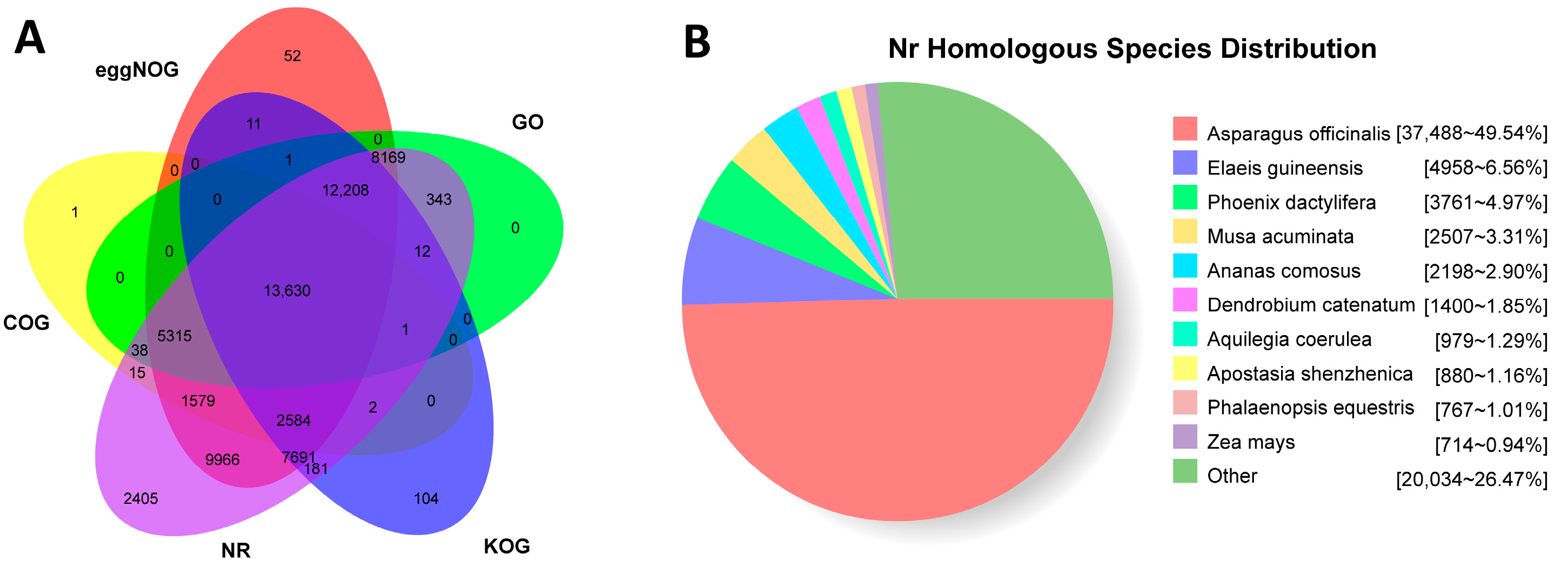
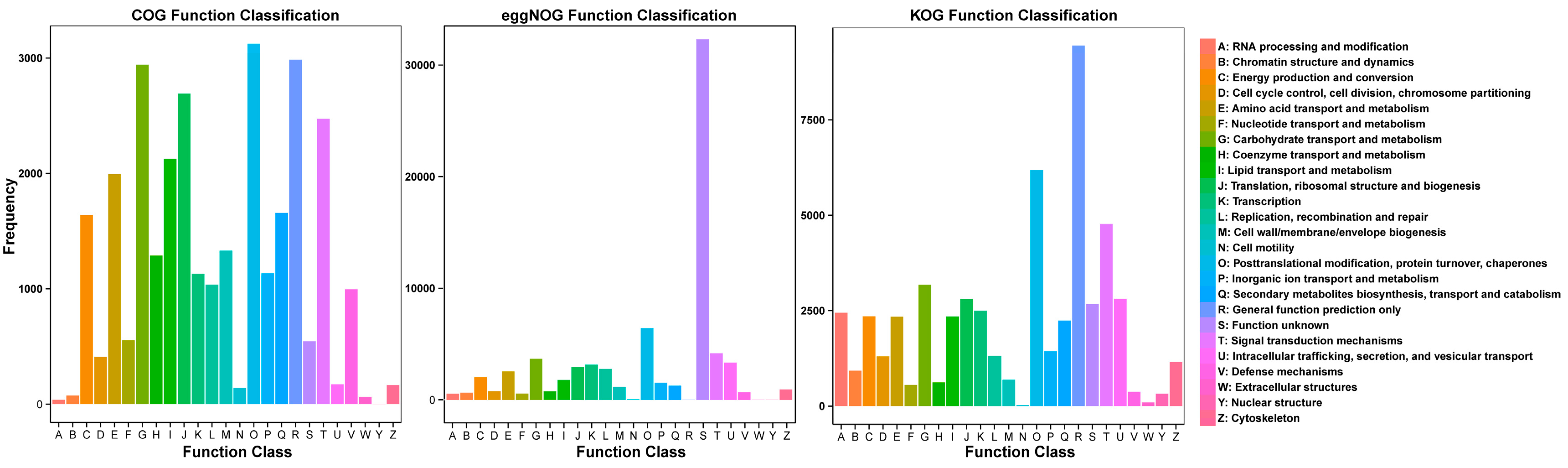
3.3. Functional Annotation of Non-Redundant Transcripts
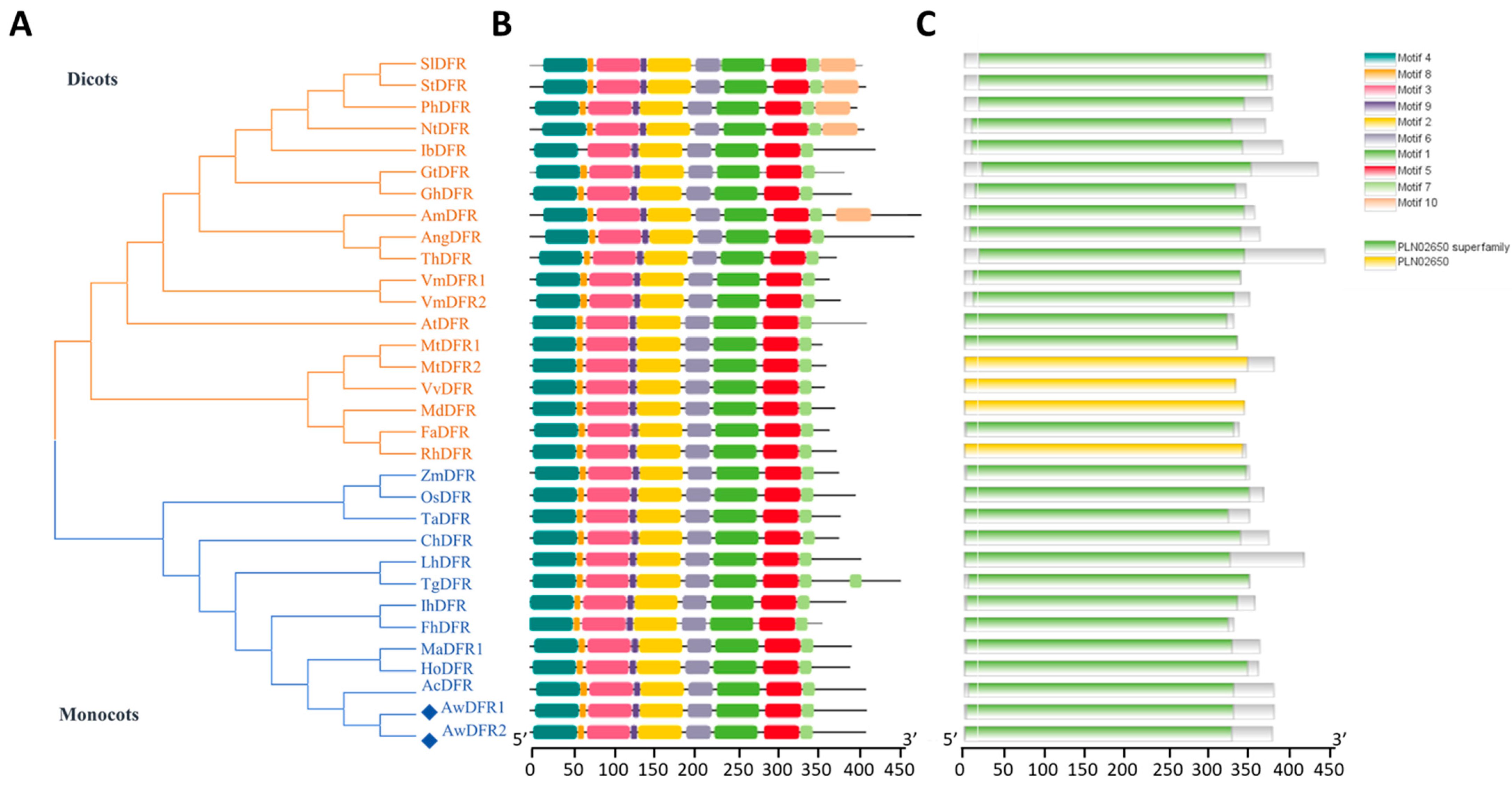
3.4. Identification and Expression Level of AwDFRs
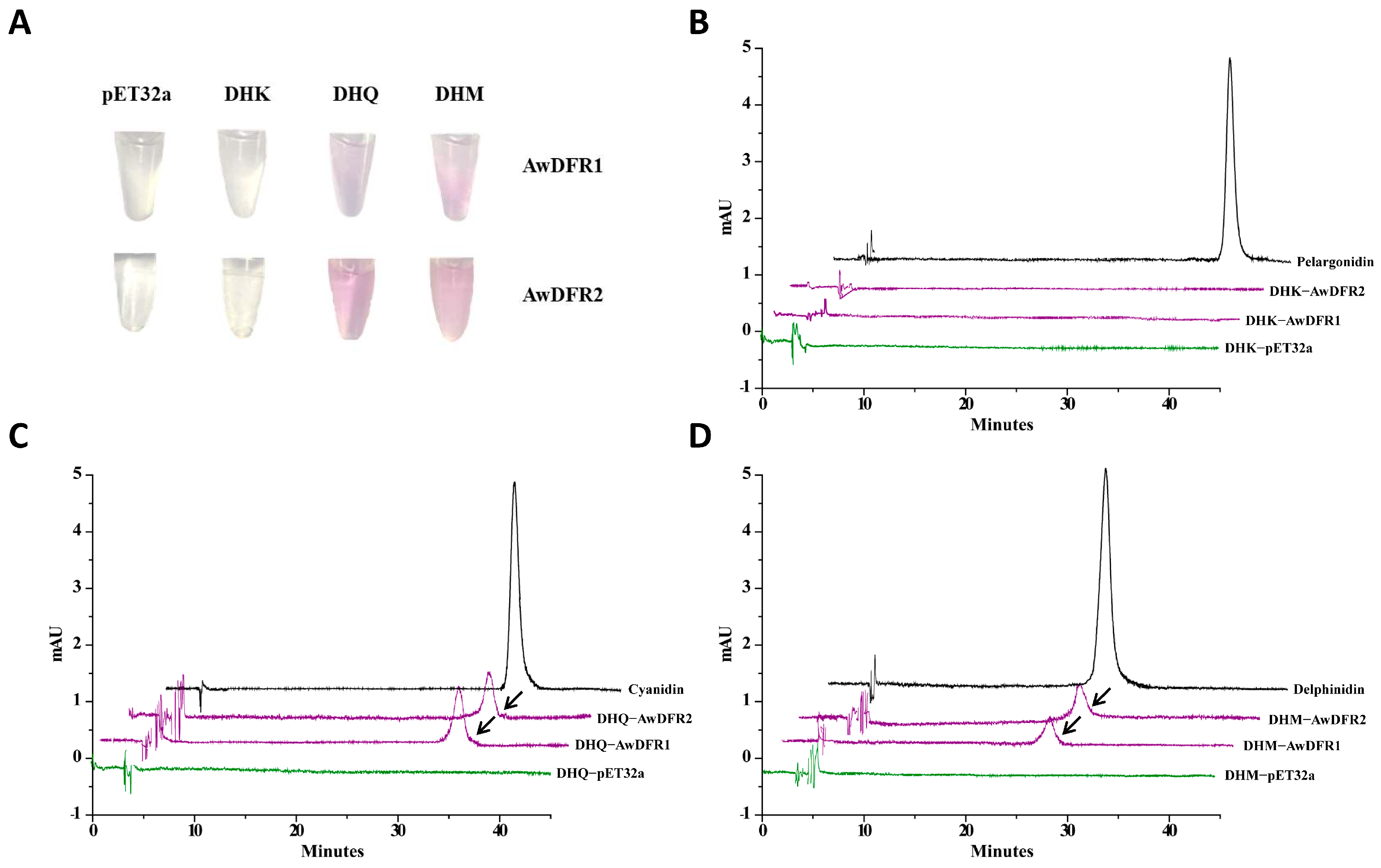
3.5. Enzyme Activity Analysis of Recombinant AwDFRs
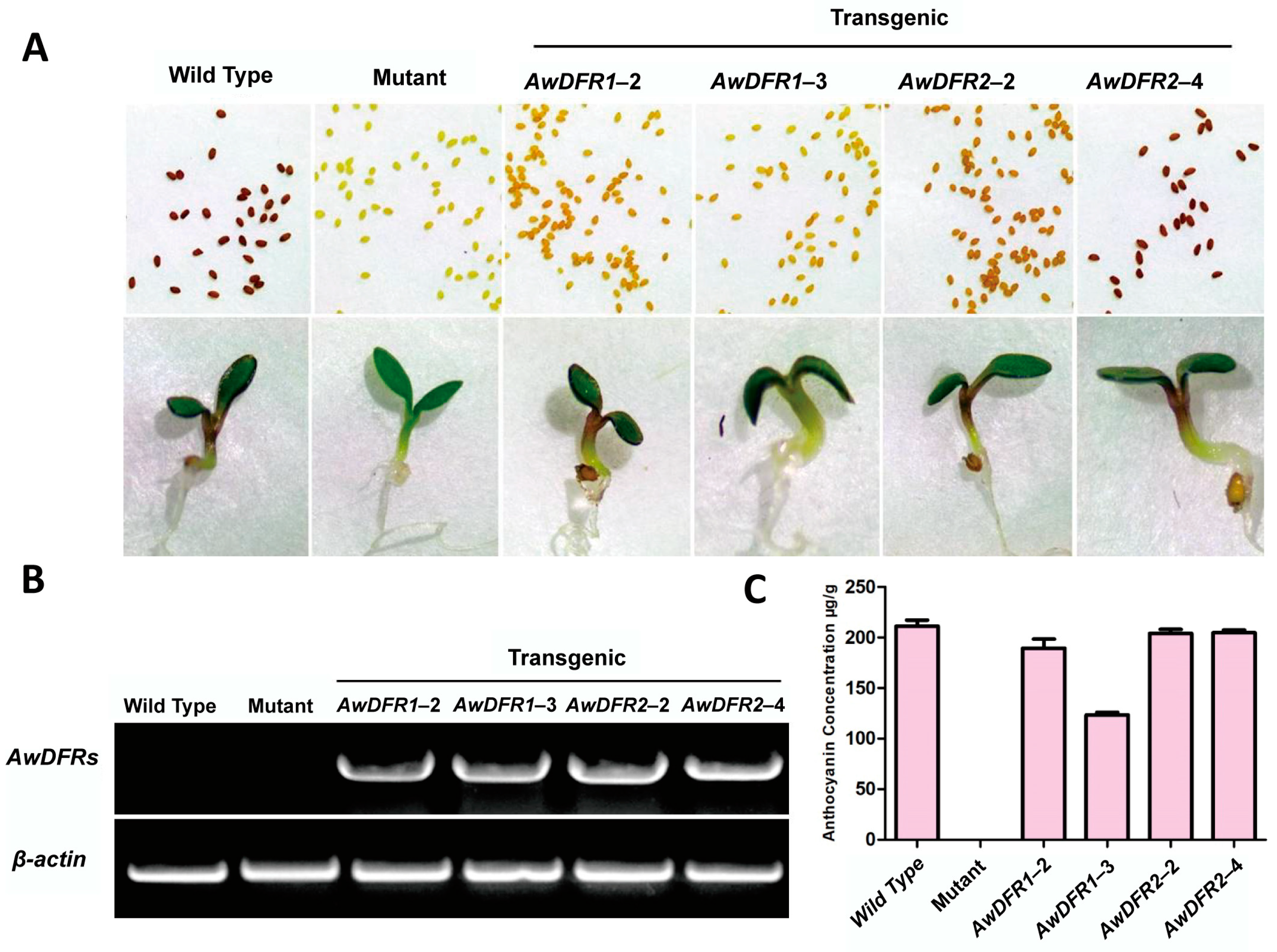
3.6. Transgenic Confirmation of AwDFRs in Arabidopsis
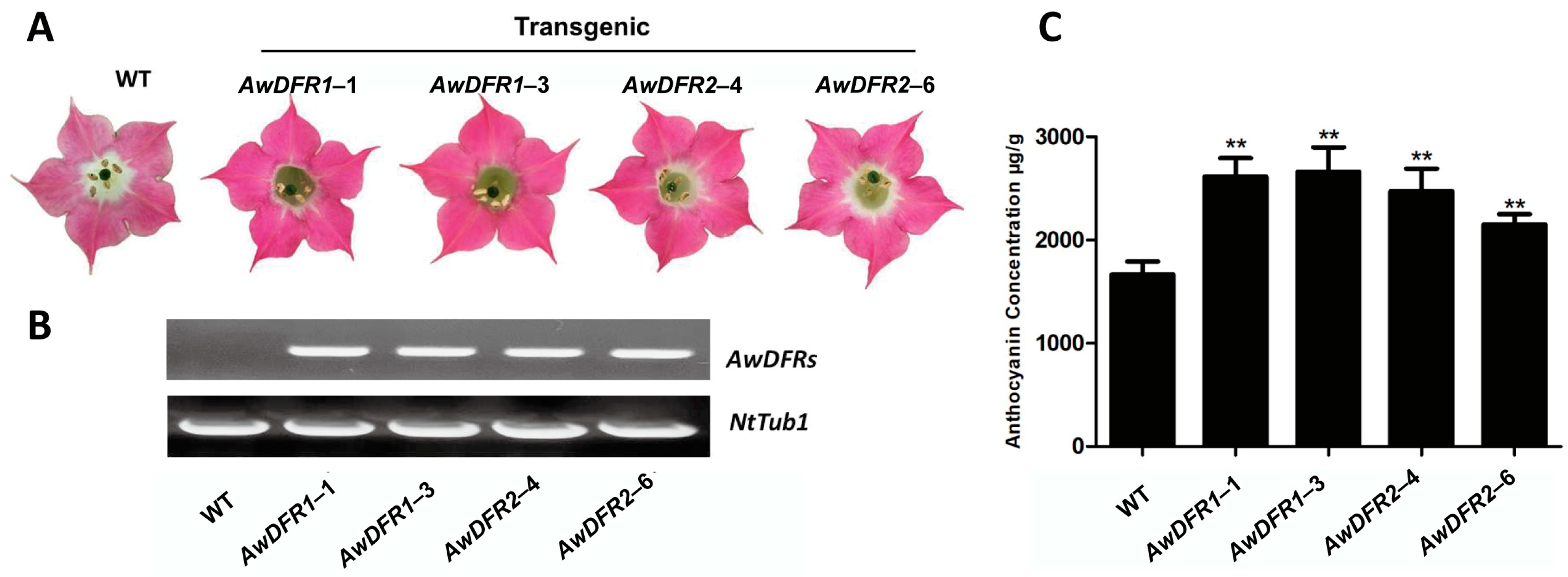
3.7. Transgenic Confirmation of AwDFRs in Tobacco
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhao, J.; Huang, M.; Ni, R.; Peng, T.; Chen, X.; Nan, H. The complete chloroplast genome of Allium wallichii Kunth (Amaryllidaceae). Mitochondrial DNA Part B Resour. 2023, 8, 1054–1058. [Google Scholar] [CrossRef]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colourful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colourful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Chen, X.; Liu, W.; Huang, X.; Fu, H.; Wang, Q.; Wang, Y.; Cao, J. Arg-type dihydroflavonol 4-reductase genes from the fern Dryopteris erythrosora play important roles in the biosynthesis of anthocyanins. PLoS ONE 2020, 15, e0232090. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, T. Recent progress in anti-obesity and anti-diabetes effect of berries. Antioxidants 2016, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Koning, R.A.; Grayson, I.; Weckbecker, C. The effects of bilberries, blackcurrants and their constituent anthocyanins on heart health in humans. Agro Food Industry Hi-Tech. 2015, 26, 16–20. [Google Scholar]
- Ruan, H.; Shi, X.; Gao, L.; Rashid, A.; Li, Y.; Lei, T.; Dai, X.; Xia, T.; Wang, Y. Functional analysis of the dihydroflavonol 4-reductase family of Camellia sinensis: Exploiting key amino acids to reconstruct reduction activity. Hortic. Res. 2022, 9, uhac098. [Google Scholar] [CrossRef]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Wang, S.; Zhang, F.; Niu, Y.; Wang, D. Cloning and temporal-spatial expression analysis of dfr gene from Scutellaria baicalensis with different colors. Sheng Wu Gong. Cheng Xue Bao 2021, 37, 1312–1323. [Google Scholar] [PubMed]
- Feng, X.; Zhang, Y.; Wang, H.; Tian, Z.; Xin, S.; Zhu, P. The dihydroflavonol 4-reductase BoDFR1 drives anthocyanin accumulation in pink-leaved ornamental kale. Theor. Appl. Genet. 2021, 134, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Halbwirth, H.; Kahl, S.; Jager, W.; Reznicek, G.; Forkmann, G.; Stich, K. Synthesis of (C-14)-labeled 5-deoxyflavonoids and their application in the study of dihydroflavonol/leucoanthocyanidin interconversion by dihydroflavonol 4-reductase. Plant Sci. 2006, 170, 587–595. [Google Scholar] [CrossRef]
- Stafford, H.A.; Lester, H.H. Enzymic and nonenzymic reduction of (+)-dihydroquercetin to its 3,4,-diol. Plant Physiol. 1982, 70, 695–698. [Google Scholar] [CrossRef]
- O’Reilly, C.; Shepherd, N.S.; Pereira, A.; Schwarz-Sommer, Z.; Bertram, I.; Robertson, D.S.; Peterson, P.; Saedler, H. Molecular cloning of the a1 locus of Zea mays using the transposable elements En and Mu1. EMBO J. 1985, 4, 877–882. [Google Scholar] [CrossRef]
- Xie, D.Y.; Jackson, L.A.; Cooper, J.D.; Ferreira, D.; Paiva, N.L. Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula. Plant Physiol. 2004, 134, 979–994. [Google Scholar] [CrossRef] [PubMed]
- Shimada, N.; Sasaki, R.; Sato, S.; Kaneko, T.; Tabata, S.; Aoki, T. A comprehensive analysis of six dihydroflavonol 4-reductases encoded by a gene cluster of the Lotus japonicus genome. J. Exp. Bot. 2005, 56, 2573–2585. [Google Scholar] [CrossRef]
- Singh, K.; Kumar, S.; Yadav, S.K.; Ahuja, P.S. Characterization of dihydroflavonol 4-reductase cDNA in tea [Camellia sinensis (L.) O. Kuntze]. Plant Biotechnol. Rep. 2009, 3, 95–101. [Google Scholar] [CrossRef]
- Li, Y.Q.; Liu, X.; Cai, X.; Shan, X.; Gao, R.; Yang, S.; Han, T.; Wang, S.; Wang, L.; Gao, X. Dihydroflavonol 4-reductase genes from Freesia hybrida play important and partially overlapping roles in the biosynthesis of flavonoids. Front. Plant Sci. 2017, 8, 428. [Google Scholar] [CrossRef]
- Ni, J.; Ruan, R.; Wang, L.; Jiang, Z.; Gu, X.; Chen, L.; Xu, M. Functional and correlation analyses of dihydroflavonol-4-reductase genes indicate their roles in regulating anthocyanin changes in Ginkgo biloba. Ind. Crop. Prod. 2020, 152, 112546. [Google Scholar] [CrossRef]
- Miosic, S.; Thill, J.; Milosevic, M.; Gosch, C.; Pober, S.; Molitor, C.; Halbwirth, H. Dihydroflavonol 4-reductase genes encode enzymes with contrasting substrate specificity and show divergent gene expression profiles in Fragaria species. PLoS ONE 2014, 9, e112707. [Google Scholar] [CrossRef] [PubMed]
- Gosch, C.; Nagesh, K.M.; Thill, J.; Miosic, S.; Plaschil, S.; Milosevic, M.; Halbwirth, H. Isolation of dihydroflavonol 4-reductase cDNA clones from Angelonia x angustifolia and heterologous expression as GST fusion protein in Escherichia coli. PLoS ONE 2014, 9, e107755. [Google Scholar] [CrossRef]
- Johnson, E.T.; Ryu, S.; Yi, H.; Shin, B.; Cheong, H.; Choi, G. Alteration of a single amino acid changes the substrate specificity of dihydrof lavonol 4-reductase. Plant J. 2001, 25, 325–333. [Google Scholar] [CrossRef]
- Johnson, E.T.; Yi, H.; Shin, B.; Oh, B.J.; Cheong, H.; Choi, G. Cymbidium hybrida dihydroflavonol 4-reductase does not efficiently reduce dihydrokaempferol to produce orange pelargonidin-type anthocyanins. Plant J. 1999, 19, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Petit, P.; Granier, T.; d’Estaintot, B.L.; Manigand, C.; Bathany, K.; Schmitter, J.M.; Lauvergeat, V.; Hamdi, S.; Gallois, B. Crystal structure of grape dihydroflavonol 4-reductase, a key enzyme in flavonoid biosynthesis. J. Mol. Biol. 2007, 368, 1345–1357. [Google Scholar] [CrossRef] [PubMed]
- Roslan, N.D.; Yusop, J.M.; Baharum, S.N.; Othman, R.; Mohamed-Hussein, Z.A.; Ismail, I.; Zainal, Z. Flavonoid biosynthesis genes putatively identified in the aromatic plant Polygonum minus via expressed sequences tag (EST) analysis. Int. J. Mol. Sci. 2012, 13, 2692–2706. [Google Scholar] [CrossRef]
- Li, W.Z.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Felipe, A.S.; Robert, M.W.; Panagiotis, I.; Evgenia, V.K.; Evgeny, M.Z. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar]
- Li, J.; Ma, W.; Zeng, P.; Wang, J.; Geng, B.; Yang, J.; Cui, Q. LncTar: A tool for predicting the RNA targets of long noncoding RNAs. Brief. Bioinform. 2015, 16, 806. [Google Scholar] [CrossRef]
- Sun, W.; Sun, S.; Xu, H.; Wang, Y.; Chen, Y.; Xu, X.; Yi, Y.; Ju, Z. Characterization of Two Key Flavonoid 3-O-Glycosyltransferases Involved in the Formation of Flower Color in Rhododendron Delavayi. Front. Plant Sci. 2022, 13, 863482. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Zhou, N.; Feng, C.; Sun, S.; Tang, M.; Tang, X.; Ju, Z.; Yi, Y. Functional analysis of a dihydroflavonol 4-reductase gene in Ophiorrhiza japonica (OjDFR1) reveals its role in the regulation of anthocyanin. PeerJ 2021, 9, e12323. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for agrobacterium mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusionproteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 4, 2019–2025. [Google Scholar] [CrossRef] [PubMed]
- Fanali, C.; Dugo, L.; D’Orazio, G.; Lirangi, M.; Dacha, M.; Dugo, P.; Luigi, M. Analysis of anthocyanins in commercial fruit juices by using nano-liquid chromatography- electrospray-mass spectrometry and high-performance liquid chromatography with UV-visdetector. J. Separ. Sci. 2001, 34, 150–159. [Google Scholar] [CrossRef]
- Finkers, R.; van Kaauwen, M.; Ament, K.; Burger-Meijer, K.; Egging, R.; Huits, H.; Kodde, L.; Kroon, L.; Shigyo, M.; Sato, S.; et al. Insights from the first genome assembly of onion (Allium cepa). G3-Genes Genomes Genet. 2021, 11, 243. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhu, S.; Li, N.; Cheng, Y.; Zhao, J.; Qiao, X.; Lu, L.; Liu, S.; Wang, Y.; Liu, C.; et al. A chromosome-level genome assembly of garlic (Allium sativum) provides insights into genome evolution and allicin biosynthesis. Mol. Plant 2020, 13, 1328–1339. [Google Scholar] [CrossRef] [PubMed]
- Lei, T.; Huang, J.; Ruan, H.; Qian, W.; Fang, Z.; Gu, C.; Zhang, N.; Liang, Y.; Wang, Z.; Gao, L.; et al. Competition between FLS and DFR regulates the distribution of flavonols and proanthocyanidins in Rubus chingii Hu. Front. Plant Sci. 2023, 14, 1134993. [Google Scholar] [CrossRef] [PubMed]
- Zhao, A.G.; Ding, R.W.; Wang, C.; Chen, C.; Wang, D.M. Insights into the catalytic and regulatory mechanisms of dihydroflavonol 4-reductase, a key enzyme of anthocyanin synthesis in Zanthoxylum bungeanum. Tree Physiol. 2023, 43, 169–184. [Google Scholar]
- Zhao, D.; Tao, J.; Han, C.; Ge, J. Flower color diversity revealed by differential expression of flavonoid biosynthetic genes and flavonoid accumulation in herbaceous peony (Paeonia lactiflora Pall.). Mol. Biol. Rep. 2012, 39, 11263–11275. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuka, T.; Nishihara, M.; Mishiba, K.; Yamamura, S. Temporal expression of flavonoid biosynthesis-related genes regulates flower pigmentation in gentian plants. Plant Sci. 2005, 168, 1309–1318. [Google Scholar] [CrossRef]
- Sun, W.; Zhou, N.; Wang, Y.; Sun, S.; Zhang, Y.; Ju, Z.; Yi, Y. Characterization and functional analysis of RdDFR1 regulation on flower color formation in Rhododendron delavayi. Plant Physiol. Biochem. 2021, 169, 203–210. [Google Scholar] [CrossRef]
- Mori, S.; Otani, M.; Kobayashi, H.; Nakano, M. Isolation and characterization of the dihydroflavonol 4-reductase gene in the monocotyledonous ornamental Agapanthus praecox ssp. orientalis (Leighton) Leighton. Sci. Hortic. 2014, 166, 24–30. [Google Scholar] [CrossRef]
- Liu, X.; Xiang, M.; Fan, Y.; Yang, C.; Zeng, L.; Zhang, Q.; Chen, M.; Liao, Z. A rootpreferential DFR-like gene encoding dihydrokaempferol reductase involved in anthocyanin biosynthesis of purple-fleshed sweet potato. Front. Plant Sci. 2017, 8, 279. [Google Scholar]
- Liu, H.; Lou, Q.; Ma, J. Cloning and functional characterization of dihydroflavonol 4-reductase gene involved in anthocyanidin biosynthesis of grape hyacinth. Int. J. Mol. Sci. 2019, 20, 4743. [Google Scholar] [CrossRef] [PubMed]
- Forkmann, G.; Ruhnau, B. Distinct substrate specificity of dihydroflavonol 4-reductase from flowers of Petunia hybrida. Z. Für Naturforschung C 1987, 42, 1146–1148. [Google Scholar] [CrossRef]
- Wang, H.; Fan, W.; Li, H.; Yang, J.; Huang, J.; Zhang, P. Functional characterization of Dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses. PLoS ONE 2013, 8, e78484. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Gou, J.Q.; Jia, Z.C.; Yang, L.; Sun, Y.M.; Xiao, X.Y.; Song, F.; Luo, K.M. Molecular cloning and characterization of two genes encoding dihydroflavonol 4- reductase from Populus trichocarpa. PLoS ONE 2012, 7, e30364. [Google Scholar] [CrossRef]




| Number | Anthocyanins | Content of Anthocyanins (μg/g) | ||
|---|---|---|---|---|
| Stage 1 | Stage 2 | Stage 3 | ||
| 1 | Cyanidin-3-O-rutinoside | 4.1877 | 4.3255 | 4.2816 |
| 2 | Cyanidin-3-O-(6-O-p-coumaroyl)-glucoside | 1.1055 | 3.0440 | 3.5191 |
| 3 | Cyanidin-3-O-(6-O-malonyl-beta-D-glucoside) | 11.9047 | 14.5685 | 28.1693 |
| 4 | Cyanidin-3-O-glucoside | 15.4419 | 49.9805 | 41.9956 |
| 5 | Cyanidin-3,5-O-diglucoside | 68.8578 | 60.8605 | 42.7762 |
| 6 | Pelargonidin-3,5-O-diglucoside | 3.2711 | 3.6893 | 1.6777 |
| 7 | Pelargonidin-3-O-(6-O-malonyl-beta-D-glucoside) | 1.3095 | 2.3213 | 3.8407 |
| 8 | Pelargonidin-3-O-glucoside | 1.2388 | 4.4071 | 3.0832 |
| 9 | Peonidin-3-O-glucoside | 1.1826 | 2.6445 | 3.8103 |
| 10 | Peonidin-3-O-(6-O-malonyl-beta-D-glucoside) | 14.2415 | 22.0668 | 29.4758 |
| 11 | Peonidin-3,5-O-diglucoside | 24.3333 | 20.0501 | 12.6799 |
| 12 | Petunidin-3-O-glucoside | 1.1258 | 1.1350 | 0.9218 |
| 13 | Petunidin-3-O-(6-O-malonyl-beta-D-glucoside) | 1.2707 | 2.2378 | 1.4461 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ju, Z.; Liang, L.; Shi, H.; Zheng, Y.; Zhao, W.; Sun, W.; Pang, Y. Full-Length Transcriptome Analysis and Characterization of DFRs Involved in the Formation of Anthocyanin in Allium wallichii. Horticulturae 2024, 10, 1068. https://doi.org/10.3390/horticulturae10101068
Ju Z, Liang L, Shi H, Zheng Y, Zhao W, Sun W, Pang Y. Full-Length Transcriptome Analysis and Characterization of DFRs Involved in the Formation of Anthocyanin in Allium wallichii. Horticulturae. 2024; 10(10):1068. https://doi.org/10.3390/horticulturae10101068
Chicago/Turabian StyleJu, Zhigang, Lin Liang, Hongxi Shi, Yaqiang Zheng, Wenxuan Zhao, Wei Sun, and Yuxin Pang. 2024. "Full-Length Transcriptome Analysis and Characterization of DFRs Involved in the Formation of Anthocyanin in Allium wallichii" Horticulturae 10, no. 10: 1068. https://doi.org/10.3390/horticulturae10101068
APA StyleJu, Z., Liang, L., Shi, H., Zheng, Y., Zhao, W., Sun, W., & Pang, Y. (2024). Full-Length Transcriptome Analysis and Characterization of DFRs Involved in the Formation of Anthocyanin in Allium wallichii. Horticulturae, 10(10), 1068. https://doi.org/10.3390/horticulturae10101068






