Genome-Wide Association Analyses Defined the Interplay between Two Major Loci Controlling the Fruit Texture Performance in a Norwegian Apple Collection (Malus × domestica Borkh.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Phenotypic Assessment
2.3. Dynamic Aspects of Fruit Texture
2.4. Genotyping
2.5. Statistical Analyses and Phenotype–Genotype Associations
3. Results
3.1. Subselection of Individuals
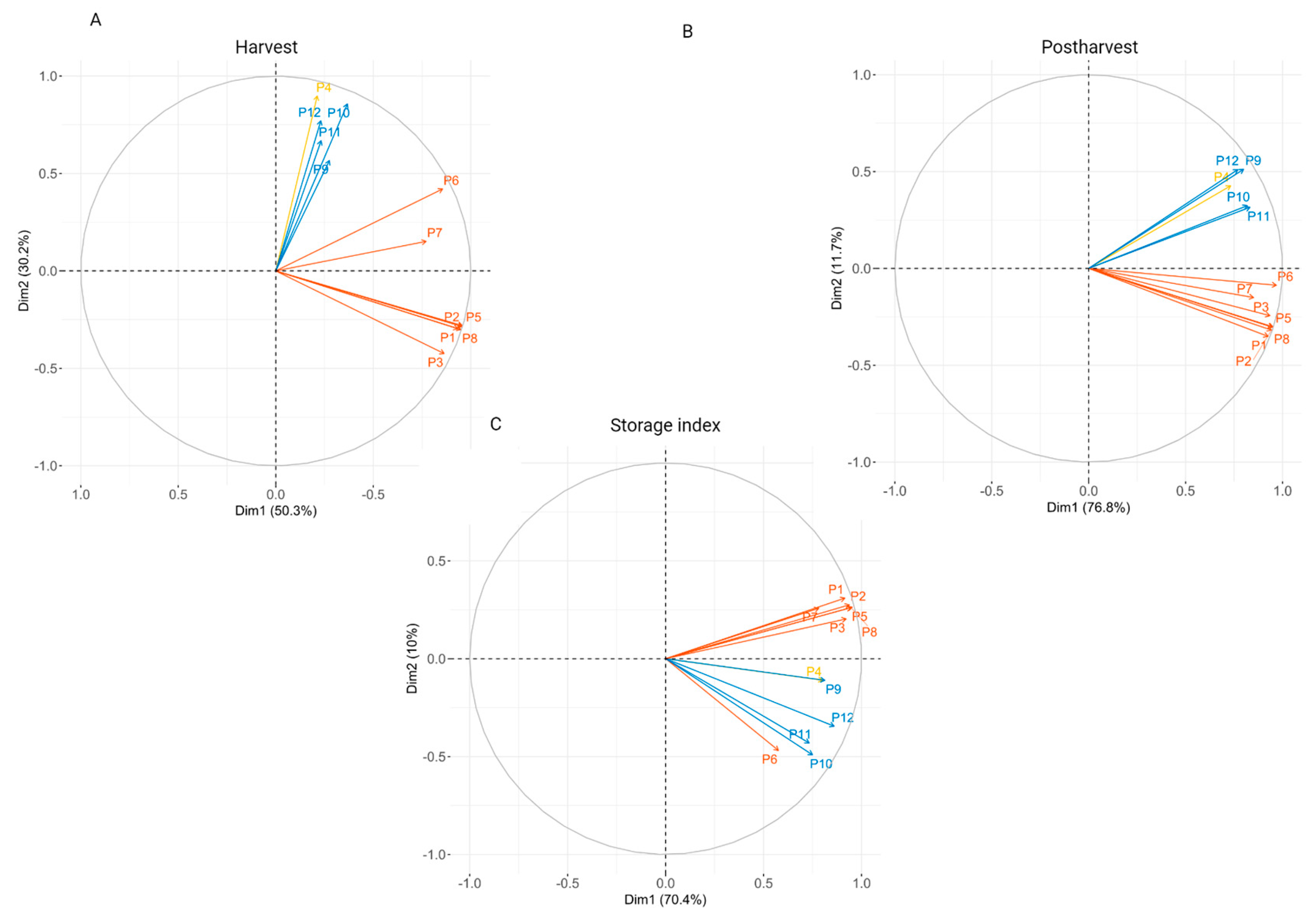
3.2. Fruit Texture Assessment and Principal Component Analysis
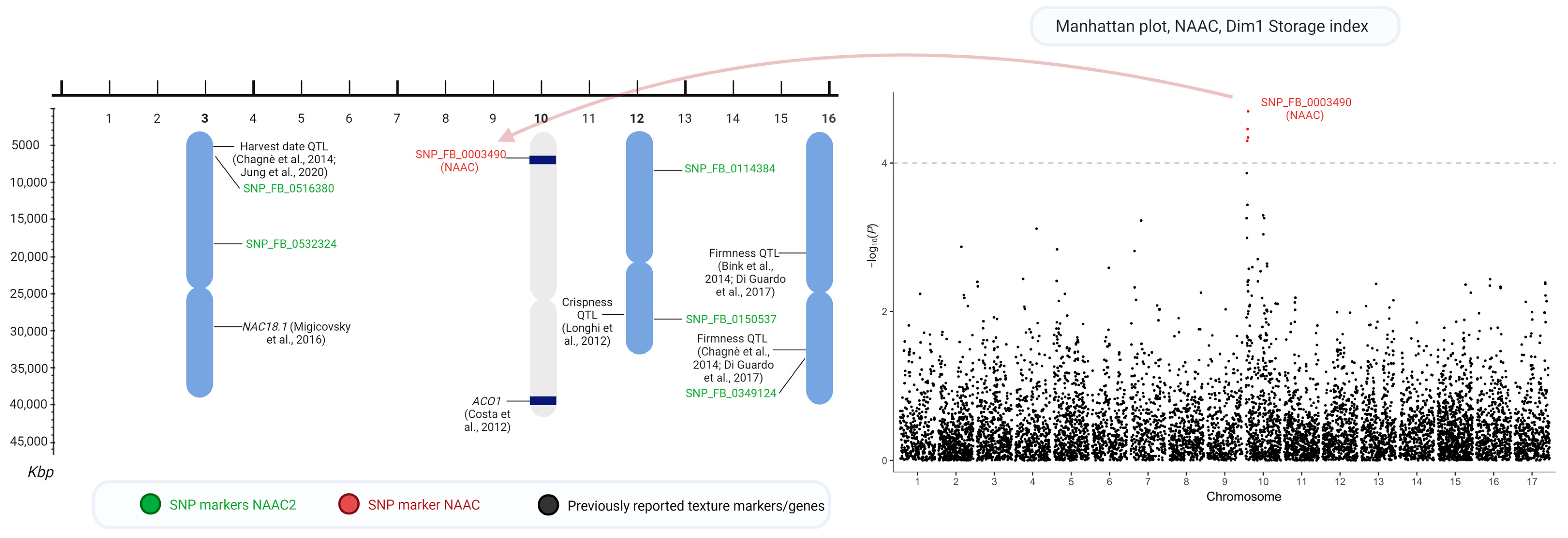
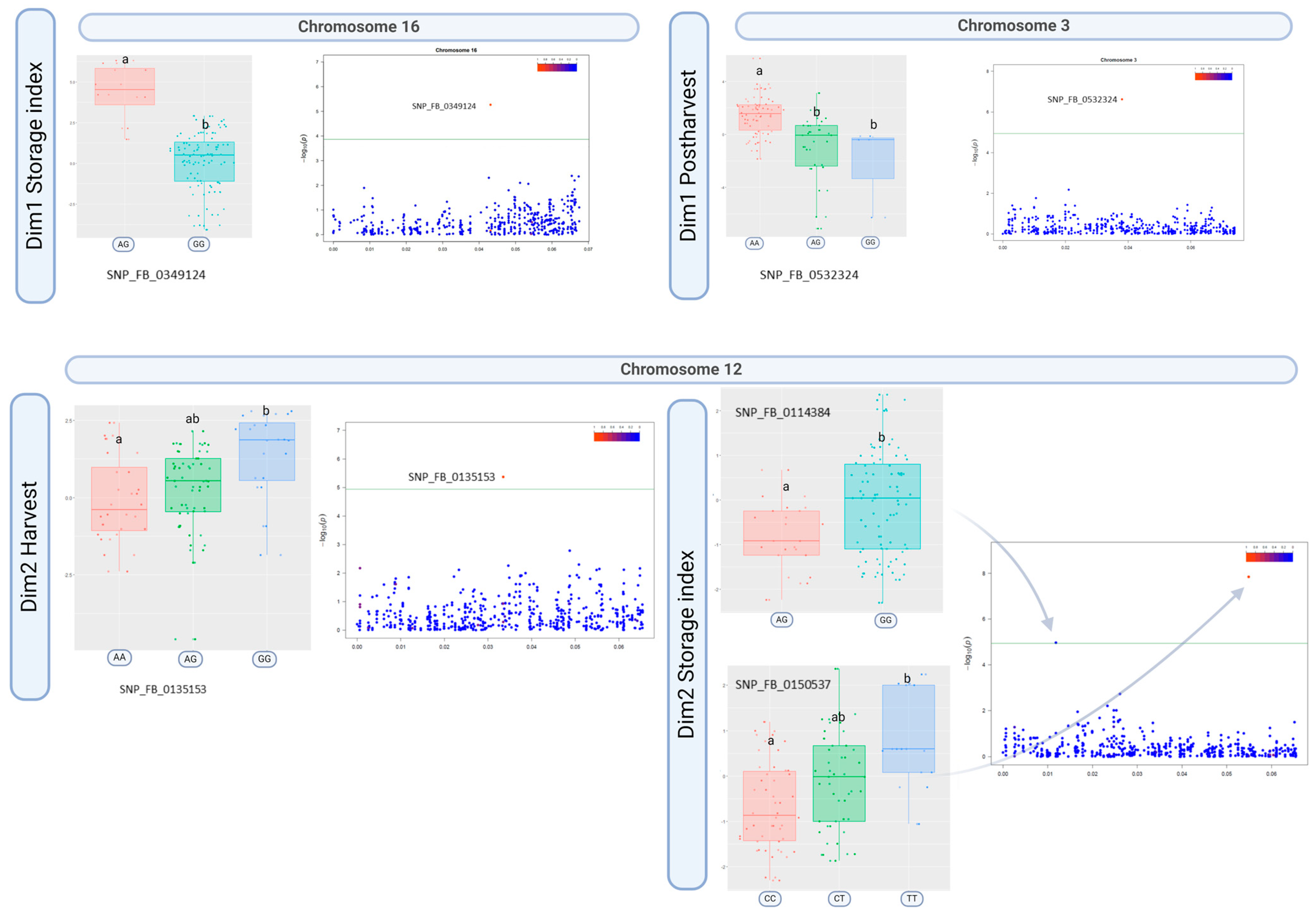
3.3. Phenotype–Genotype Associations
4. Discussion
4.1. Novel SNPs Associated with Texture Properties Mapped on Chromosome 12
4.2. Signals on Chromosome 3 Associated with the Storage Index
4.3. Breeding for Texture in Norwegian Apples
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- LMD. Meld. St. 11 Strategi for auka sjølvforsyning av jordbruksvarer og plan for opptrapping av inntektsmoglegheitene i jordbruket. In Ministry of Agriculture and Food; Tryggings- og serviceorganisasjonen til departementa: Oslo, Norway, 2023; p. 58. [Google Scholar]
- OFG. Frukt- og grøntstatistikk 2023. Available online: www.frukt.no (accessed on 30 September 2024).
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Nybom, H.; Ahmadi-Afzadi, M.; Garkava-Gustavsson, L.; Sehic, J.; Tahir, I. Selection for improved fruit texture and storability in apple. In Proceedings of the XXVIII International Horticultural Congress on Science and Horticulture for People (IHC2010): International Symposium, Leuven, Belgium, 22–27 August 2010; pp. 849–854. [Google Scholar]
- El-Ramady, H.R.; Domokos-Szabolcsy, É.; Abdalla, N.A.; Taha, H.S.; Fári, M. Postharvest Management of Fruits and Vegetables Storage. In Sustainable Agriculture Reviews: Volume 15; Lichtfouse, E., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 65–152. [Google Scholar] [CrossRef]
- Galvez-Lopez, D.; Laurens, F.; Quéméner, B.; Lahaye, M. Variability of cell wall polysaccharides composition and hemicellulose enzymatic profile in an apple progeny. Int. J. Biol. Macromol. 2011, 49, 1104–1109. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.; Cappellin, L.; Longhi, S.; Guerra, W.; Magnago, P.; Porro, D.; Soukoulis, C.; Salvi, S.; Velasco, R.; Biasioli, F.; et al. Assessment of apple (Malus × domestica Borkh.) fruit texture by a combined acoustic-mechanical profiling strategy. Postharvest Biol. Technol. 2011, 61, 21–28. [Google Scholar] [CrossRef]
- Costa, F.; Van de Weg, W.E.; Stella, S.; Dondini, L.; Pratesi, D.; Musacchi, S.; Sansavini, S. Map position and functional allelic diversity of Md-Exp7, a new putative expansin gene associated with fruit softening in apple (Malus × domestica Borkh.) and pear (Pyrus communis). Tree Genet. Genomes 2008, 4, 575–586. [Google Scholar] [CrossRef]
- Tadiello, A.; Longhi, S.; Moretto, M.; Ferrarini, A.; Tononi, P.; Farneti, B.; Busatto, N.; Vrhovsek, U.; Molin, A.D.; Avanzato, C.; et al. Interference with ethylene perception at receptor level sheds light on auxin and transcriptional circuits associated with the climacteric ripening of apple fruit (Malus × domestica Borkh.). Plant J. 2016, 88, 963–975. [Google Scholar] [CrossRef]
- Baumgartner, I.O.; Kellerhals, M.; Costa, F.; Dondini, L.; Pagliarani, G.; Gregori, R.; Tartarini, S.; Leumann, L.; Laurens, F.; Patocchi, A. Development of SNP-based assays for disease resistance and fruit quality traits in apple (Malus × domestica Borkh.) and validation in breeding pilot studies. Tree Genet. Genomes 2016, 12, 35. [Google Scholar] [CrossRef]
- Migicovsky, Z.; Yeats, T.H.; Watts, S.; Song, J.; Forney, C.F.; Burgher-MacLellan, K.; Somers, D.J.; Gong, Y.; Zhang, Z.; Vrebalov, J.; et al. Apple Ripening Is Controlled by a NAC Transcription Factor. Front. Genet. 2021, 12, 671300. [Google Scholar] [CrossRef]
- Jung, M.; Keller, B.; Roth, M.; Aranzana, M.J.; Auwerkerken, A.; Guerra, W.; Al-Rifaï, M.; Lewandowski, M.; Sanin, N.; Rymenants, M. Genetic architecture and genomic predictive ability of apple quantitative traits across environments. Hortic. Res. 2022, 9, uhac028. [Google Scholar] [CrossRef]
- Larsen, B.; Migicovsky, Z.; Jeppesen, A.A.; Gardner, K.M.; Toldam-Andersen, T.B.; Myles, S.; Ørgaard, M.; Petersen, M.A.; Pedersen, C. Genome-Wide Association Studies in Apple Reveal Loci for Aroma Volatiles, Sugar Composition, and Harvest Date. Plant Genome 2019, 12, 180104. [Google Scholar] [CrossRef]
- Watts, S.; Migicovsky, Z.; Myles, S. Large-scale apple GWAS reveals NAC18.1 as a master regulator of ripening traits. Fruit Res. 2023, 3, 32. [Google Scholar] [CrossRef]
- Nybom, H. Advances in understanding texture development in apples. In Improving the Quality of Apples; Costa, F., Ed.; Burleigh Dodds Science Publishing Limited: Cambridge, UK, 2023; Volume 142, p. 230. [Google Scholar]
- Laurens, F.; Aranzana, M.J.; Arus, P.; Bassi, D.; Bink, M.; Bonany, J.; Caprera, A.; Corelli-Grappadelli, L.; Costes, E.; Durel, C.-E.; et al. An integrated approach for increasing breeding efficiency in apple and peach in Europe. Hortic. Res. 2018, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.M.; Haley, C.S.; Thompson, R. Marker-Assisted Introgression in Backcross Breeding Programs. Genetics 1996, 144, 1923–1932. [Google Scholar] [CrossRef] [PubMed]
- Eberhart, S.A. Factors effecting efficiencies of breeding methods. Afr. Soils 1970, 15, 655–680. [Google Scholar]
- Lush, J.L. Animal Breeding Plans; Iowa State College Press: Ames, IA, USA, 1943; pp. viii + 437. [Google Scholar]
- Kostick, S.A.; Norelli, J.L.; Teh, S.L.; Evans, K.M. Quantitative variation and heritability estimates of fire blight resistance in a pedigree-connected apple germplasm set. J. Plant Pathol. 2021, 103, 65–75. [Google Scholar] [CrossRef]
- Kouassi, A.B.; Durel, C.-E.; Costa, F.; Tartarini, S.; van de Weg, E.; Evans, K.; Fernandez-Fernandez, F.; Govan, C.; Boudichevskaja, A.; Dunemann, F.; et al. Estimation of genetic parameters and prediction of breeding values for apple fruit-quality traits using pedigreed plant material in Europe. Tree Genet. Genomes 2009, 5, 659–672. [Google Scholar] [CrossRef]
- Gilpin, L.; Røen, D.; Schubert, M.; Davik, J.; Rumpunen, K.; Gardli, K.A.; Hjeltnes, S.H.; Alsheikh, M. Genetic Characterization of the Norwegian Apple Collection. Horticulturae 2023, 9, 575. [Google Scholar] [CrossRef]
- Bianco, L.; Cestaro, A.; Sargent, D.J.; Banchi, E.; Derdak, S.; Di Guardo, M.; Salvi, S.; Jansen, J.; Viola, R.; Gut, I. Development and validation of a 20K single nucleotide polymorphism (SNP) whole genome genotyping array for apple (Malus × domestica Borkh). PLoS ONE 2014, 9, e110377. [Google Scholar] [CrossRef]
- Farneti, B.; Di Guardo, M.; Khomenko, I.; Cappellin, L.; Biasioli, F.; Velasco, R.; Costa, F. Genome-wide association study unravels the genetic control of the apple volatilome and its interplay with fruit texture. J. Exp. Bot. 2017, 68, 1467–1478. [Google Scholar] [CrossRef]
- Costa, F.; Cappellin, L.; Fontanari, M.; Longhi, S.; Guerra, W.; Magnago, P.; Gasperi, F.; Biasioli, F. Texture dynamics during postharvest cold storage ripening in apple (Malus × domestica Borkh.). Postharvest Biol. Technol. 2012, 69, 54–63. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing: Vienna, Austria, 2023.
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses 2020, R Package Version 1.0.7. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 20 August 2024).
- Harrell, F.E., Jr.; Harrell, M.F.E., Jr. Package ‘hmisc’. CRAN2018 2019, 2019, 235–236. [Google Scholar]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. arXiv 2014, arXiv:1406.5823. [Google Scholar]
- Vanderzande, S.; Howard, N.P.; Cai, L.; Da Silva Linge, C.; Antanaviciute, L.; Bink, M.C.; Kruisselbrink, J.W.; Bassil, N.; Gasic, K.; Lezzoni, A. High-quality, genome-wide SNP genotypic data for pedigreed germplasm of the diploid outbreeding species apple, peach, and sweet cherry through a common workflow. PLoS ONE 2019, 14, e0210928. [Google Scholar] [CrossRef] [PubMed]
- Howard, N.P.; Troggio, M.; Durel, C.-E.; Muranty, H.; Denancé, C.; Bianco, L.; Tillman, J.; van de Weg, E. Integration of Infinium and Axiom SNP array data in the outcrossing species Malus × domestica and causes for seemingly incompatible calls. BMC Genom. 2021, 22, 246. [Google Scholar] [CrossRef] [PubMed]
- Di Pierro, E.A.; Gianfranceschi, L.; Di Guardo, M.; Koehorst-van Putten, H.J.J.; Kruisselbrink, J.W.; Longhi, S.; Troggio, M.; Bianco, L.; Muranty, H.; Pagliarani, G.; et al. A high-density, multi-parental SNP genetic map on apple validates a new mapping approach for outcrossing species. Hortic. Res. 2016, 3, 16057. [Google Scholar] [CrossRef] [PubMed]
- Howard, N.P.; van de Weg, E.; Bedford, D.S.; Peace, C.P.; Vanderzande, S.; Clark, M.D.; Teh, S.L.; Cai, L.; Luby, J.J. Elucidation of the ‘Honeycrisp’ pedigree through haplotype analysis with a multi-family integrated SNP linkage map and a large apple (Malus × domestica) pedigree-connected SNP data set. Hortic. Res. 2017, 4, 17003. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Jung, S.; Lee, T.; Cheng, C.H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef]
- Bink, M.C.A.M.; Jansen, J.; Madduri, M.; Voorrips, R.E.; Durel, C.E.; Kouassi, A.B.; Laurens, F.; Mathis, F.; Gessler, C.; Gobbin, D.; et al. Bayesian QTL analyses using pedigreed families of an outcrossing species, with application to fruit firmness in apple. Theor. Appl. Genet. 2014, 127, 1073–1090. [Google Scholar] [CrossRef]
- Chagné, D.; Dayatilake, D.; Diack, R.; Oliver, M.; Ireland, H.; Watson, A.; Gardiner, S.E.; Johnston, J.W.; Schaffer, R.J.; Tustin, S. Genetic and environmental control of fruit maturation, dry matter and firmness in apple (Malus × domestica Borkh.). Hortic. Res. 2014, 1, 14046. [Google Scholar] [CrossRef]
- Di Guardo, M.; Bink, M.C.A.M.; Guerra, W.; Letschka, T.; Lozano, L.; Busatto, N.; Poles, L.; Tadiello, A.; Bianco, L.; Visser, R.G.F.; et al. Deciphering the genetic control of fruit texture in apple by multiple family-based analysis and genome-wide association. J. Experimental Botany 2017, 68, 1451–1466. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Roth, M.; Aranzana, M.J.; Auwerkerken, A.; Bink, M.; Denancé, C.; Dujak, C.; Durel, C.-E.; Font i Forcada, C.; Cantin, C.M.; et al. The apple REFPOP—A reference population for genomics-assisted breeding in apple. Hortic. Res. 2020, 7, 189. [Google Scholar] [CrossRef] [PubMed]
- Longhi, S.; Hamblin, M.T.; Trainotti, L.; Peace, C.P.; Velasco, R.; Costa, F. A candidate gene based approach validates Md-PG1 as the main responsible for a QTL impacting fruit texture in apple (Malus x domesticaBorkh). BMC Plant Biology 2013, 13, 37. [Google Scholar] [CrossRef] [PubMed]
- Migicovsky, Z.; Gardner, K.M.; Money, D.; Sawler, J.; Bloom, J.S.; Moffett, P.; Chao, C.T.; Schwaninger, H.; Fazio, G.; Zhong, G.-Y.; et al. Genome to Phenome Mapping in Apple Using Historical Data. Plant Genome 2016, 9. [Google Scholar] [CrossRef]
- Daccord, N.; Celton, J.-M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef]
- Dujak, C.; Coleto-Alcudia, V.; Aranzana, M.J. Genomic analysis of fruit size and shape traits in apple: Unveiling candidate genes through GWAS analysis. Hortic. Res. 2023, 11, uhad270. [Google Scholar] [CrossRef]
- McClure, K.A.; Gong, Y.; Song, J.; Vinqvist-Tymchuk, M.; Campbell Palmer, L.; Fan, L.; Burgher-MacLellan, K.; Zhang, Z.; Celton, J.-M.; Forney, C.F.; et al. Genome-wide association studies in apple reveal loci of large effect controlling apple polyphenols. Hortic. Res. 2019, 6, 107. [Google Scholar] [CrossRef]
- Roth, M.; Muranty, H.; Di Guardo, M.; Guerra, W.; Patocchi, A.; Costa, F. Genomic prediction of fruit texture and training population optimization towards the application of genomic selection in apple. Hortic. Res. 2020, 7, 148. [Google Scholar] [CrossRef]
- Kostick, S.A.; Bernardo, R.; Luby, J.J. Genomewide selection for fruit quality traits in apple: Breeding insights gained from prediction and postdiction. Hortic. Res. 2023, 10, uhad088. [Google Scholar] [CrossRef]
- King, G.J.; Lynn, J.R.; Dover, C.J.; Evans, K.M.; Seymour, G.B. Resolution of quantitative trait loci for mechanical measures accounting for genetic variation in fruit texture of apple (Malus pumila Mill.). Theor. Appl. Genet. 2001, 102, 1227–1235. [Google Scholar] [CrossRef]
- Bus, V.G.M.; Bowen, J.K.; Patocchi, A.; Broggini, G.A.L.; Kumar, S.; Laurens, F.N.D. Breeding fruit cultivars with durable disease resistance. In Integrated Management of Diseases and Insect Pests of Tree Fruit; Xu, X., Ed.; Burleigh Dodds Science Publishing Limited: Cambridge, UK, 2019; pp. 233–274. [Google Scholar] [CrossRef]
- Howard, N.P.; van de Weg, E.; Tillman, J.; Tong, C.B.S.; Silverstein, K.A.T.; Luby, J.J. Two QTL characterized for soft scald and soggy breakdown in apple (Malus × domestica) through pedigree-based analysis of a large population of interconnected families. Tree Genet. Genomes 2017, 14, 2. [Google Scholar] [CrossRef]
- Longhi, S.; Moretto, M.; Viola, R.; Velasco, R.; Costa, F. Comprehensive QTL mapping survey dissects the complex fruit texture physiology in apple (Malus × domestica Borkh.). J. Exp. Bot. 2012, 63, 1107–1121. [Google Scholar] [CrossRef] [PubMed]
- Liebhard, R.; Kellerhals, M.; Pfammatter, W.; Jertmini, M.; Gessler, C. Mapping quantitative physiological traits in apple (Malus × domestica Borkh.). Plant Mol. Biol. 2003, 52, 511–526. [Google Scholar] [CrossRef] [PubMed]
- Sagar, M.; Chervin, C.; Roustan, J.-P.; Bouzayen, M.; Zouine, M. Under-expression of the Auxin Response Factor Sl-ARF4 improves post-harvest behavior of tomato fruits. Plant Signal. Behav. 2013, 8, e25647. [Google Scholar] [CrossRef]
- Urrestarazu, J.; Muranty, H.; Denancé, C.; Leforestier, D.; Ravon, E.; Guyader, A.; Guisnel, R.; Feugey, L.; Aubourg, S.; Celton, J.-M.; et al. Genome-Wide Association Mapping of Flowering and Ripening Periods in Apple. Front. Plant Sci. 2017, 8, 1923. [Google Scholar] [CrossRef]
- Brown, S. Apple. In Fruit Breeding, Handbook of Plant Breeding; Badenes, M.L., Byrne, D.H., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 329–367. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gilpin, L.; Costa, F.; Røen, D.; Alsheikh, M. Genome-Wide Association Analyses Defined the Interplay between Two Major Loci Controlling the Fruit Texture Performance in a Norwegian Apple Collection (Malus × domestica Borkh.). Horticulturae 2024, 10, 1049. https://doi.org/10.3390/horticulturae10101049
Gilpin L, Costa F, Røen D, Alsheikh M. Genome-Wide Association Analyses Defined the Interplay between Two Major Loci Controlling the Fruit Texture Performance in a Norwegian Apple Collection (Malus × domestica Borkh.). Horticulturae. 2024; 10(10):1049. https://doi.org/10.3390/horticulturae10101049
Chicago/Turabian StyleGilpin, Liv, Fabrizio Costa, Dag Røen, and Muath Alsheikh. 2024. "Genome-Wide Association Analyses Defined the Interplay between Two Major Loci Controlling the Fruit Texture Performance in a Norwegian Apple Collection (Malus × domestica Borkh.)" Horticulturae 10, no. 10: 1049. https://doi.org/10.3390/horticulturae10101049
APA StyleGilpin, L., Costa, F., Røen, D., & Alsheikh, M. (2024). Genome-Wide Association Analyses Defined the Interplay between Two Major Loci Controlling the Fruit Texture Performance in a Norwegian Apple Collection (Malus × domestica Borkh.). Horticulturae, 10(10), 1049. https://doi.org/10.3390/horticulturae10101049






