Comparative Proteomics Analysis of Primulina serrulata Leaves Reveals New Insight into the Formation of White Veins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Chl Determination
2.2. Protein Extraction, Digestion, and TMT Labeling
2.3. High pH Phase Separation
2.4. MS analysis
2.5. Protein Identification and Quantification
2.6. Bioinformatics Analysis
3. Results
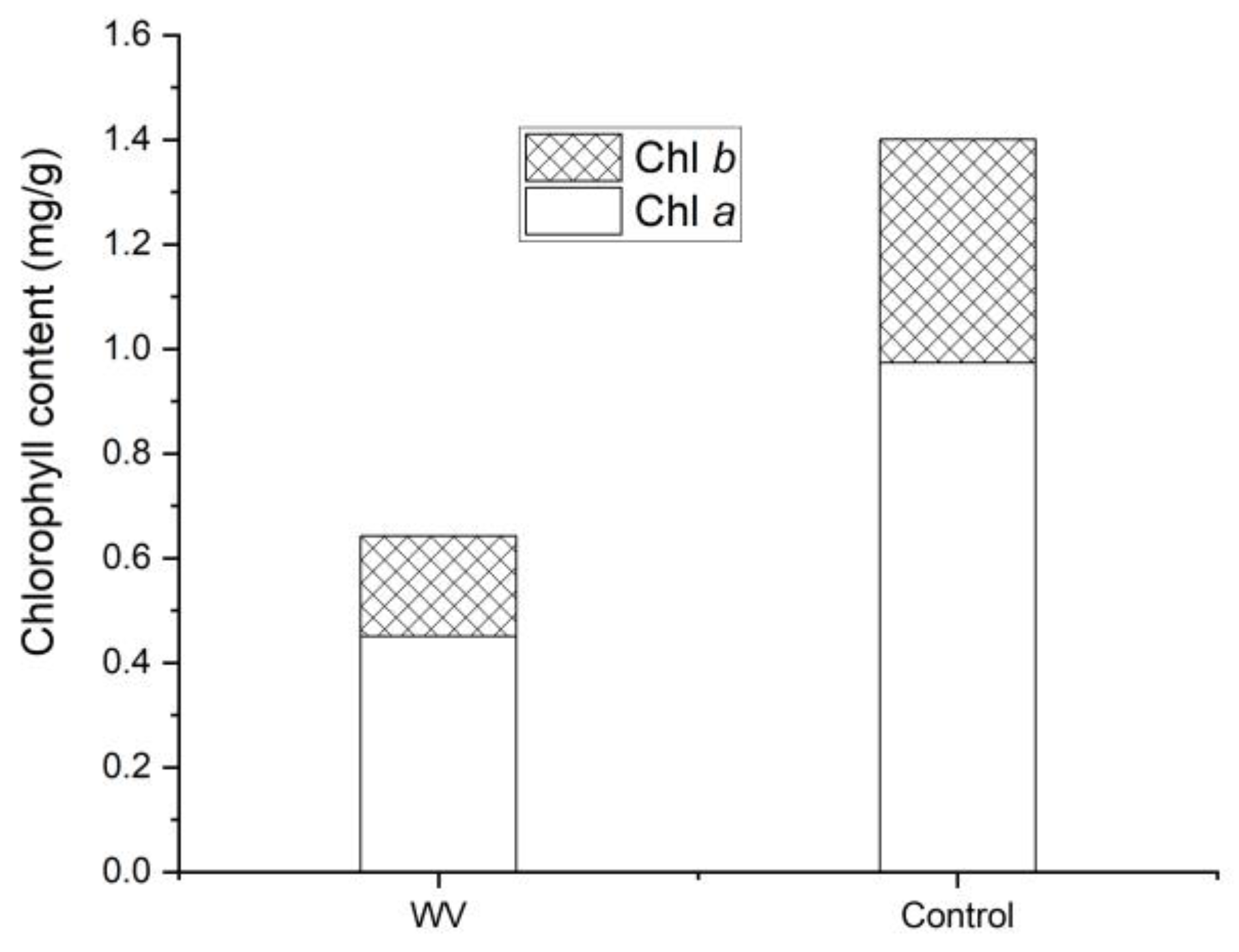
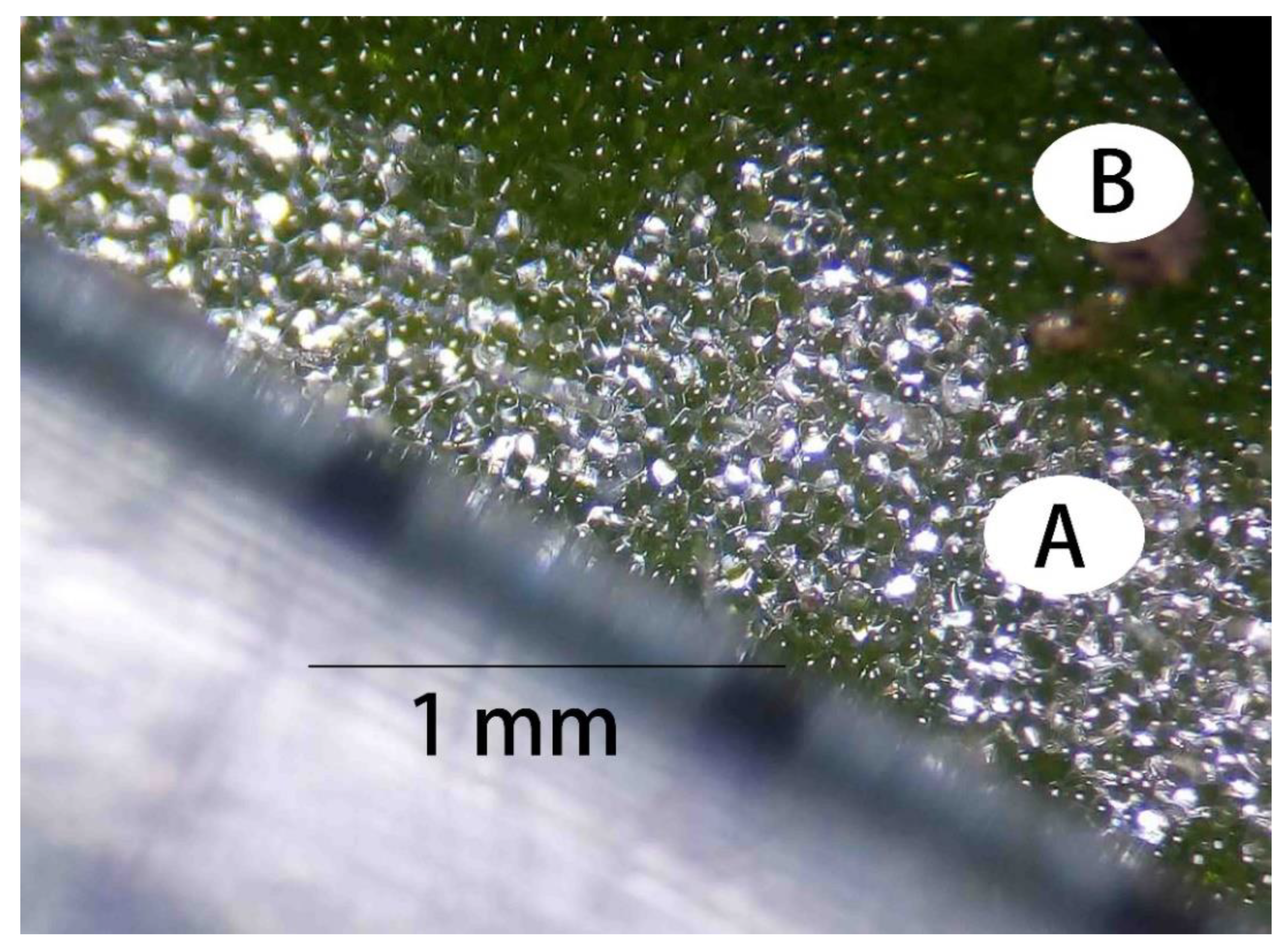
3.1. Chl Contents in WV Plants and the Control
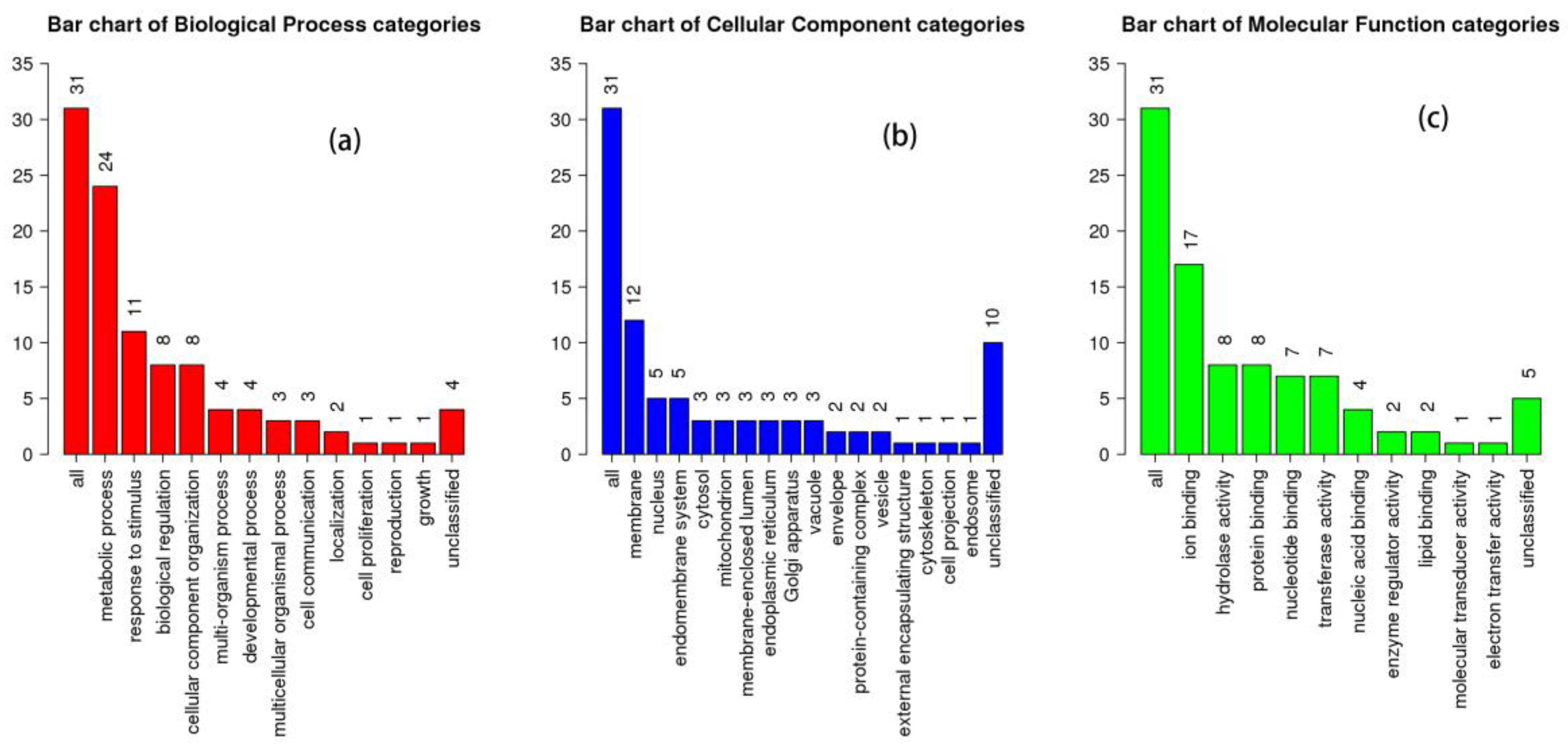
3.2. Functional Analysis of Total Proteins
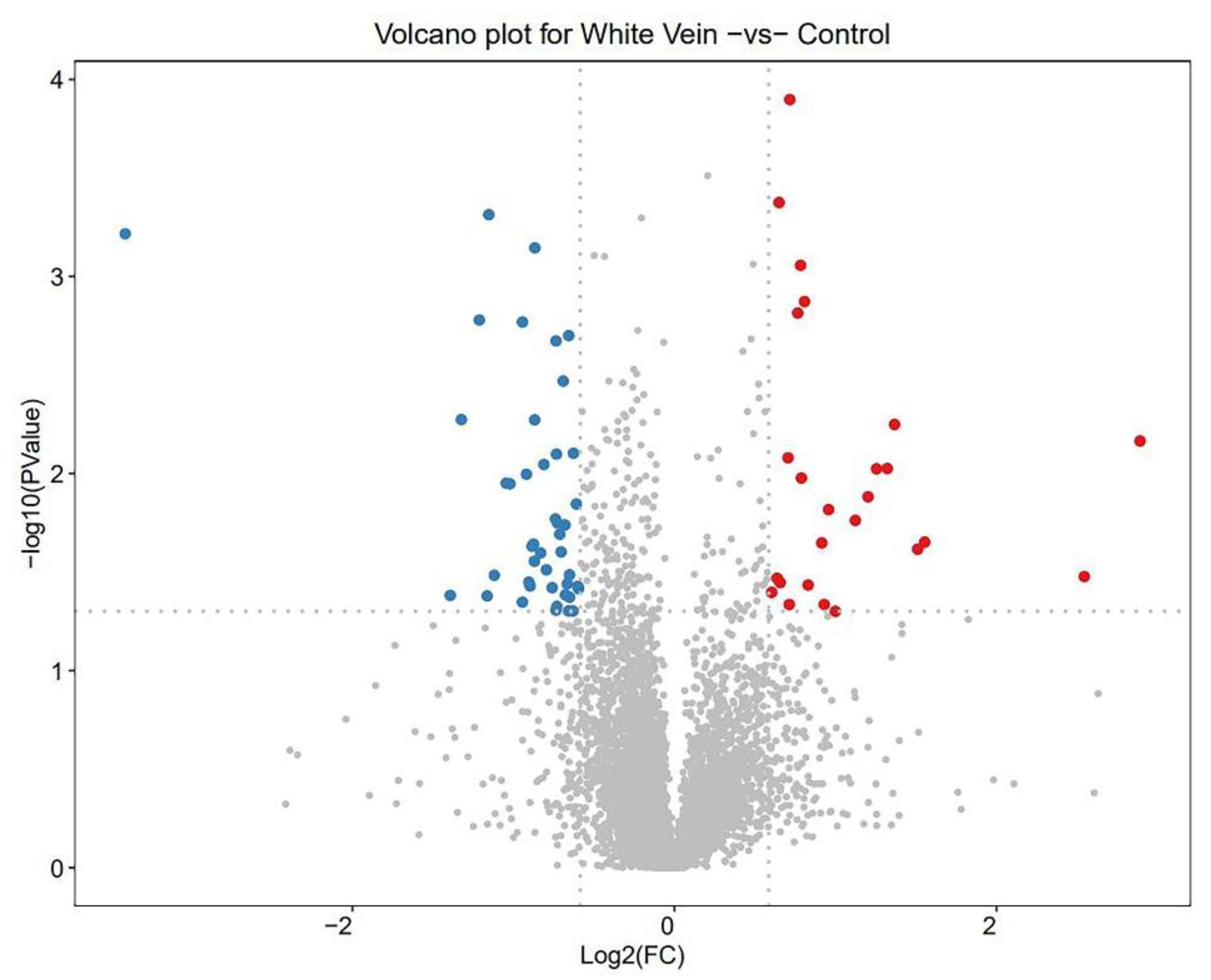
3.3. Quantity of DEPs
3.4. Functional Enrichment Analysis of the DEPs
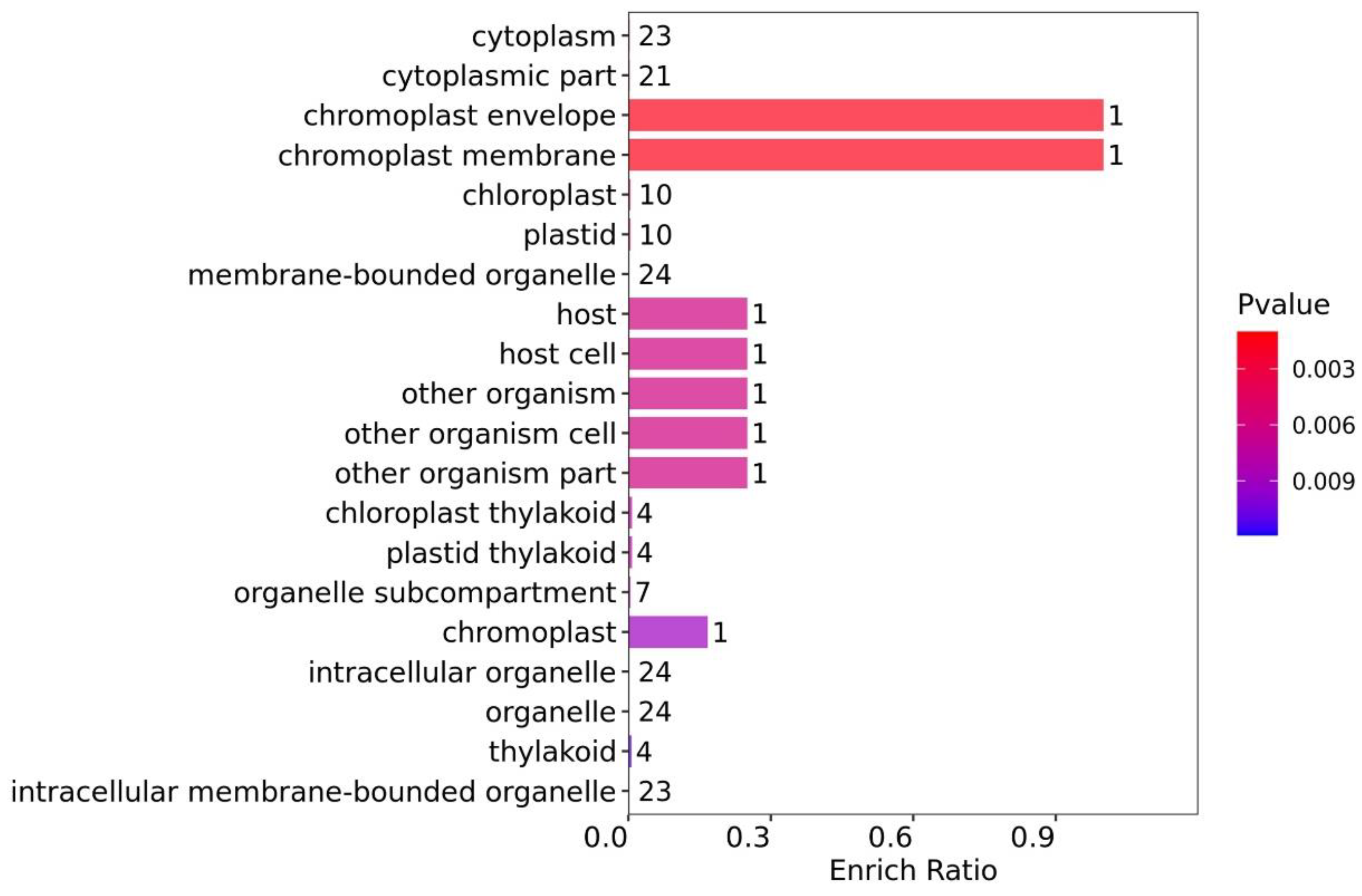
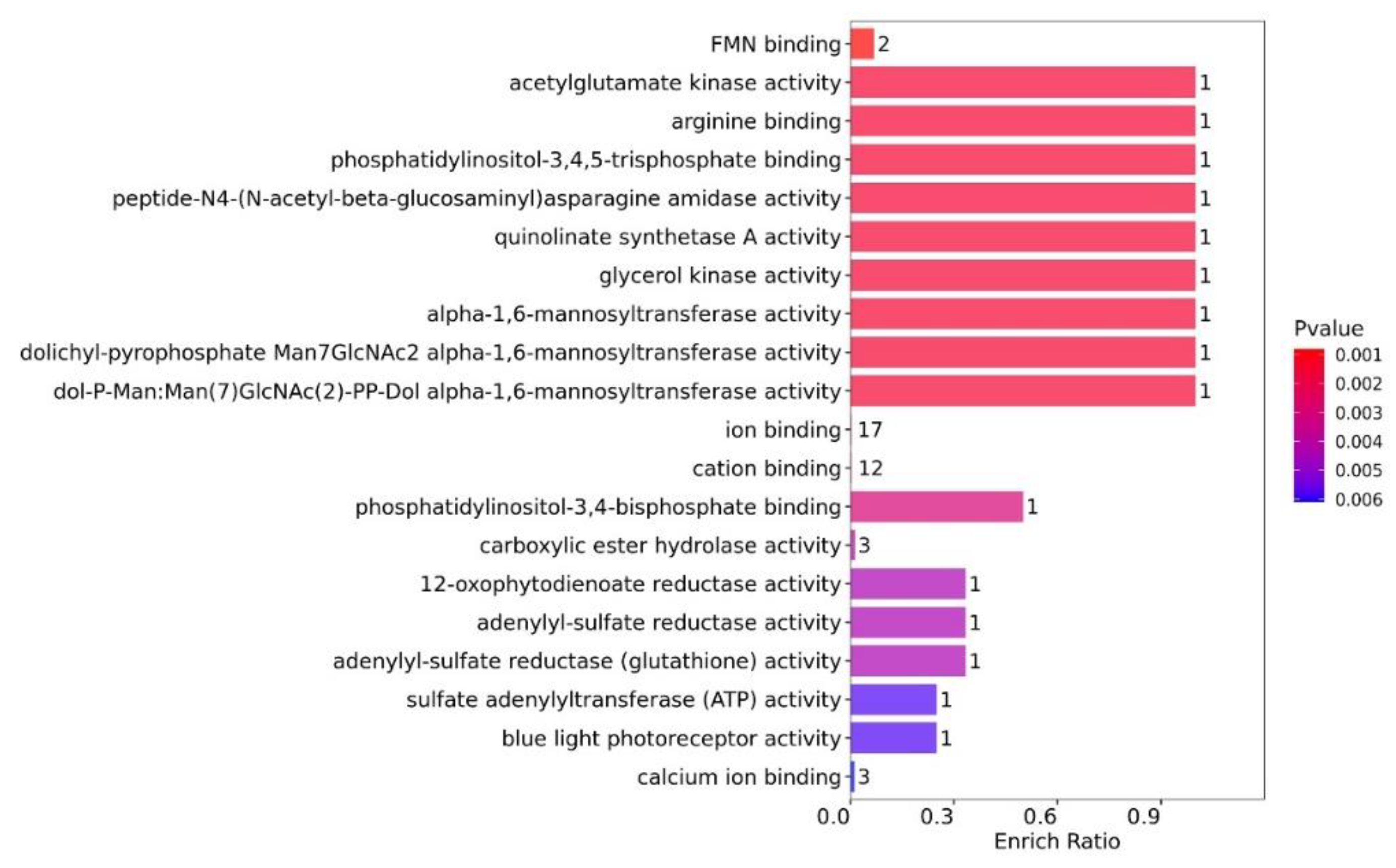
3.4.1. Functional GO Enrichment Analysis
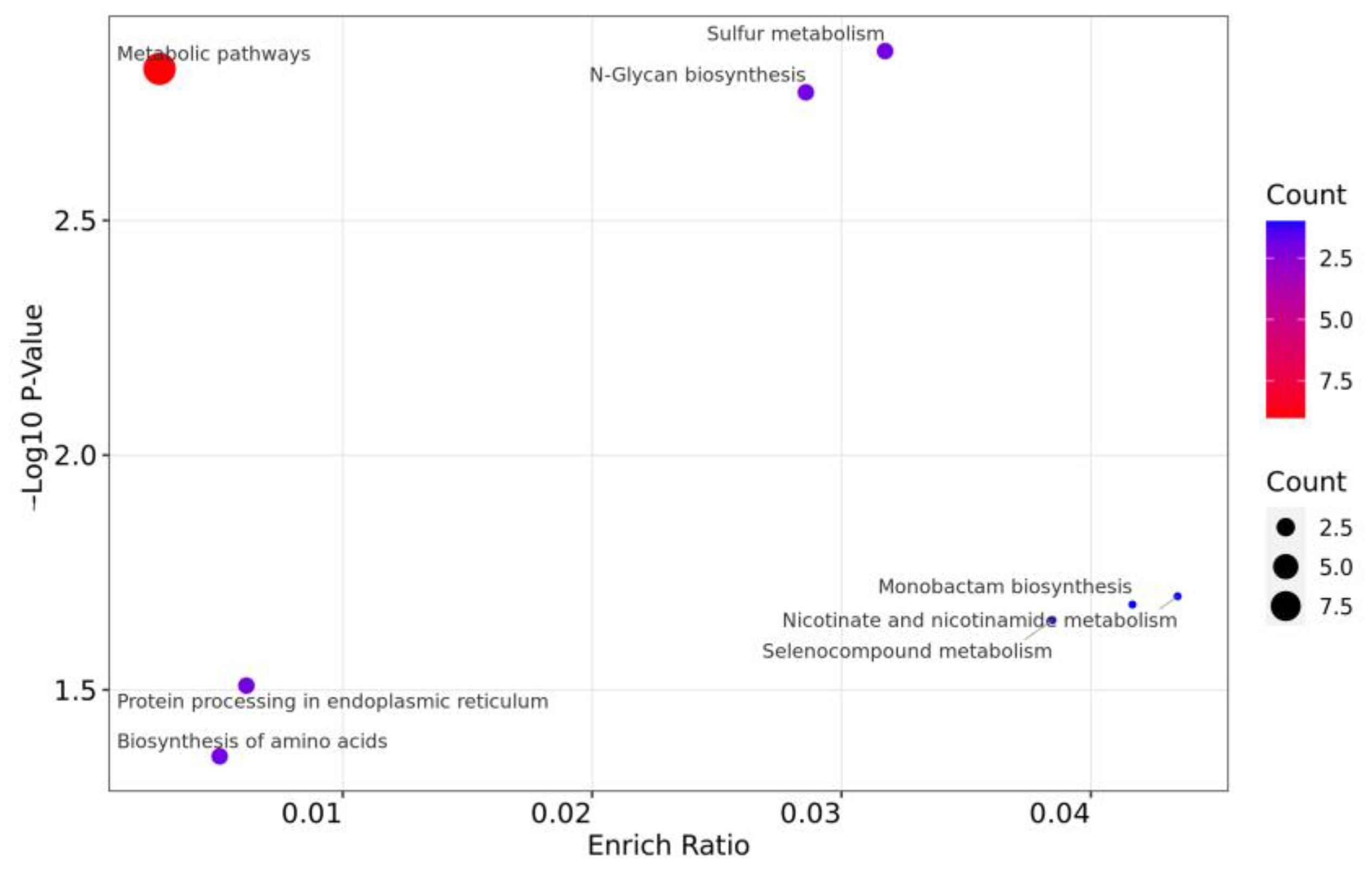
3.4.2. Functional KEGG Enrichment Analysis
3.5. Protein Interaction Network Analysis
4. Discussion
4.1. Proteins Affecting the Formation of Chloroplast and Chl
4.2. Proteins Related to Stress Resistance
4.3. Proteins Related to Biomacromolecule Modification
4.4. Proteins Related to Iron and/or Sulfur Increase in WV Leaves
4.5. DEPs Affecting Cell-Wall Formation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hedtke, B.; Wagner, I.; Börner, T.; Hess, W. Inter-organellar crosstalk in higher plants: Impaired chloroplast development affects mitochondrial gene and transcript levels. Plant J. Cell Mol. Biol. 1999, 19, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Toshoji, H.; Katsumata, T.; Takusagawa, M.; Yusa, Y.; Sakai, A. Effects of chloroplast dysfunction on mitochondria: White sectors in variegated leaves have higher mitochondrial DNA levels and lower dark respiration rates than green sectors. Protoplasma 2011, 249, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Ma, J.; He, Y.; Lin, Z.; Li, X.; Yu, S.; Li, R.; Jiang, F.; Li, X.; Huang, Z.; et al. High-throughput sequencing analysis revealed the regulation patterns of small RNAs on the development of A. comosus var. bracteatus leaves. Sci. Rep. 2018, 8, 1947. [Google Scholar] [CrossRef] [PubMed]
- Aluru, M.; Bae, H.; Wu, D.; Rodermel, S. The Arabidopsis immutans mutation affects plastid differentiation and the morphogenesis of white and green sectors in variegated plants. Plant Physiol. 2001, 127, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Foudree, A.; Putarjunan, A.; Kambakam, S.; Nolan, T.; Fussell, J.; Pogorelko, G.; Rodermel, S. The Mechanism of Variegation in immutans Provides Insight into Chloroplast Biogenesis. Front. Plant Sci. 2012, 3, 260. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hung, C.; Qiu, J.; Chen, J.; Kittur, F.; Oldham, C.; Henny, R.; Burkey, K.; Fan, L.; Xie, J. Accumulation of high OPDA level correlates with reduced ROS and elevated GSH benefiting white cell survival in variegated leaves. Sci. Rep. 2017, 7, 44158. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Y.; He, L.T.; Lai, J.L.; He, S.T.; Wu, Y.X.; Zheng, Y.S. The variation of chlorophyll biosynthesis and the structure in different color leaves of Bambusa multiplex ‘Silverstripe’. J. For. Environ. 2017, 37, 385–391. [Google Scholar]
- Wei, Y.L.; Shi, Y.F.; Wu, J.L. Rice (Oryza sativa L.) leaf color mutants. J. Nucl. Agric. Sci. 2011, 25, 1169–1178. [Google Scholar]
- Li, S.M.; Wang, S.; Wang, P.; Gao, L.L.; Yang, R.T.; Li, Y. Label-free comparative proteomic and physiological analysis provides insight into leaf color variation of the golden-yellow leaf mutant of Lagerstroemia indica. J. Proteom. 2020, 228, 103942. [Google Scholar] [CrossRef]
- Xu, M.Y.; He, P.; Lai, W.; Chen, L.H.; Ge, L.L.; Liu, S.Q.; Yang, Y.J.; Advances in Molecular Mechanism of Plant Leaf Color Variation. Molecular Plant Breeding 2021. Available online: https://kns.cnki.net/kcms/detail/46.1068.S.20210112.1619.016.html (accessed on 22 October 2023).
- Dong, X.Y.; Huang, L.B.; Chen, Q.S.; Lyu, Y.Z.; Sun, H.N.; Liang, Z.H. Physiological and anatomical differences and differentially expressed genes reveal yellow leaf coloration in Shumard Oak. Plants 2020, 9, 169. [Google Scholar] [CrossRef]
- Li, C.F.; Xu, Y.; Ma, J.Q.; Jin, J.Q.; Huang, D.; Yao, M.; Ma, C.L.; Chen, L. Biochemical and transcriptomic analyses reveal different metabolite biosynthesis profiles among three color and developmental stages in ‘Anji Baicha’ (Camellia sinensis). BMC Plant Biol. 2016, 16, 195. [Google Scholar] [CrossRef] [PubMed]
- Du, W.K.; Yuan, S.X.; Hu, F.R. Research Progress on Molecular Mechanisms of the Leaf Color Mutation. Mol. Plant Breed. 2019, 17, 1888–1897. [Google Scholar]
- Jiang, H.; Deng, T.; Lv, X.-Y.; Zhang, R.-B.; Wen, F. Primulina serrulata (Gesneriaceae), a new species from southeastern Guizhou, China. PhytoKeys 2019, 132, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.L.; Qu, W.J.; Li, X.F. Experimental Guidance of Plant Biology, 4th ed.; Higher Education Press: Beijing, China, 2009. [Google Scholar]
- Tolley, N.J. Functional Analysis of a Reticulon Protein from Arabidopsis thaliana; Department of Biological Sciences, University of Warwick: Coventry, UK, 2010. [Google Scholar]
- Tarasenko, V.I.; Garnik, E.Y.; Katyshec, A.I.; Subota, I.Y.; Konstantinov, Y.M. Disruption of Arabidopsis reticulon gene RTNLB16 results in chloroplast dysfunction and oxidative stress. J. Stress Physiol. Biochem. 2012, 8, S21. [Google Scholar]
- Knopf, R.R.; Feder, A.; Mayer, K.; Lin, A.; Rozenberg, M.; Schaller, A.; Adam, Z. Rhomboid proteins in the chloroplast envelope affect the level of allene oxide synthase in Arabidopsis thaliana. Plant J. 2012, 72, 559–571. [Google Scholar] [CrossRef] [PubMed]
- De la Rosa, M.A.; Navarro, J.A.; Dıaz-Quintana, A.; De la Cerda, B.; Molina-Heredia, F.P.; Balme, A.; del S Murdoch, P.; Dıaz-Moreno, I.; Durán, R.V.; Hervás, M. An evolutionary analysis of the reaction mechanisms of photosystem I reduction by cytochrome c6 and plastocyanin. Bioelectrochemistry 2002, 55, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Zheng, Y. Phototropin is partly involved in blue-light-mediated stem elongation, flower initiation, and leaf expansion: A comparison of phenotypic responses between wild Arabidopsis and its phototropin mutants. Environ. Exp. Bot. 2020, 171, 103967. [Google Scholar] [CrossRef]
- Millner, P.A. Are guanine nucleotide-binding proteins involved in regulation of thylakoid protein kinase activity? FEBS Lett. 1987, 226, 155–160. [Google Scholar] [CrossRef]
- Romerro, L.C.; Lam, E. Guanine nucleotide binding protein involvement in early steps of phytochrome-regulated gene expression. Proc. Natl. Acad. Sci. USA 1993, 90, 1465–1469. [Google Scholar] [CrossRef]
- Shih, M.-D.; Hoekstra, F.A.; Hsing, Y.-I.C. Late embryogenesis abundant proteins. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2008; pp. 211–255. [Google Scholar]
- Wang, F.-B.; Wan, C.-Z.; Niu, H.-F.; Qi, M.-Y.; Gang, L.; Zhang, F.; Hu, L.-B.; Ye, Y.-X.; Wang, Z.-X.; Pei, B.-L. OsMas1, a novel maspardin protein gene, confers tolerance to salt and drought stresses by regulating ABA signaling in rice. J. Integr. Agric. 2023, 22, 341–359. [Google Scholar] [CrossRef]
- Liu, C.; Niu, G.; Zhang, H.; Sun, Y.; Sun, S.; Yu, F.; Lu, S.; Yang, Y.; Li, J.; Hong, Z. Trimming of N-glycans by the Golgi-localized α-1, 2-mannosidases, MNS1 and MNS2, is crucial for maintaining RSW2 protein abundance during salt stress in Arabidopsis. Mol. Plant 2018, 11, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Völker, U.; Engelmann, S.; Maul, B.; Riethdorf, S.; Völker, A.; Schmid, R.; Mach, H.; Hecker, M. Analysis of the induction of general stress proteins of Bacillus subtilis. J. Microbiol. 1994, 140, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Traylor-Knowles, N.; Rose, N.H.; Sheets, E.A.; Palumbi, S.R. Early transcriptional responses during heat stress in the coral Acropora hyacinthus. Biol. Bull. 2017, 232, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Margaria, P.; Abbà, S.; Palmano, S. Novel aspects of grapevine response to phytoplasma infection investigated by a proteomic and phospho-proteomic approach with data integration into functional networks. BMC Genom. 2013, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Manara, A.; DalCorso, G.; Leister, D.; Jahns, P.; Baldan, B.; Furini, A. At SIA 1 AND At OSA 1: Two Abc1 proteins involved in oxidative stress responses and iron distribution within chloroplasts. New Phytol. 2014, 201, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Tafesse, F.G.; Guimaraes, C.P.; Maruyama, T.; Carette, J.E.; Lory, S.; Brummelkamp, T.R.; Ploegh, H.L. GPR107, a G-protein-coupled Receptor Essential for Intoxication by Pseudomonas aeruginosa Exotoxin A, Localizes to the Golgi and Is Cleaved by Furin. J. Biol. Chem. 2014, 289, 24005–24018. [Google Scholar] [CrossRef]
- Qiao, K.; Tian, Y.; Hu, Z.; Chai, T. Wheat cell number regulator CNR10 enhances the tolerance, translocation, and accumulation of heavy metals in plants. Environ. Sci. Technol. 2018, 53, 860–867. [Google Scholar] [CrossRef]
- Suzuki, T.; Park, H.; Hollingsworth, N.M.; Sternglanz, R.; Lennarz, W.J. PNG1, a yeast gene encoding a highly conserved peptide: N-glycanase. J. Cell Biol. 2000, 149, 1039–1052. [Google Scholar] [CrossRef]
- Farmer, E.E.; Goossens, A. Jasmonates: What ALLENE OXIDE SYNTHASE does for plants. J. Exp. Bot. 2019, 70, 3373–3378. [Google Scholar] [CrossRef]
- De Lorenzo, G.; Ferrari, S. Polygalacturonase-inhibiting proteins in defense against phytopathogenic fungi. Curr. Opin. Plant Biol. 2002, 5, 295–299. [Google Scholar] [CrossRef]
- Shi, H.; Liu, W.; Yao, Y.; Wei, Y.; Chan, Z. Alcohol dehydrogenase 1 (ADH1) confers both abiotic and biotic stress resistance in Arabidopsis. Plant Sci. 2017, 262, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Anduro, G.; Ceniceros-Ojeda, E.A.; Casados-Vázquez, L.E.; Bencivenni, C.; Sierra-Beltrán, A.; Murillo-Amador, B.; Tiessen, A. Genome-wide analysis of the beta-glucosidase gene family in maize (Zea mays L. var B73). Plant Mol. Biol. 2011, 77, 159–183. [Google Scholar] [CrossRef] [PubMed]
- Zielińska-Dawidziak, M. Plant ferritin—A source of iron to prevent its deficiency. Nutrients 2015, 7, 1184–1201. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B. Molecular Biology of the Cell; Garland Science: New York, NY, USA, 2017. [Google Scholar]
- Campbell, W.H. Nitrate reductase structure, function and regulation: Bridging the gap between biochemistry and physiology. Annu. Rev. Plant Biol. 1999, 50, 277–303. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.J.; Alben, J.O.; Gennis, R.B. Spectroscopic evidence for a heme-heme binuclear center in the cytochrome bd ubiquinol oxidase from Escherichia coli. Proc. Natl. Acad. Sci. USA 1993, 90, 5863–5867. [Google Scholar] [CrossRef]
- Anand, A.; Srivastava, P.K. A molecular description of acid phosphatase. Appl. Biochem. 2012, 167, 2174–2197. [Google Scholar] [CrossRef]
- Marsh, H., Jr.; Evans, H.; Matrone, G. Investigations of the role of iron in chlorophyll metabolism. II. Effect of iron deficiency on chlorophyll synthesis. Plant Physiol. 1963, 38, 638. [Google Scholar] [CrossRef]
- Granick, S. Evolution of Heme and Chlorophyll, Evolving Genes and Proteins; Elsevier: Amsterdam, The Netherlands, 1965; pp. 67–88. [Google Scholar]
- Christensen, G.; Zane, G.; Kazakov, A.; Li, X.; Rodionov, D.; Novichkov, P.; Dubchak, I.; Arkin, A.; Wall, J. Rex (encoded by DVU_0916) in Desulfovibrio vulgaris Hildenborough is a repressor of sulfate adenylyl transferase and is regulated by NADH. J. Bacteriol. 2015, 197, 29–39. [Google Scholar] [CrossRef]
- Rosier, K.; McDevitt, M.T.; Smet, J.; Floyd, B.J.; Verschoore, M.; Marcaida, M.J.; Bingman, C.A.; Lemmens, I.; Peraro, M.D.; Tavernier, J. Prolyl endopeptidase-like is a (thio) esterase involved in mitochondrial respiratory chain function. Iscience 2021, 24, 103460. [Google Scholar] [CrossRef]
- Kopriva, S.; Koprivova, A. Plant adenosine 5′-phosphosulphate reductase: The past, the present, and the future. J. Exp. Bot. 2004, 55, 1775–1783. [Google Scholar] [CrossRef]
- Choudens, S.O.-D.; Loiseau, L.; Sanakis, Y.; Barras, F.; Fontecave, M. Quinolinate synthetase, an iron–sulfur enzyme in NAD biosynthesis. FEBS Lett. 2005, 579, 3737–3743. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Assmann, S.M. The glycolytic enzyme, phosphoglycerate mutase, has critical roles in stomatal movement, vegetative growth, and pollen production in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 5179–5189. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Li, Q.; Yin, Q.; Wang, J.; Zhang, B.; Gan, S.; Wu, A.-M. Identification of novel miRNAs and their target genes in Eucalyptus grandis. Tree Genet. Genomes 2018, 14, 1–9. [Google Scholar] [CrossRef]
- Costacurta, A.; Vanderleyden, J. Synthesis of phytohormones by plant-associated bacteria. Crit. Rev. Microbiol. 1995, 21, 1–18. [Google Scholar] [CrossRef]
- Zhou, H.-W.; Ben-Arie, R.; Lurie, S. Pectin esterase, polygalacturonase and gel formation in peach pectin fractions. Phytochemistry 2000, 55, 191–195. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dou, Q.-L.; Xie, D.-J.; Deng, T.; Chen, M.-F.; Qian, Z.-M.; Wang, S.-S.; Zhang, R.-B. Comparative Proteomics Analysis of Primulina serrulata Leaves Reveals New Insight into the Formation of White Veins. Horticulturae 2024, 10, 19. https://doi.org/10.3390/horticulturae10010019
Dou Q-L, Xie D-J, Deng T, Chen M-F, Qian Z-M, Wang S-S, Zhang R-B. Comparative Proteomics Analysis of Primulina serrulata Leaves Reveals New Insight into the Formation of White Veins. Horticulturae. 2024; 10(1):19. https://doi.org/10.3390/horticulturae10010019
Chicago/Turabian StyleDou, Quan-Li, Da-Jun Xie, Tan Deng, Mo-Fang Chen, Zheng-Min Qian, Shuang-Shuang Wang, and Ren-Bo Zhang. 2024. "Comparative Proteomics Analysis of Primulina serrulata Leaves Reveals New Insight into the Formation of White Veins" Horticulturae 10, no. 1: 19. https://doi.org/10.3390/horticulturae10010019
APA StyleDou, Q.-L., Xie, D.-J., Deng, T., Chen, M.-F., Qian, Z.-M., Wang, S.-S., & Zhang, R.-B. (2024). Comparative Proteomics Analysis of Primulina serrulata Leaves Reveals New Insight into the Formation of White Veins. Horticulturae, 10(1), 19. https://doi.org/10.3390/horticulturae10010019





