Analysis of the Diversity Presented by Vitis vinifera L. in the Volcanic Island of La Gomera (Canary Archipelago, Spain) Using Simple Sequence Repeats (SSRs) as Molecular Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Extraction and Purification
2.3. Microsatellites
2.4. DNA Amplification
2.5. Amplified Fragments Length Measurement
2.6. Data Analysis
3. Results
3.1. SSR Polymorphism
3.2. Grapevine Variety Analysis
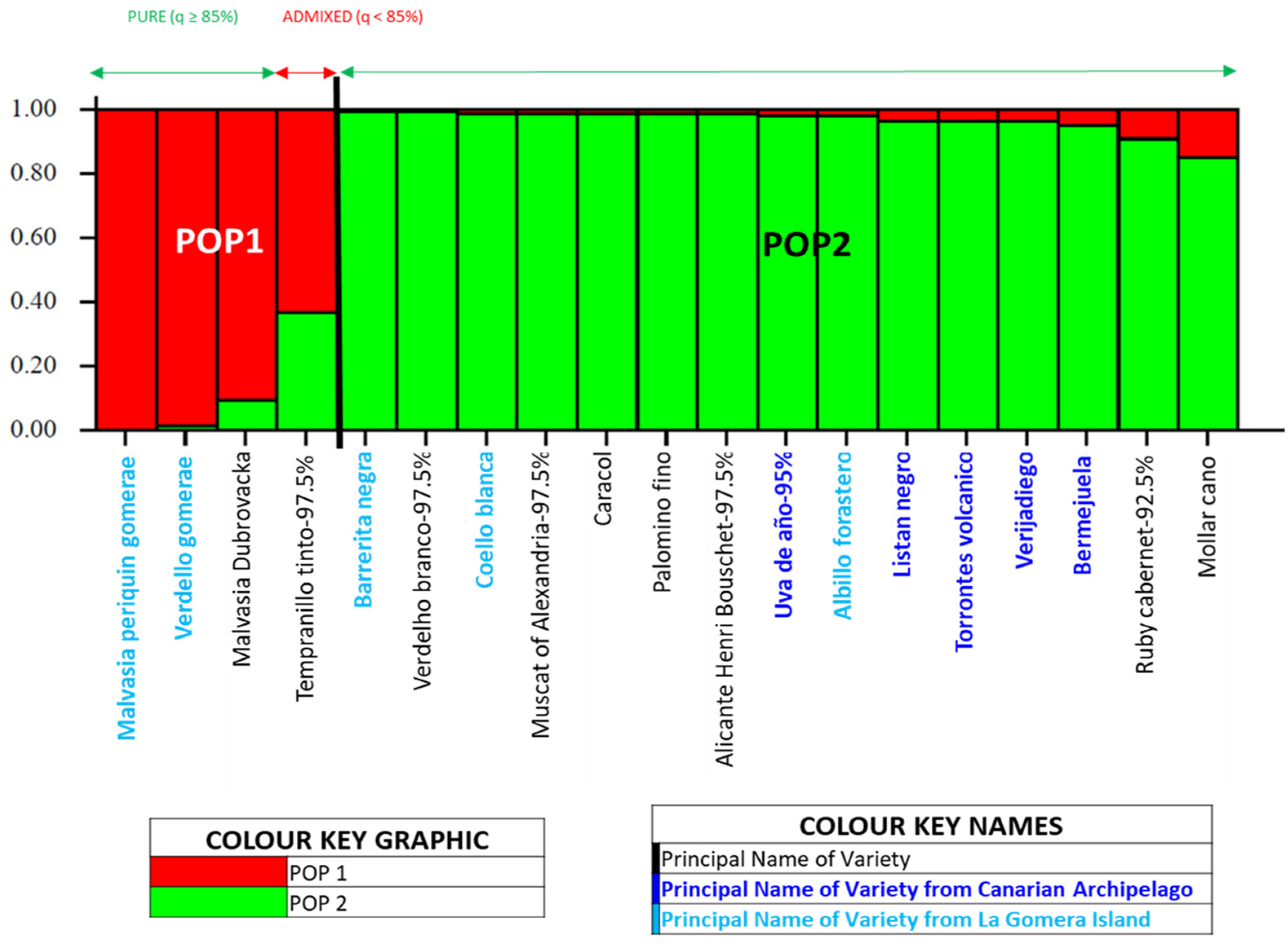
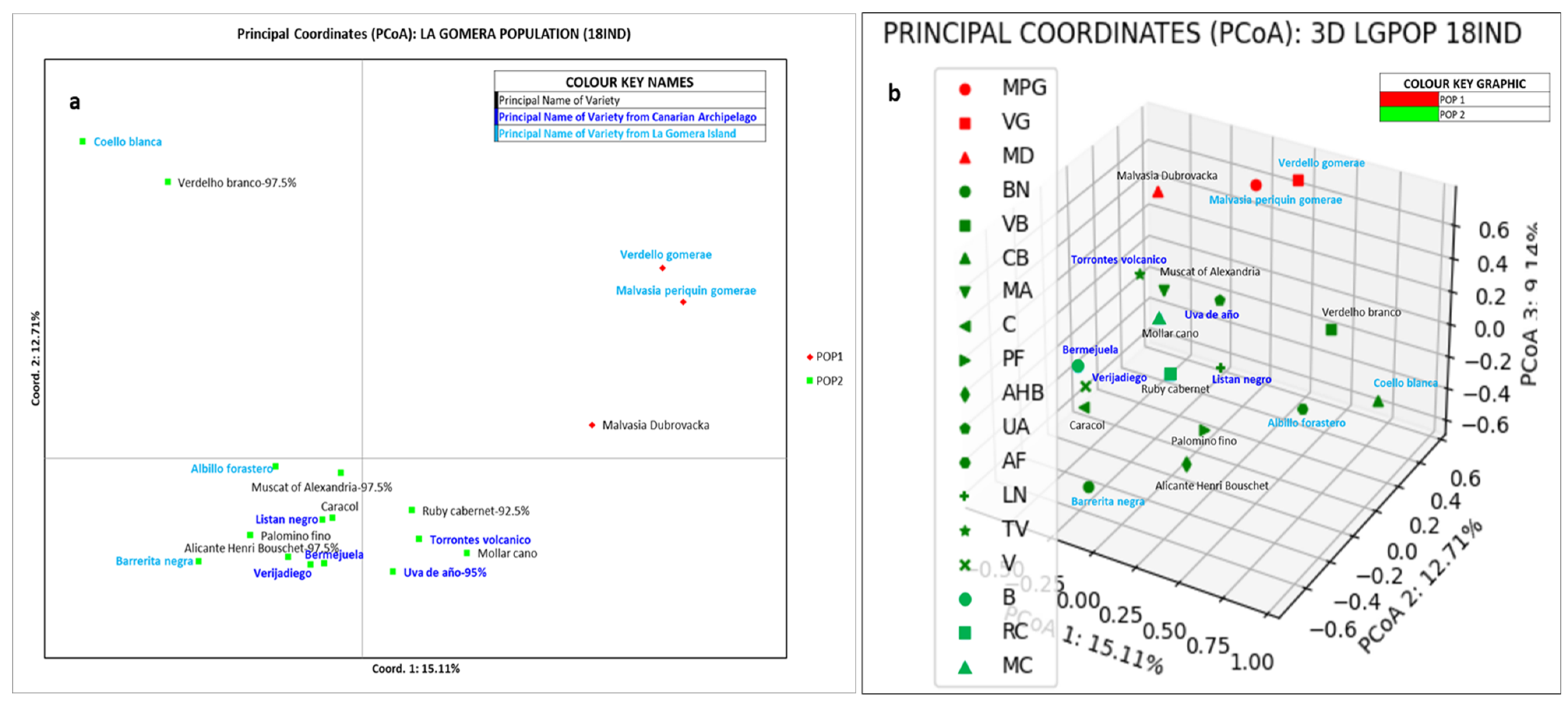
3.3. Genetic Structure of the Grapevine Population of the Island of La Gomera
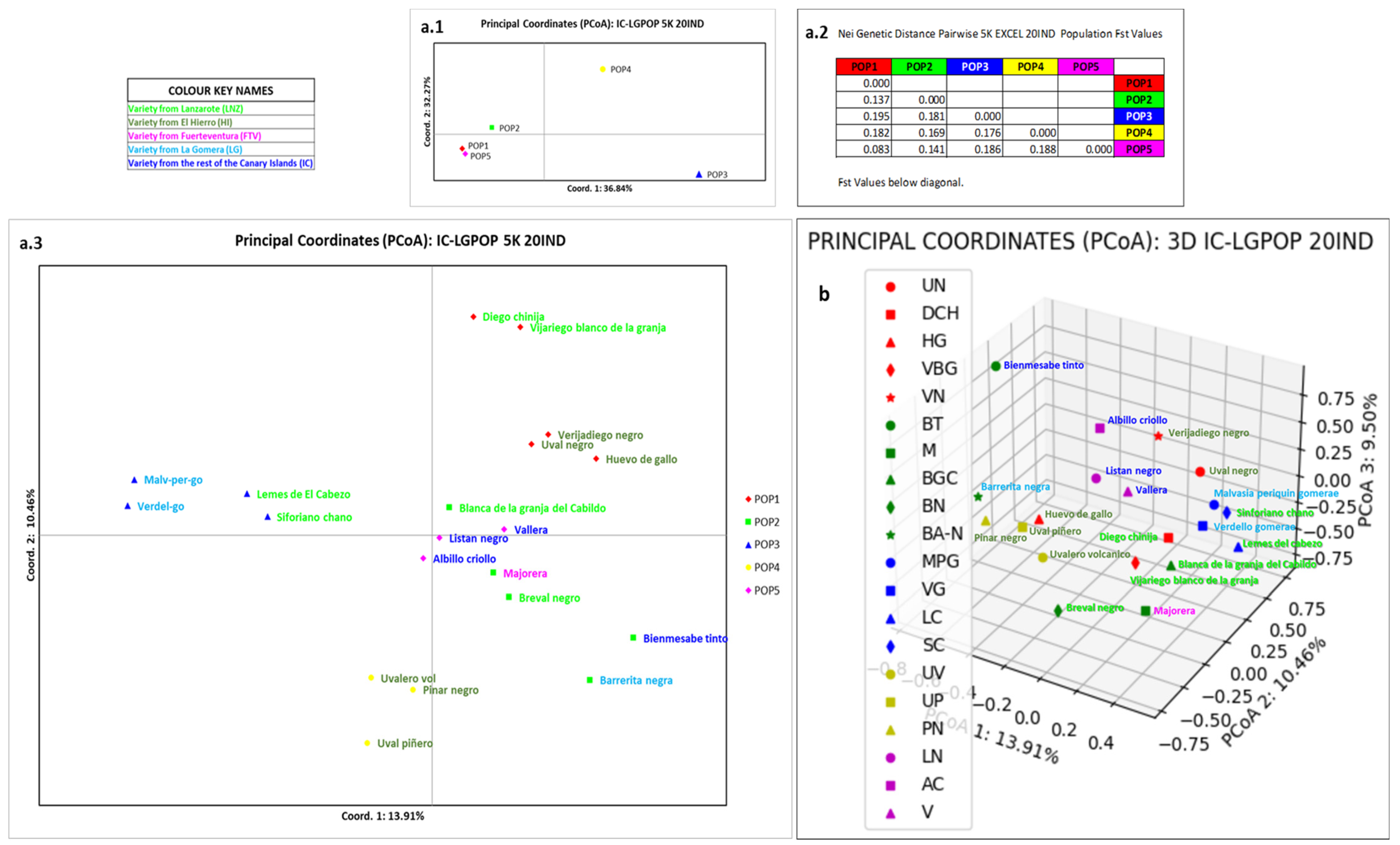
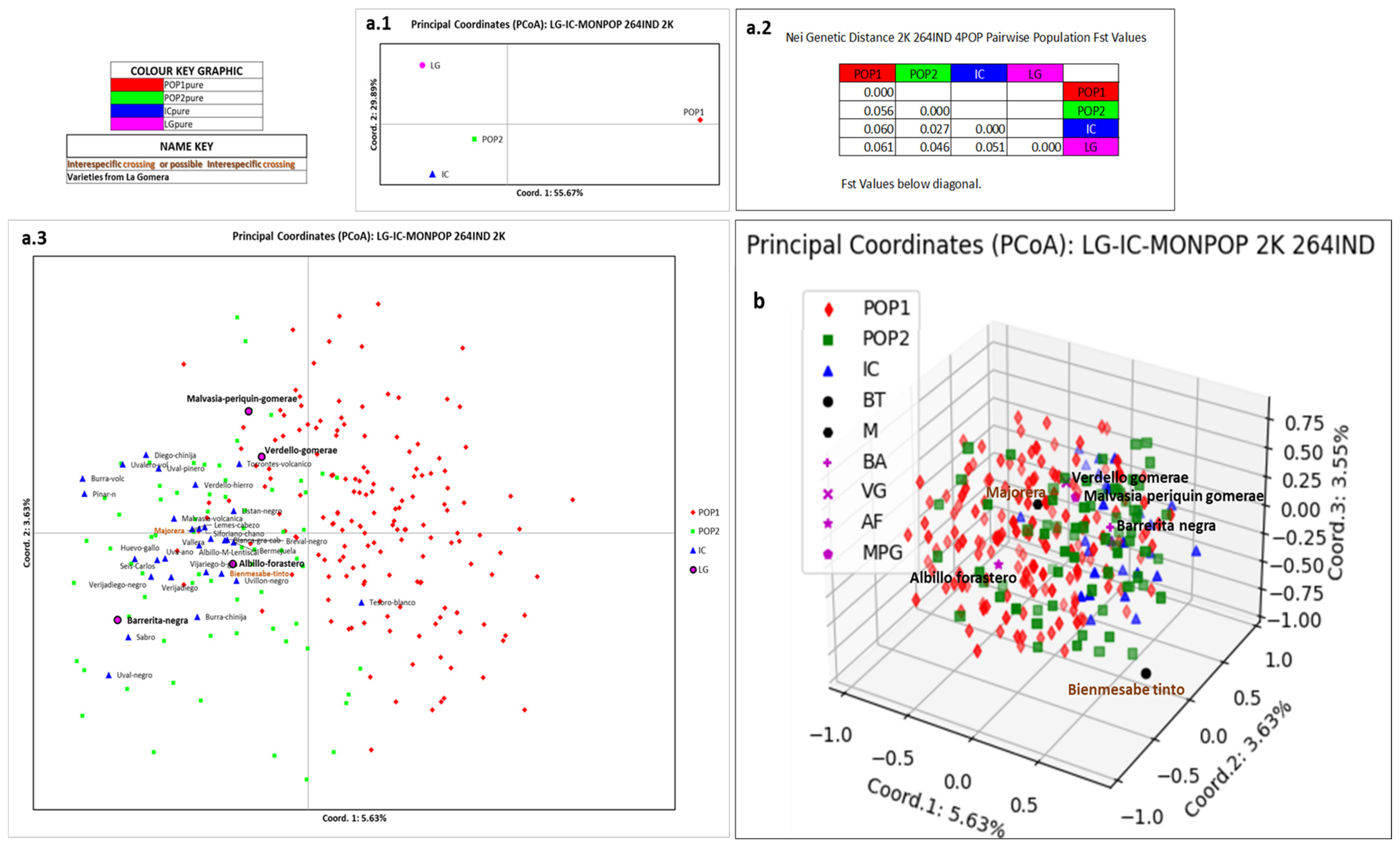
3.4. Relation of La Gomera Grapevine Population with respect to the Canary Archipelago Population
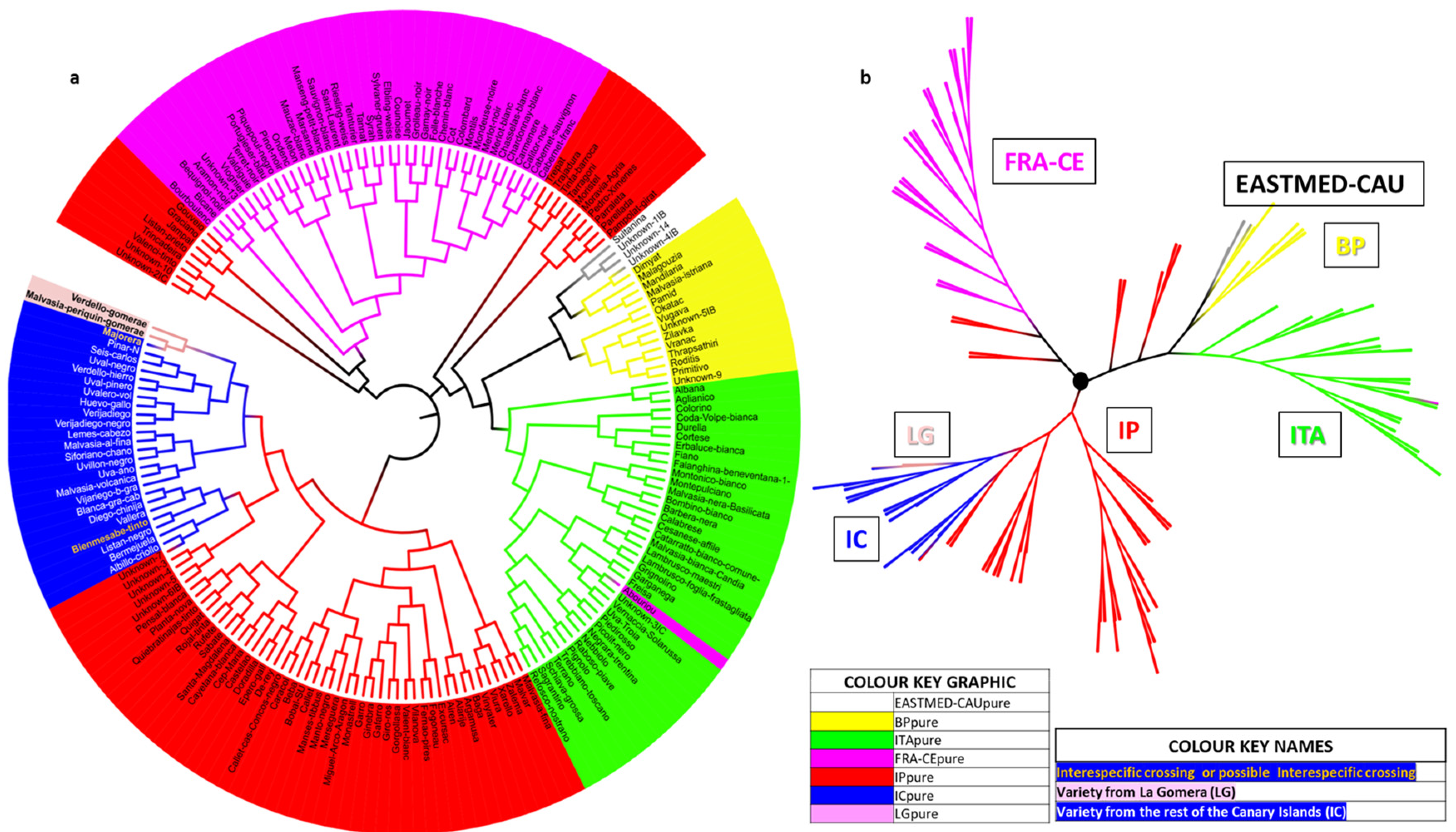
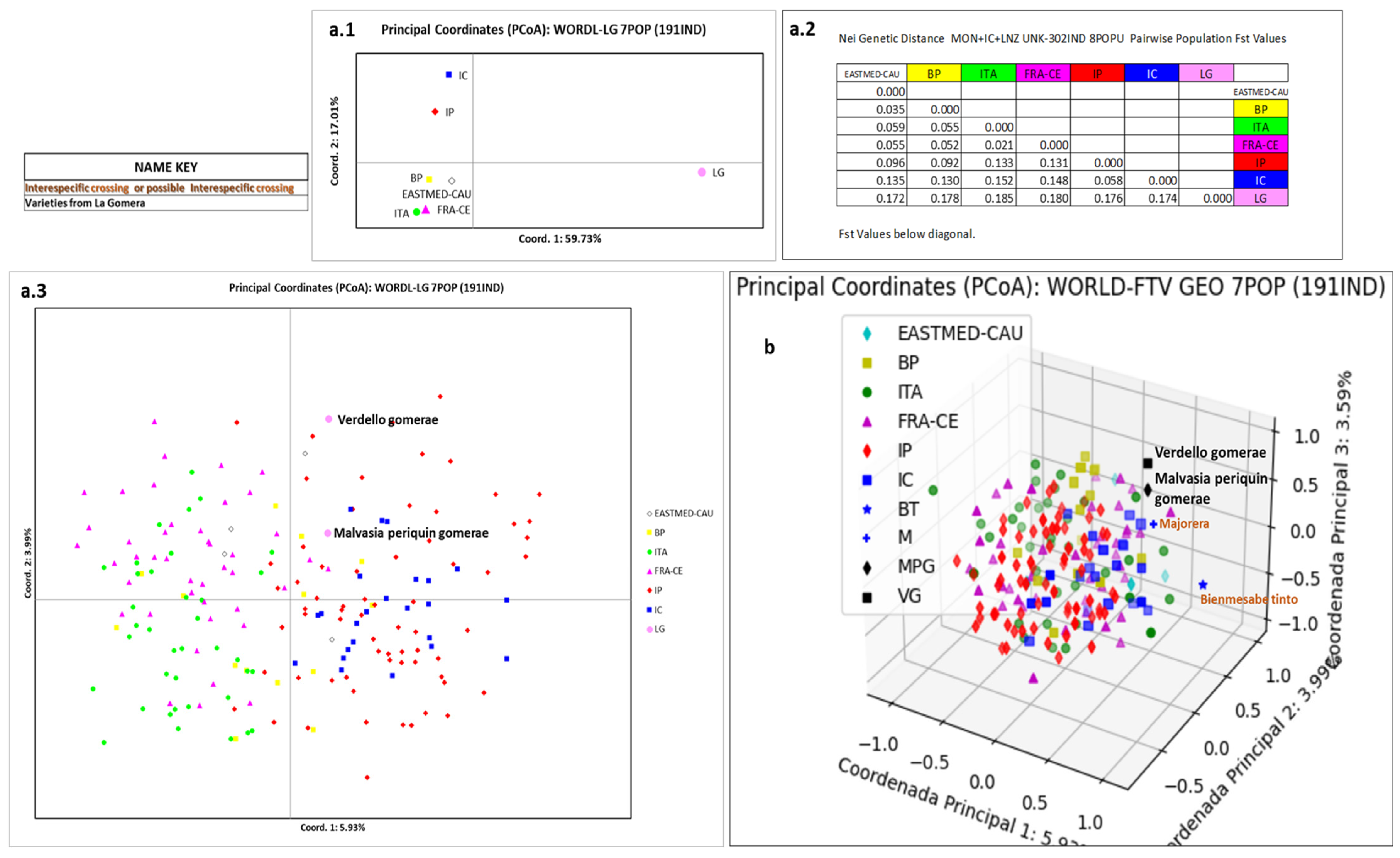
3.5. Relation of La Gomera Grapevine Population with Respect to the World Population
4. Discussion
4.1. SSR Polymorphism
4.2. Grapevine Variety Analysis
4.3. Genetic Structure of the Grapevine Population of the Island of La Gomera
4.4. Relation of La Gomera Grapevine Population with Respect to the Canary Archipelago Population
4.5. Relation of La Gomera Grapevine Population with Respect to the World Population
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wolkovich, E.M.; García de Cortázar-Atauri, I.; Morales-Castilla, I.; Nicholas, K.A.; Lacombe, T. From Pinot to Xinomavro in the world’s future wine-growing regions. Nat. Clim. Chang. 2018, 8, 29–37. [Google Scholar] [CrossRef]
- NASS. 2021. Available online: https://quickstats.nass.usda.gov/results/9E7FD873-AD79-3190-8C62-0B7B559A0F68 (accessed on 23 April 2023).
- Análisis Annual del Sector Vitivinícola Mundial en 2021. Organización Internacional de la Viña y el Vino (OIV). Available online: https://www.oiv.int/sites/default/files/documents/OIV_Analisis_anual_del_sector_vitivinicola_mundial_en_2021.pdf (accessed on 17 October 2023).
- INE. 2021. Available online: https://www.ine.es/jaxiT3/Datos.htm?t=2910 (accessed on 7 July 2023).
- Wikimedia Commons. 2014. Available online: https://commons.wikimedia.org/w/index.php?curid=33566121#file (accessed on 7 July 2023).
- NASA. 2011. Available online: https://www.flickr.com/photos/gsfc/6630087415/in/photostream/ (accessed on 7 July 2023).
- Arco, M.J.; Atiénzar, E.; Rosario, M.C.; Arco, M.M.; González, C.; Arco, M.C. El Menceyato de Icod en el poblamiento de Tenerife: D.; Gaspar, Las Palomas y Los Guanches. Sobre el poblamiento y las estrategias de alimentación vegetal entre los Guanches. Eres 2000, 9, 67–129. [Google Scholar]
- Hidalgo, J.; Hidalgo, L. La Filoxera. In Tratado de Viticultura, 2nd ed.; Mundi-Prensa: Madrid, Spain, 2019; Volume 1. [Google Scholar]
- Climate Data. 2021. Available online: https://es.climate-data.org/europe/espana/la-gomera-10273/ (accessed on 7 July 2023).
- Elías, L.V. El Paisaje del Viñedo en las Islas Canarias; PASOS: Revista de turismo y Patrimonio Cultural; Asociación Canaria de Antropología: Tenerife, Spain, 2013; Volume 11, p. 152. Available online: http://www.pasosonline.org/Publicados/pasosoedita/PSEdita11.pdf (accessed on 7 July 2023).
- Instituto Canario de Estadística. 2021. Available online: https://www3.gobiernodecanarias.org/istac/statistical-visualizer/visualizer/data.html?resourceType=dataset&agencyId=ISTAC&resourceId=E01135A_000004&version=1.0# (accessed on 8 July 2023).
- Consejo Regulador de la Gomera. 2020. Available online: https://vinoslagomera.com/ (accessed on 8 July 2023).
- Pliego de Condiciones de la Denominación de Origen Protegida de Vinos “La Gomera”. 2013. Available online: https://vinoslagomera.com/wp-content/uploads/2013/03/Pliego_de_condiciones_DOP_de_vinos_La_Gomera.pdf (accessed on 8 July 2023).
- Rodriguez-Torres, I. Albillo Forastero. In Variedades de vid Cultivadas en Canarias. Descriptores Morfológicos. Caracterización Morfológica, Molecular, Agronómica y Enológica, 2nd ed.; Instituto Canario de Investigaciones Agrarias, Ed.; Gobierno de Canarias: Santa Cruz de Tenerife, Spain, 2018; Available online: https://www.icia.es/icia/download/Publicaciones/Variedades_Vid_Canarias.pdf (accessed on 8 July 2023).
- Maul, E.; Röckel, R. Vitis International Variety Catalogue (VIVC): A cultivar database referenced by genetic profiles and morphology. In BIO Web of Conferences; EDP Sciences: Hulis, France, 2015; Volume 5, p. 01009. [Google Scholar] [CrossRef]
- Crespan, M. Autochthonous varieties and more used vines, link to genetic. In Proceedings of the 2nd OENOVITI International Symposium, Session I: Genetic Characterization, Phenotyping and Pedigrees, Geisenheim, Germany, 3–5 November 2014; pp. 7–11. Available online: https://www.calameo.com/books/003503752f575f124dc11 (accessed on 11 July 2023).
- This, P.; Lacombe, T.; Thomas, M.R. Historical origins and genetic diversity of wine grapes. Trends Genet. 2006, 22, 511–519. [Google Scholar] [CrossRef]
- Lopes, M.S.; Sefc, K.M.; Eiras-Dias, E.; Steinkellner, H.; Laimer Câmara Machado, M.; Câmara Machado, A. The use of microsatellites for germplasm management in a Portuguese grapevine collection. Theor. Appl. Genet. 1999, 99, 733–739. [Google Scholar] [CrossRef]
- Sefc, K.M.; Lefort, F.; Grando, M.S.; Scott, K.D.; Steinkellner, H.; Thomas, M.R. Microsatellite Markers for Grapevine: Tools for Cultivar Identification and Pedigree Reconstruction. In Microsatellite Markers for Grapevine: A State of the Art; Roubelakis-Angelakis, K.A., Ed.; Molecular Biology & Biotechnology of the Grapevine; Springer: Dordrecht, The Netherlands, 2001. [Google Scholar] [CrossRef]
- Marsal, G.; Baiges, I.; Canals, J.M.; Zamora, F.; Fort, F. A fast, efficient method for extracting DNA from leaves, stems, and seeds of Vitis vinifera L. Am. J. Enol. Vitic. 2011, 62, 376–381. [Google Scholar] [CrossRef]
- Marsal, G.; Boronat, N.; Canals, J.M.; Zamora, F.; Fort, F. Comparison of the efficiency of some of the most usual DNA extraction methods for woody plants in different tissues of Vitis vinifera L. J. Int. Sci. Vigne Vin 2013, 47, 227–237. [Google Scholar] [CrossRef]
- Fort, F.; Hayoun, L.; Valls, J.; Canals, J.M.; Arola, L.; Zamora, F. A new and simple method for rapid extraction and isolation of high-quality RNA from grape (Vitis vinifera) berries. J. Sci. Food Agric. 2008, 88, 179–184. [Google Scholar] [CrossRef]
- Thomas, M.R.; Scott, N.S. Microsatellite repeats in grapevine reveal DNA polymorphisms when analyzed as sequence-tagged sites (STSs). Theor. Appl. Genet. 1993, 86, 985–990. [Google Scholar] [CrossRef]
- Bowers, J.E.; Dangl, G.S.; Vignani, R.; Meredith, C.P. Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome 1996, 39, 628–633. [Google Scholar] [CrossRef]
- Bowers, J.E.; Dangl, G.S.; Meredith, C.P. Development and characterization of additional microsatellite DNA markers for grape. Am. J. Enol. Vitic. 1999, 50, 243–246. [Google Scholar] [CrossRef]
- Sefc, K.M.; Regner, F.; Turetschek, E.; Glössl, J.; Steinkellner, H. Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome 1999, 42, 367–373. [Google Scholar] [CrossRef]
- Scott, K.D.; Eggler, P.; Seaton, G.; Rosseto, M.; Abblet, E.M.; Lee, L.S.; Henry, R.J. Analysis of SSRs derived from grape ESTs. Theor. Appl. Genet. 2000, 100, 723–726. [Google Scholar] [CrossRef]
- Lefort, F.; Kyvelos, C.; Zervou, M.; Edwards, K.; Roubelakis-Angelakis, K. Characterization of new microsatellite loci from Vitis vinifera and their conservation in some Vitis species and hybrids. Mol. Ecol. Resour. 2002, 2, 20–21. [Google Scholar] [CrossRef]
- Cipriani, G.; Spadotto, A.; Jurman, I.; Di Gaspero, G.; Crespan, M.; Meneghetti, S.; Frare, E.; Vignani, R.; Cresti, M.; Morgante, M.; et al. The SSR-based molecular profile of 1005 grapevine (Vitis vinifera L.) accessions uncovers new synonymy and parentages and reveals a large admixture amongst varieties of different geographic origins. Theor. Appl. Genet. 2010, 121, 1569–1585. [Google Scholar] [CrossRef]
- Dalbó, M.A.; Ye, G.N.; Weeden, N.F.; Steinkellner, H.; Sefc, K.M.; Reisch, B.I. A gene-controlling sex in grapevines is placed on a molecular marker-based genetic map. Genome 2000, 43, 333–340. [Google Scholar] [CrossRef]
- This, P.; Jung, A.; Boccacci, P.; Borrego, J.; Botta, R.; Costantini, L.; Crespan, M.; Dangl, G.S.; Eisenheld, C.; Ferreira-Monteiro, F.; et al. Development of a standard set of microsatellite reference alleles for the identification of grape cultivars. Theor. Appl. Genet. 2004, 109, 1448–1458. [Google Scholar] [CrossRef]
- Paetkau, D.; Calvert, W.; Stirling, I.; Strobeck, C. Microsatellite analysis of population structure in Canadian polar bears. Mol. Ecol. 1995, 4, 347–354. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Paetkau, D.; Slade, R.; Burden, M.; Estoup, A. Genetic assignment methods for the direct, real-time estimation of migration rate: A simulation-based exploration of accuracy and power. Mol. Ecol. 2004, 13, 55–65. [Google Scholar] [CrossRef]
- VanderPlas, J. Three-Dimensional Plotting in Matplotlib. In Python Data Science Handbook; O’Reilly Media, Inc.: Sevastopol, CA, USA, 2016; Available online: https://jakevdp.github.io/PythonDataScienceHandbook/04.12-three-dimensional-plotting.html (accessed on 21 August 2023).
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic threes. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Fort, F.; Lin-Yang, Q.; Suárez-Abreu, L.R.; Sancho-Galán, P.; Canals, J.M.; Zamora, F. Study of Molecular Biodiversity and Population Structure of Vitis vinifera L. ssp. vinifera on the Volcanic Island of El Hierro (Canary Islands, Spain) by Using Microsatellite Markers. Horticulturae 2023, 9, 1297. [Google Scholar] [CrossRef]
- Fort, F.; Marsal, G.; Mateo-Sanz, J.M.; Pena, V.; Canals, J.M.; Zamora, F. Molecular characterisation of the current cultivars of Vitis vinifera L. in Lanzarote (Canary Islands, Spain) reveals nine individuals which correspond to eight new varieties and two new sports. Oeno One 2022, 56, 281–295. [Google Scholar] [CrossRef]
- Rodriguez-Torres, I. Variedades de vid Cultivadas en Canarias. Descriptores Morfológicos. Caracterización Morfológica, Molecular, Agronómica y Enológica, 1st ed.; Instituto Canario de Investigaciones Agrarias, Gobierno de Canarias: Santa Cruz de Tenerife, Spain, 2013. [Google Scholar]
- Bacilieri, R.; Lacombe, T.; Cunff, L.L.; Di Vecchi-Staraz, M.; Laucou, V.; Genna, B.; Perós, J.P.; This, P.; Boursiquot, J.M. Genetic structure in cultivated grapevines is linked to geography and human selection. BMC Plant Biol. 2013, 13, 25. [Google Scholar] [CrossRef] [PubMed]
- Fort, F.; Lin-Yang, Q.; Valls, C.; Sancho-Galán, P.; Canals, J.M.; Zamora, F. Characterisation and Identification of Vines from Fuerteventura (Canary Volcanic Archipelago (Spain)) Using Simple Sequence Repeat Markers. Horticulturae 2023, 9, 1301. [Google Scholar] [CrossRef]
- Marsal, G.; Mateo, J.M.; Canals, J.M.; Zamora, F.; Fort, F. SSR analysis of 338 accessions planted in Penedes (Spain) reveals 28 unreported molecular profiles of Vitis vinifera L. Am. J. Enol. Vitic. 2016, 67, 466–470. [Google Scholar] [CrossRef]
- Marsal, G.; Bota, J.; Martorell, A.; Canals, J.M.; Zamora, F.; Fort, F. Local cultivars of Vitis vinifera L. in Spanish Islands: Balearic Archipelago. Sci. Hortic. 2017, 226, 122–132. [Google Scholar] [CrossRef]
- Marsal, G.; Mendez, J.J.; Mateo-Sanz, J.M.; Ferrer, S.; Canals, J.M.; Zamora, F.; Fort, F. Molecular characterization of Vitis vinifera L. local cultivars from volcanic areas (the Canary Islands and Madeira) using SSR markers. Oeno One 2019, 4, 667–680. [Google Scholar] [CrossRef]
- Maul, E.; Röckel, F. Vitis International Variety Catalogue. 2015. Available online: http://www.vivc.de (accessed on 7 July 2023).
- Zerolo, J.; Cabello, F.; Espino, A.; Borrego, J.; Ibañez, J.; Rodriguez-Torres, I.; Muñoz-Organero, G.; Rubio, C.; Hernández, M. Variedades de Vid de Cultivo Tradicional en Canarias, 1st ed.; Instituto Canario de Calidad Agroalimentaria, Gobierno de Canarias: Santa Cruz de Tenerife, Spain, 2006. [Google Scholar]
- Macías, A.M. Colonización y viticultura. El caso de las Canarias, 1350–1550. In Douro: Estudos y Documentos; Revistas da Facultade de Letras da Universidade do Porto: Porto, Portugal, 2002; Volume 13, Available online: https://ojs.letras.up.pt/index.php/dou/article/view/12586 (accessed on 7 July 2023).
- Macías, A. El paisaje vitícola de Canarias. Cinco siglos de historia. Ería 2005, 68, 351–364. [Google Scholar] [CrossRef]
- Aliquo, G.; Torres, R.; Lacombe, T.; Boursiquot, J.M.; Laucou, V.; Gualpa, J.; Fanzone, M.; Sari, S.; Pérez-Peña, J.; Prieto, J.A. Identity and parentage of some South American grapevine cultivars present in Argentina. Aust. J. Grape Wine Res. 2017, 23, 452–460. [Google Scholar] [CrossRef]
- Moita, A.; Santos, R.; Catarina, A. Unraveling the origin of Vitis vinifera L. Verdelho. Aust. J. Grape Wine Res. 2018, 24, 450–460. [Google Scholar] [CrossRef]
- Avramidou, E.V.; Masaoutis, I.; Pitsoli, T.D.; Kapazoglou, A.; Pikraki, M.; Trantas, E.A.; Nikolantonakis, M.; Doulis, A.G. Analysis of Wine-Producing Vitis vinifera L. Biotypes, Autochthonous to Crete (Greece), Employing Ampelographic and Microsatellite Markers. Life 2023, 13, 220. [Google Scholar] [CrossRef]
- Costantini, L.; Monaco, A.; Vouillamoz, J.F.; Forlani, M.; Grando, M.S. Genetic relationships among local Vitis vinifera cultivars from Campania (Italy). Vitis 2005, 44, 25–34. [Google Scholar] [CrossRef]
- Ibañez, J.; De Andrés, M.T.; Molino, A.; Borrego, J. Genetic study of key Spanish grapevine varieties using microsatellite. analysis. Am. J. Enol. Vitic. 2003, 54, 22–30. [Google Scholar] [CrossRef]
- Vélez, M.D.; Ibáñez, J. Evaluation of the uniformity and stability of Microsatellite markers in grapevine. Acta Hortic. 2009, 827, 163–168. [Google Scholar] [CrossRef]
- Cabezas, A.; Ibañez, J.; Lijavetzky, D.; Vélez, D.; Bravo, G.; Rodríguez, V.; Carreño, I.; Jermakow, A.M.; Carreño, J.; Ruiz-García, L.; et al. A 48 SNP set for grapevine cultivar identification. BMC Plant Biol. 2011, 11, 153. [Google Scholar] [CrossRef]
- Riaz, S.; Garisson, K.E.; Dangl, G.S.; Boursiquot, J.M.; Meredith, C.P. Genetic divergence and chimerism within ancient asexually propagated winegrape cultivars. J. Am. Soc. Hortic. Sci. 2002, 127, 508–514. [Google Scholar] [CrossRef]
- Pelsy, F. Molecular and cellular mechanisms of diversity within grapevine varieties. Heredity 2010, 104, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Grigoriou, A.; Tsaniklidis, G.; Hagidimitriou, M.; Nikoloudakis, N. The Cypriot indigenous grapevine germplasm is a multi-clonal varietal mixture. Plants 2020, 9, 1034. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Garcia, R.; Ruiz-García, L.; Bolling, L.; Ocete, R.; López, M.A.; Arnold, C.; Ergul, A.; Söylemezoğlu, G.; Uzun, H.I.; Cabello, F.; et al. Multiple origins of cultivated grapevine (Vitis vinifera L. ssp. sativa) based on chloroplast DNA polymorphisms. Mol. Ecol. 2006, 15, 3707–3714. [Google Scholar] [CrossRef] [PubMed]
- Casanova, J.; Mozas, P.; Ortiz, J.M. Ampelography and microsatellite DNA analysis of autochthonous and endangered grapevine cultivars in the province of Huesca (Spain). Span. J. Agric. Res. 2011, 9, 790–800. [Google Scholar] [CrossRef]
- Emanuelli, F.; Lorenzi, S.; Grzeskowiak, L.; Catalano, V.; Stefanini, M.; Troggio, M.; Myles, S.; Martinez-Zapater, J.M.; Zyprian, E.; Moreira, F.M.; et al. Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol. 2013, 13, 39. [Google Scholar] [CrossRef]
- Dong, Y.; Duan, S.; Xia, Q.; Liang, Z.; Dong, X.; Margaryan, K.; Musayev, M.; Goryslavets, S.; Zdunić, G.; Bert, P.-F.; et al. Dual domestications and origin of traits in grapevine evolution. Science 2023, 379, 892–901. [Google Scholar] [CrossRef]
- García-Muñoz, S.; Lacombe, T.; De Andrés, M.T.; Gaforio, L.; Muñoz-Organero, G.; Laucou, V.; This, P.; Cabello, F. Grape varieties (Vitis vinifera L.) from the Balearic Islands: Genetic characterization and relationship with Iberian Peninsula and Mediterranean Basin. Genet. Resour. Crop Evol. 2012, 59, 589–605. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fort, F.; Lin-Yang, Q.; Valls, C.; Sancho-Galán, P.; Canals, J.M.; Zamora, F. Analysis of the Diversity Presented by Vitis vinifera L. in the Volcanic Island of La Gomera (Canary Archipelago, Spain) Using Simple Sequence Repeats (SSRs) as Molecular Markers. Horticulturae 2024, 10, 14. https://doi.org/10.3390/horticulturae10010014
Fort F, Lin-Yang Q, Valls C, Sancho-Galán P, Canals JM, Zamora F. Analysis of the Diversity Presented by Vitis vinifera L. in the Volcanic Island of La Gomera (Canary Archipelago, Spain) Using Simple Sequence Repeats (SSRs) as Molecular Markers. Horticulturae. 2024; 10(1):14. https://doi.org/10.3390/horticulturae10010014
Chicago/Turabian StyleFort, Francesca, Qiying Lin-Yang, Carla Valls, Pau Sancho-Galán, Joan Miquel Canals, and Fernando Zamora. 2024. "Analysis of the Diversity Presented by Vitis vinifera L. in the Volcanic Island of La Gomera (Canary Archipelago, Spain) Using Simple Sequence Repeats (SSRs) as Molecular Markers" Horticulturae 10, no. 1: 14. https://doi.org/10.3390/horticulturae10010014
APA StyleFort, F., Lin-Yang, Q., Valls, C., Sancho-Galán, P., Canals, J. M., & Zamora, F. (2024). Analysis of the Diversity Presented by Vitis vinifera L. in the Volcanic Island of La Gomera (Canary Archipelago, Spain) Using Simple Sequence Repeats (SSRs) as Molecular Markers. Horticulturae, 10(1), 14. https://doi.org/10.3390/horticulturae10010014







