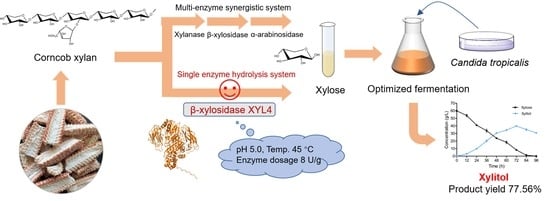
Discovery of a Novel β-xylosidase with Xylanase Activity and Its Application in the Production of Xylitol from Corncob Xylan
Abstract
1. Introduction
2. Results and Discussion
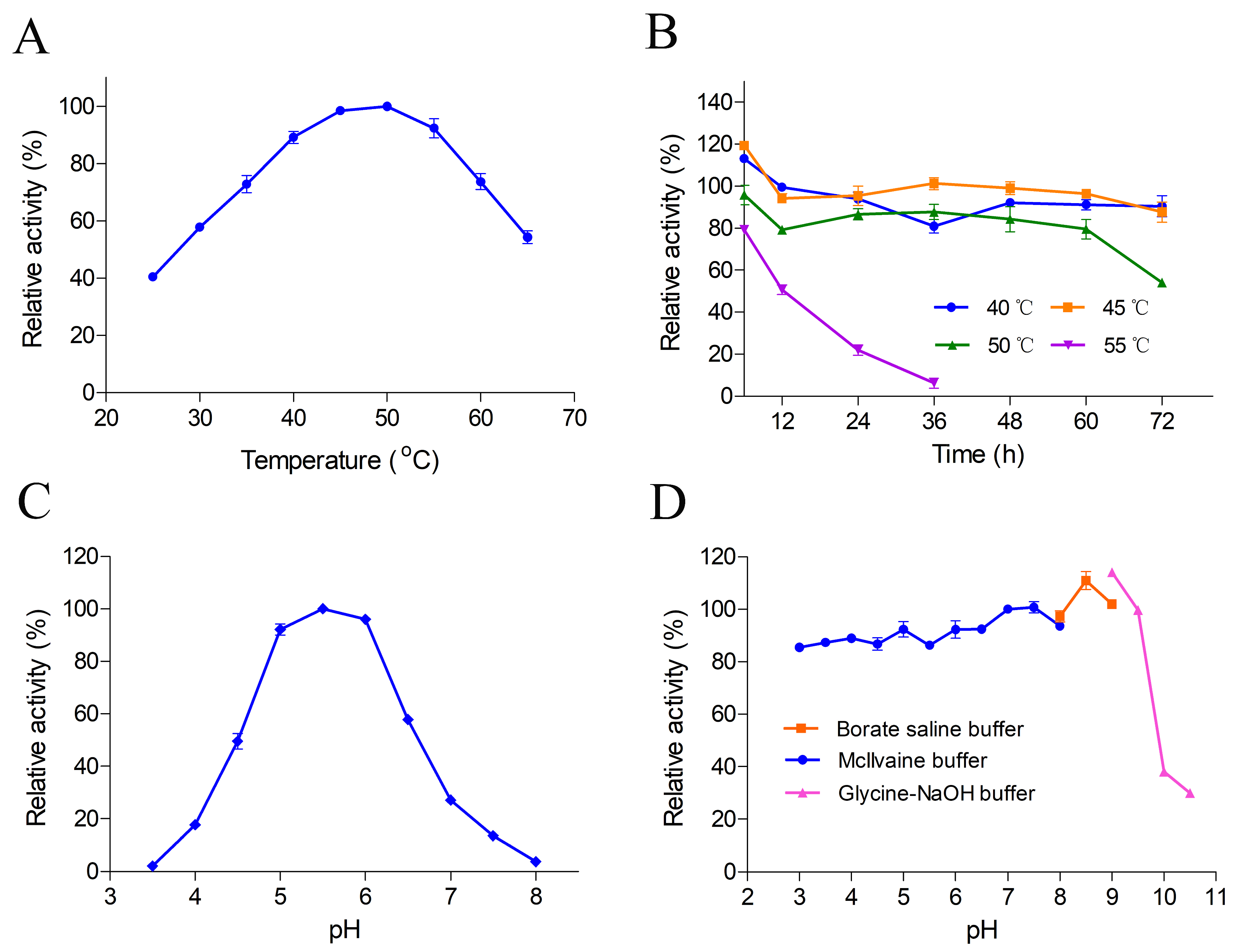
2.1. Enzymatic Properties of Recombinant Protein
2.2. Xylan Substrate Specificity
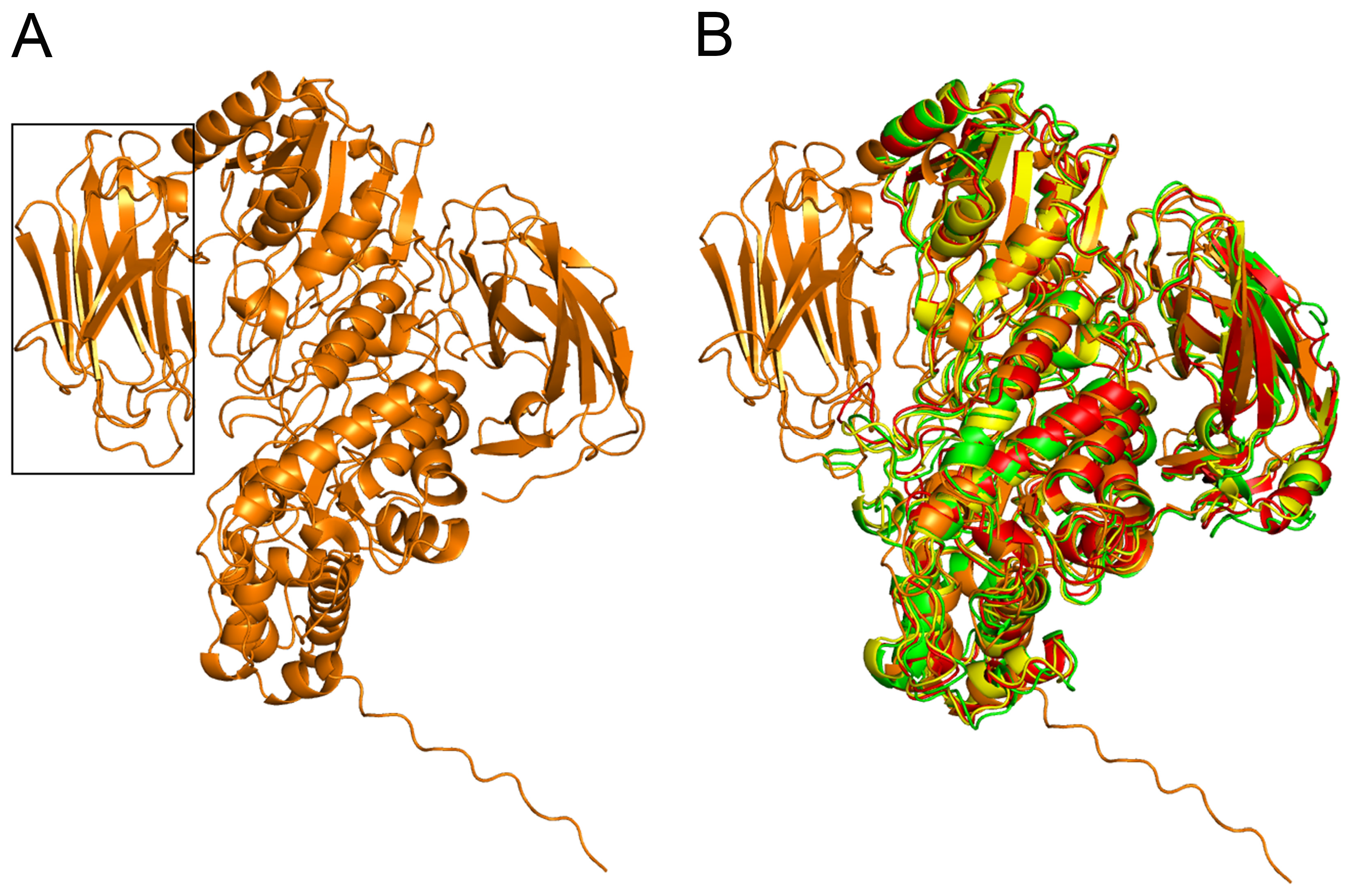
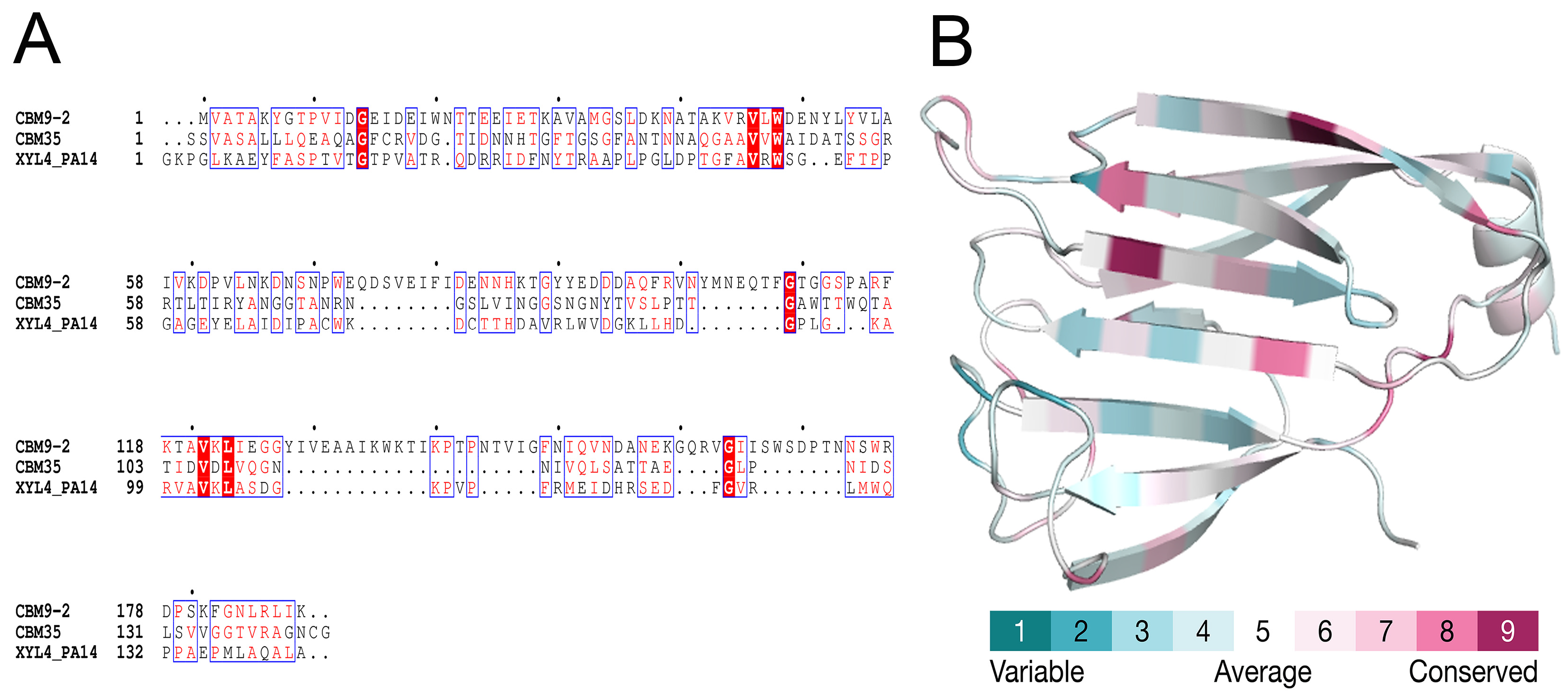
2.3. Comparative Modeling and Structural Analysis
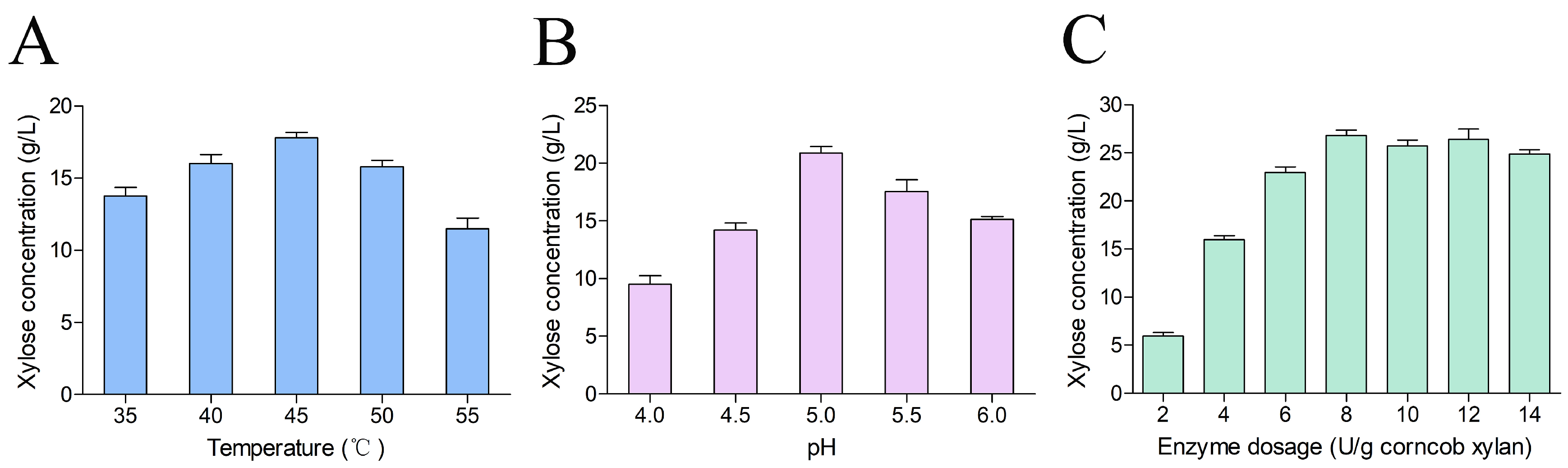
2.4. Optimization of Hydrolysis Conditions for Corncob Xylan
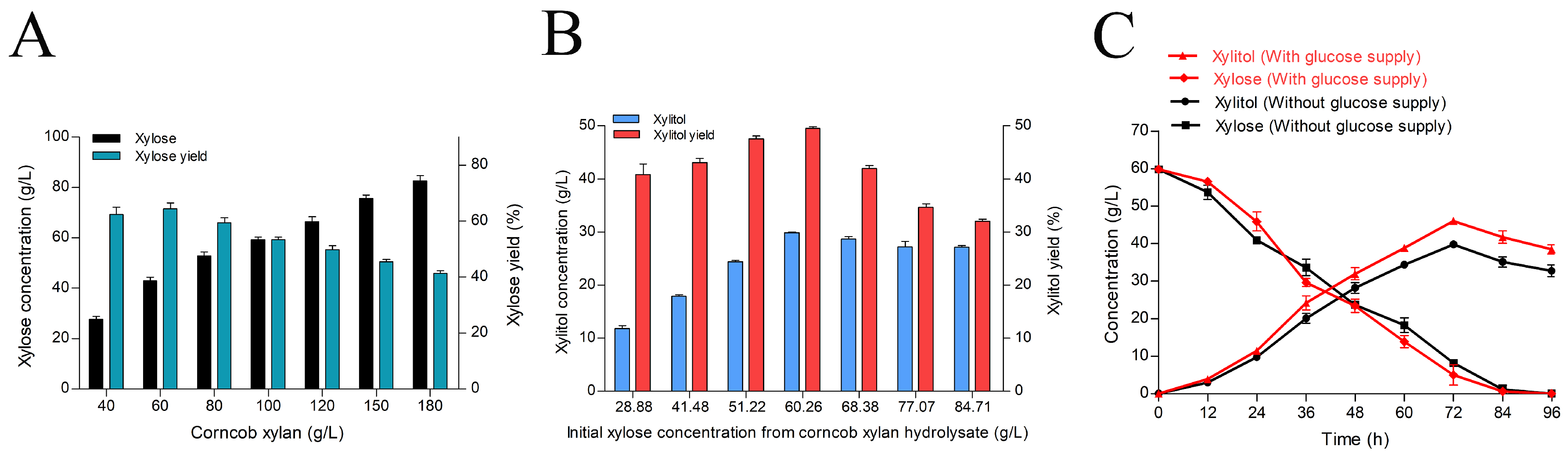
2.5. Production of Xylitol from Enzymatic Corncob Xylan Hydrolysate
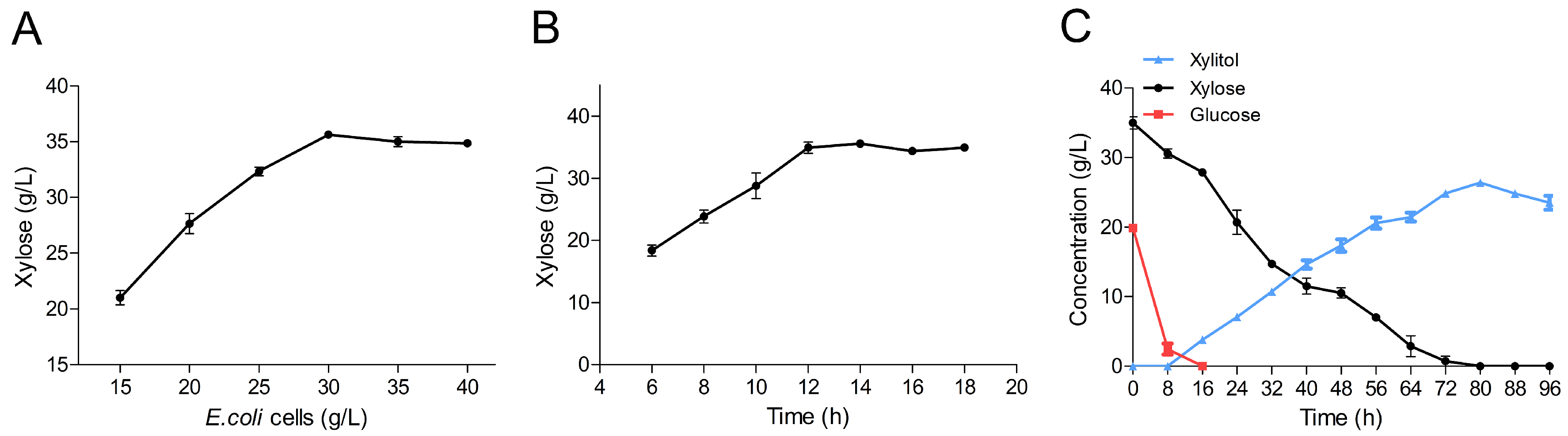
2.6. Production of Xylitol via Whole-Cell Catalysis
3. Materials and Methods
3.1. Strains, Plasmids and Chemicals
3.2. Production of Recombinant Protein
3.3. Enzyme Activity Assays and Biochemical Properties
3.4. Substrate Specificity Assays
3.5. Bioinformatic Analysis
3.6. Optimization of Hydrolysis Conditions for Corncob Xylan
3.7. Production of Xylitol from Enzymatic Corncob Xylan Hydrolysate
3.8. Production of Xylitol by Whole-Cell Catalysis
3.9. Analytical Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Victor, D.G.; Leape, J. Global climate agreement: After the talks. Nature 2015, 527, 439–441. [Google Scholar] [CrossRef]
- Biely, P.; Singh, S.; Puchart, V. Towards enzymatic breakdown of complex plant xylan structures: State of the art. Biotechnol. Adv. 2016, 34, 1260–1274. [Google Scholar] [CrossRef]
- Yuan, Q.; Zhang, H.; Qian, Z.; Yang, X. Pilot-plant production of xylo-oligosaccharides from corncob by steaming, enzymatic hydrolysis and nanofiltration. J. Chem. Technol. Biot. 2004, 79, 1073–1079. [Google Scholar] [CrossRef]
- Martins, M.P.; Ventorim, R.Z.; Coura, R.R.; Maitan-Alfenas, G.P.; Alfenas, R.F.; Guimarães, V. The β-xylosidase from Ceratocystis fimbriata RM35 improves the saccharification of sugarcane bagasse. Biocatal. Agric. Biotechnol. 2018, 13, 291–298. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, Z.; Liu, Y.; You, X.; Yang, S.; Yan, Q. Biochemical characterization of a novel exo-oligoxylanase from Paenibacillus barengoltzii suitable for monosaccharification from corncobs. Biotechnol. Biofuels 2019, 12, 190. [Google Scholar] [CrossRef]
- Li, Z.; Guo, X.; Feng, X.; Li, C. An environment friendly and efficient process for xylitol bioconversion from enzymatic corncob hydrolysate by adapted Candida tropicalis. Chem. Eng. J. 2015, 263, 249–256. [Google Scholar] [CrossRef]
- Cintra, L.C.; Fernandes, A.G.; de Oliveira, I.C.M.; Siqueira, S.J.L.; Costa, I.G.O.; Colussi, F.; Jesuíno, R.S.A.; Ulhoa, C.J.; de Faria, F. Characterization of a recombinant xylose tolerant β-xylosidase from Humicola grisea var. thermoidea and its use in sugarcane bagasse hydrolysis. Int. J. Biol. Macromol. 2017, 105, 262–271. [Google Scholar] [CrossRef]
- Qing, Q.; Wyman, C. Supplementation with xylanase and β-xylosidase to reduce xylo-oligomer and xylan inhibition of enzymatic hydrolysis of cellulose and pretreated corn stover. Biotechnol. Biofuels 2020, 4, 18. [Google Scholar] [CrossRef]
- Kim, S.K.; Russell, J.; Cha, M.; Himmel, M.E.; Bomble, Y.J.; Westpheling, J. Co-expression of a β-D-xylosidase from Thermotoga maritima and a family 10 xylanase from Acidothermus cellulolyticus significantly improves the xylan degrading activity of the Caldicellulosiruptor bescii exoproteome. Appl. Environ. Microb. 2021, 87, e0052421. [Google Scholar] [CrossRef]
- Terrasan, C.R.F.; Trobo-Maseda, L.; Moreno-Pérez, S.; Carmona, E.C.; Pessela, B.C.; Guisan, J. Co-immobilization and stabilization of xylanase, β-xylosidase and α-L-arabinofuranosidase from Penicillium janczewskii for arabinoxylan hydrolysis. Process Biochem. 2016, 51, 614–623. [Google Scholar] [CrossRef]
- Kambourova, M.; Mandeva, R.; Fiume, I.; Maurelli, L.; Morana, A. Hydrolysis of xylan at high temperature by co-action of the xylanase from Anoxybacillus flavithermus BC and the beta-xylosidase/alpha-arabinosidase from Sulfolobus solfataricus Oalpha. J. Appl. Microbiol. 2010, 102, 1586–1593. [Google Scholar] [CrossRef]
- Zafar, A.; Hamid, A.; Peng, L.; Wang, Y.; Aftab, M.N. Enzymatic hydrolysis of lignocellulosic biomass using a novel, thermotolerant recombinant xylosidase enzyme from Clostridium clariflavum: A potential addition for biofuel industry. RSC Adv. 2022, 12, 14917–14931. [Google Scholar] [CrossRef]
- Kwak, S.; Jo, J.H.; Yun, E.J.; Jin, Y.S.; Seo, J.H. Production of biofuels and chemicals from xylose using native and engineered yeast strains. Biotechnol. Adv. 2019, 37, 271–283. [Google Scholar] [CrossRef]
- Zappaterra, F.; Tupini, C.; Summa, D.; Cristofori, V.; Costa, S.; Trapella, C.; Lampronti, I.; Tamburini, E. Xylitol as a hydrophilization moiety for a biocatalytically synthesized ibuprofen prodrug. Int. J. Mol. Sci. 2022, 23, 2026. [Google Scholar] [CrossRef]
- Hyvönen, L.; Koivistoinen, P.; Voirol, F. Food technological evaluation of xylitol. Adv. Food Nutr. Res. 1982, 28, 373–403. [Google Scholar]
- Salli, K.; Lehtinen, M.J.; Tiihonen, K.; Ouwehand, A.C. Xylitol’s health benefits beyond dental health: A comprehensive review. Nutrients 2019, 11, 1813. [Google Scholar] [CrossRef]
- Martins, M.L.; Leite, K.F.; Magno, M.B.; Masterson, D.; Vicente-Gomila, J.M.; Cavalcanti, Y.W.; Maia, L.C.; Fonseca-Gonçalves, A. The xylitol applicability and its effects in health area aorldwide: A bibliometric analysis based on randomized controlled trials. Pesqui. Bras. Em Odontopediatria E Clínica Integr. 2022, 22, e210046. [Google Scholar] [CrossRef]
- Lomba, L.; Garralaga, M.P.; Werner, Á.; Giner, B.; Baptista, P.M.; Sánchez-Romero, N. Ibuprofen solubility and cytotoxic study of deep eutectic solvents formed by xylitol, choline chloride and water. J. Drug Deliv. Sci. Technol. 2023, 82, 104327. [Google Scholar] [CrossRef]
- de Albuquerque, T.L.; da Silva, I.J.; de Macedo, G.R.; Rocha, M. Biotechnological production of xylitol from lignocellulosic wastes: A review. Process Biochem. 2014, 49, 1779–1789. [Google Scholar] [CrossRef]
- Nyyssölä, A.; Pihlajaniemi, A.; Palva, A.; Weymarn, N.; Leisola, M. Production of xylitol from D-xylose by recombinant Lactococcus lactis. J. Biotechnol. 2005, 118, 55–66. [Google Scholar] [CrossRef]
- Xu, B.; Dai, L.; Zhang, W.; Yang, Y.; Wu, Q.; Li, J.; Tang, X.; Zhou, J.; Ding, J.; Han, N. Characterization of a novel salt-, xylose-and alkali-tolerant GH43 bifunctional β-xylosidase/α-L-arabinofuranosidase from the gut bacterial genome. J. Biosci. Bioeng. 2019, 128, 429–437. [Google Scholar] [CrossRef]
- Kumar, S.; Chemie, D. Purification and regulation of the synthesis of a β-xylosidase from Aspergillus nidulans. FEMS Microbiol. Lett. 2010, 235, 287–293. [Google Scholar] [CrossRef]
- Yin, Y.R.; Xian, W.; Han, M.; Zhou, E.; Liu, L.; Xiao, M.; Li, W. Expression and characterisation of a pH and salt tolerant, thermostable and xylose tolerant recombinant GH43 β-xylosidase from Thermobifida halotolerans YIM 90462T for promoting hemicellulose degradation. Antonie Van Leeuwenhoek Int. J. Gen. 2019, 112, 339–350. [Google Scholar] [CrossRef]
- Zanphorlin, L.M.; de Morais, M.A.B.; Diogo, J.A.; Domingues, M.N.; de Souza, F.H.M.; Ruller, R.; Murakami, M. Structure-guided design combined with evolutionary diversity led to the discovery of the xylose-releasing exo-xylanase activity in the glycoside hydrolase family 43. Biotechnol. Bioeng. 2019, 116, 734–744. [Google Scholar] [CrossRef]
- Boyce, A.; Walsh, G. Purification and characterisation of a thermostable β-xylosidase from Aspergillus niger van Tieghem of potential application in lignocellulosic bioethanol production. Appl. Biochem. Biotechnol. 2018, 186, 712–730. [Google Scholar] [CrossRef]
- Ndata, K.; Nevondo, W.; Cekuse, B.; van Zyl, L.J.; Trindade, M. Characterization of a highly xylose tolerant β-xylosidase isolated from high temperature horse manure compost. BMC Biotechnol. 2021, 21, 61. [Google Scholar] [CrossRef]
- Huy, N.D.; Thayumanavan, P.; Kwon, T.H.; Park, S.M. Characterization of a recombinant bifunctional xylosidase/arabinofuranosidase from Phanerochaete chrysosporium. J. Biosci. Bioeng. 2013, 116, 152–159. [Google Scholar] [CrossRef]
- Chysirichote, T.; Phaiboonsilpa, N.; Laosiripojana, N. High production of cellulase and xylanase in solid-state fermentation by Trichoderma reesei using spent copra and wheat bran in rotary bioreactor. Ind. Eng. Chem. Res. 2023, 62, 3087–3097. [Google Scholar] [CrossRef]
- de Carvalho, D.R.; Carli, S.; Meleiro, L.P.; Rosa, J.C.; de Oliveira, A.H.C.; Jorge, J.A.; Furriel, R. A halotolerant bifunctional β-xylosidase/α-L-arabinofuranosidase from Colletotrichum graminicola: Purification and biochemical characterization. Int. J. Biol. Macromol. 2018, 114, 741–750. [Google Scholar] [CrossRef]
- Patel, H.; Kumar, A.K.; Shah, A. Purification and characterization of novel bi-functional GH3 family β-xylosidase/β-glucosidase from Aspergillus niger ADH-11. Int. J. Biol. Macromol. 2018, 109, 1260–1269. [Google Scholar] [CrossRef]
- Kojima, K.; Sunagawa, N.; Mikkelsen, N.E.; Hansson, H.; Karkehabadi, S.; Samejima, M.; Sandgren, M.; Igarashi, K. Comparison of glycoside hydrolase family 3 β-xylosidases from basidiomycetes and ascomycetes reveals evolutionarily distinct xylan degradation systems. J. Biol. Chem. 2022, 298, 101670. [Google Scholar] [CrossRef] [PubMed]
- Schröder, S.P.; Boer, C.; Mcgregor, N.G.; Rowland, R.J.; Moroz, O.; Blagova, E.; Reijngoud, J.; Arentshorst, M.; Osborn, D.; Morant, M. Dynamic and functional profiling of xylan-degrading enzymes in Aspergillus Secretomes using activity-based probes. ACS Cent. Sci. 2019, 5, 1067–1078. [Google Scholar] [CrossRef] [PubMed]
- Rigden, D.J.; Mello, L.V.; Galperin, M. The PA14 domain, a conserved all-β domain in bacterial toxins, enzymes, adhesins and signaling molecules. Trends Biochem. Sci. 2004, 29, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S. The pfam protein families database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef]
- Notenboom, V.; Boraston, A.B.; Kilburn, D.G.; Rose, D. Crystal structures of the family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A in native and ligand-bound forms. Biochemistry 2001, 40, 6248–6256. [Google Scholar] [CrossRef] [PubMed]
- Montanier, C.; Bueren, A.L.; Dumon, C.; Flint, J.E.; Correia, M.A.; Prates, J.A.; Firbank, S.J.; Lewis, R.J.; Grondin, G.G.; Ghinet, M. Evidence that family 35 carbohydrate binding modules display conserved specificity but divergent function. Biochemistry 2009, 106, 3065–3070. [Google Scholar] [CrossRef]
- Gong, X.; Li, Q.; Li, T.; Li, C.; Huang, J.; Zhou, N.; Jia, X. Chemical composition and monolignin in alkali and acid treated corncob affect sugar release. Ind. Crop. Prod. 2022, 176, 114317. [Google Scholar] [CrossRef]
- Chen, H.; Liu, L. Unpolluted fractionation of wheat straw by steam explosion and ethanol extraction. Bioresour. Technol. 2007, 98, 666–676. [Google Scholar]
- Kataria, R.; Ruhal, R.; Babu, R.; Ghosh, S. Saccharification of alkali treated biomass of kans grass contributes higher sugar in contrast to acid treated biomass. Chem. Eng. J. 2013, 230, 36–47. [Google Scholar] [CrossRef]
- Li, Z.; Chen, S.; Wang, Y.; Liu, Z.; Zhang, T. Biochemical characterization of a novel halo/organic-solvents/final-products tolerant GH39 xylosidase from saline soil and its synergic action with xylanase. Int. J. Biol. Macromol. 2020, 164, 184–192. [Google Scholar] [CrossRef]
- Xu, L.; Liu, L.; Li, S.; Zheng, W.; Cui, Y.; Liu, R.; Sun, W. Xylitol production by Candida tropicalis 31949 from sugarcane bagasse hydrolysate. Sugar Tech 2019, 21, 341–347. [Google Scholar] [CrossRef]
- Prabhu, A.A.; Thomas, D.J.; Ledesma-Amaro, R.; Leeke, G.A.; Medina, A.; Verheecke-Vaessen, C.; Coulon, F.; Agrawal, D.; Kumar, V. Biovalorisation of crude glycerol and xylose into xylitol by oleaginous yeast Yarrowia lipolytica. Microb. Cell Fact. 2020, 19, 121. [Google Scholar] [CrossRef] [PubMed]
- Ko, B.S.; Kim, J. Production of xylitol from D-xylose by a xylitol dehydrogenase gene-disrupted mutant of Candida tropicalis. Appl. Environ. Microb. 2006, 72, 4207–4213. [Google Scholar] [CrossRef]
- Li, S.; Ye, Z.; Moreb, E.A.; Hennigan, J.N. Dynamic control over feedback regulatory mechanisms improves NADPH flux and xylitol biosynthesis in engineered E. coli. Metab. Eng. 2021, 64, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, A.A.; Bosakornranut, E.; Amraoui, Y.; Agrawal, D.; Kumar, V. Enhanced xylitol production using non-detoxified xylose rich pre-hydrolysate from sugarcane bagasse by newly isolated Pichia fermentans. Biotechnol. Biofuels 2020, 13, 209. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.K.; Wu, J.; Lin, Z.N.; Zhang, J.A. Aerobic and sequential anaerobic fermentation to produce xylitol and ethanol using non-detoxified acid pretreated corncob. Biotechnol. Biofuels 2014, 7, 166. [Google Scholar] [CrossRef] [PubMed]
- Misra, S.; Raghuwanshi, S.; Saxena, R. Evaluation of corncob hemicellulosic hydrolysate for xylitol production by adapted strain of Candida tropicalis. Carbohyd. Polym. 2013, 92, 1596–1601. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Sandhu, P.P.; Ahluwalia, V.; Mishra, B.B.; Yadav, S. Improved upstream processing for detoxification and recovery of xylitol produced from corncob. Bioresour. Technol. 2019, 291, 121931. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, R.; Li, Z.; Dai, D.; Li, C.; Zhou, X. A novel pathway construction in Candida tropicalis for direct xylitol conversion from corncob xylan. Bioresour. Technol. 2013, 128, 547–552. [Google Scholar] [CrossRef]
- Li, Z.; Qu, H.; Li, C.; Zhou, X. Direct and efficient xylitol production from xylan by Saccharomyces cerevisiae through transcriptional level and fermentation processing optimizations. Bioresour. Technol. 2013, 149, 413–419. [Google Scholar] [CrossRef]
- Rivas, B.; Domínguez, J.; Domínguez, H.; Enzyme, J.P.J.; Technology, M. Bioconversion of posthydrolysed autohydrolysis liquors: An alternative for xylitol production from corn cobs. Enzyme Microb. Technol. 2002, 31, 431–438. [Google Scholar] [CrossRef]
- Miller, G. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef]
| Enzyme | Organism | Family | Specific Activity (U/mg) | Ref. | |||||
|---|---|---|---|---|---|---|---|---|---|
| pNPX | Bagasse Xylan | Oat Spelt Xylan | Birchwood Xylan | Beechwood Xylan | Corncob Xylan | ||||
| XYL4 | Sphingomona elodea | GH3 | 2 | 0.73 | 0.97 | 1.99 | 2.85 | 8.16 | This study |
| rPcXyl | Phanerochaete chrysosporium | GH43 | 1797 | — | — | <0.03 | <0.1 | — | [27] |
| Bxcg | Colletotrichum graminicola | GH3 | 284 | — | — | 0.63 | 0.60 | — | [29] |
| Xylosidase | Aspergillus niger | GH3 | 61 | — | — | 5.4 | — | — | [30] |
| Microorganisms | Feedstock | Pretreatment | Xylitol Production | Ref. | |
|---|---|---|---|---|---|
| Concentration (g/L) | Xylitol Yield (%) | ||||
| Yarrowia lipolytica | Xylose and glycerol | — | 53.2 | 97 | [42] |
| Candida tropicalis BSXDH-3 | Xylose, glycerol and glucose | — | 48.6 | 98 | [43] |
| Escherichia coli | Xylose | — | 200 | — | [44] |
| Pichia fermentans | Xylose | — | 98.9 | 67 | [45] |
| Candida tropicalis W103 | Corncob | 0.5% sulfuric acid and 1.5% phosphoric acid | 17.1 | 32 | [46] |
| Candida tropicalis MTCC 6192 | Corncob | 1% sulfuric acid | 21.98 | 37 | [47] |
| Candida tropicalis | Corncob | 0.5% nitric acid | 56.5 | 62 | [48] |
| Candida tropicalis 31949 | Sugarcane bagasse | Ultrasonic-Assisted NaOH and xylanase | 62.98 | 62.98 | [41] |
| Candida tropicalis | Corncob | Xylanase and β-xylosidase | 8.18 | 45.37 | [6] |
| Candida tropicalis IT-Xol-1 | Xylan | Xylanase AtN and β-xylosidase AtL | 3.78 | 77.1 | [49] |
| Saccharomyces cerevisiae | Xylan | Xylanase and β-xylosidase | 1.94 | 71 | [50] |
| Candida tropicalis | Corncob xylan | XYL4 | 46.74 | 77.56 | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, M.; Lin, Y.; Sun, L.; Pang, H.; Wei, H.; Huang, R.; Wei, Y.; Du, L. Discovery of a Novel β-xylosidase with Xylanase Activity and Its Application in the Production of Xylitol from Corncob Xylan. Fermentation 2023, 9, 606. https://doi.org/10.3390/fermentation9070606
Liang M, Lin Y, Sun L, Pang H, Wei H, Huang R, Wei Y, Du L. Discovery of a Novel β-xylosidase with Xylanase Activity and Its Application in the Production of Xylitol from Corncob Xylan. Fermentation. 2023; 9(7):606. https://doi.org/10.3390/fermentation9070606
Chicago/Turabian StyleLiang, Meng, Yu Lin, Lixin Sun, Hao Pang, Hang Wei, Ribo Huang, Yutuo Wei, and Liqin Du. 2023. "Discovery of a Novel β-xylosidase with Xylanase Activity and Its Application in the Production of Xylitol from Corncob Xylan" Fermentation 9, no. 7: 606. https://doi.org/10.3390/fermentation9070606
APA StyleLiang, M., Lin, Y., Sun, L., Pang, H., Wei, H., Huang, R., Wei, Y., & Du, L. (2023). Discovery of a Novel β-xylosidase with Xylanase Activity and Its Application in the Production of Xylitol from Corncob Xylan. Fermentation, 9(7), 606. https://doi.org/10.3390/fermentation9070606