Abstract
The redox balance of inorganic sulfur in heavily polluted rivers might be disrupted, making sulfur reduction a major metabolic pathway of sulfate-reducing bacteria (SRB), leading to a massive accumulation of S2− and blackening the water bodies. A mixed culture microbial consortium (MCMC) of Citrobacter sp.sp1, Ochrobactrum sp.sp2, and Stenotrophomonas sp.sp3 was used to activate native sulfate-oxidizing bacteria (SOB) to augment the S2− oxidizing process. The results demonstrated that MCMC had a significant sulfur oxidation effect, with 98% S2− removal efficiency within 50 h. The sulfide species varied greatly and were all finally oxidized to SO42−. The mechanism of bio-augmentation was revealed through high throughput sequencing analysis. The MCMC could stimulate and simplify the community structure to cope with the sulfide change. The microorganisms (family level) including Enterococcaceae, Flavobacteriaceae, Comamonadaceae, Methylophilaceae, Caulobacteraceae, Rhodobacteraceae, and Burkholderiaceae were thought to be associated with sulfide metabolism through the significant microbial abundance difference in the bio-treatment group and control group. Further Pearson correlation analysis inferred the functions of different microorganisms: Comamonadaceae, Burkholderiaceae, Alcaligenaceae, Methylophilaceae, and Caulobacteraceae played important roles in S2− oxidization and SO42− accumulation; and Comamonadaceae, Burkholderiaceae, Alcaligenaceae, Methylophilaceae, Caulobacteraceae, Campylobacteraceae, Bacteriovoracaceae, and Rhodobacteraceae promoted the sulfur oxidation during the whole process.
1. Introduction
In recent decades, much S2− has been released into the rivers with the intensification of industry and anthropogenic activity, resulting in the accumulation of inorganic sulfur in natural water, especially in heavily polluted rivers [1]. Studies have found that the accumulation of S2− is one of the main causes of river black odor in China [2]. The black-odor phenomenon in water bodies not only leads to the destruction of the original ecological function but also seriously affects the regular life of the surrounding residents [3]. The blackened river is mainly formed by the combination of S2− and metal ions in the water bodies, which affects the survival and reproduction of plants and animals in the water body and leads to abnormal microbial activity [2,3]. This phenomenon has become one of the prominent water environment problems in China. Song et al. found that the S2− concentrations ranged from 3.84 to 17.93 mg/L along Dongsha River in Beijing, which was a heavily polluted river [4]. Sheng et al. also found the mean concentration of S2− could reach 3.15 mg/L in the Dihe River in China [5]. The standard of sulfide from the Ministry of Environment and Protection of the People’s Republic of China GB 3838-2002 (MEP) divided the water qualities into five levels, according to the environmental functions and protection objectives of water bodiesand the maximum concentration standard (level V) for sulfide is ≤1.0 mg/L [6]. The high concentration of S2− in a heavily polluted river largely contributes to the generation and accumulation of hydrogen sulfide and insoluble metal sulfides due to the combination of S2− and hydrogen or heavy metal ions in water bodies [7]. Hydrogen sulfide, a biologically important molecule with complex physiological functions, can cause potential inhalation hazards as well as an inhibitory effect on cellular respiration [8]. While metal sulfides such as CuS, MnS, and FeS are often suspended in the river, and further aggravate the river blackening [2]. S2− in a river comes from two pathways: one is exogenous input, and the other is endogenous release [4]. Exogenous input mainly refers to the discharge of S2−-containing wastewater from industry, agriculture, vehicular traffic, and other sources [9]. Endogenous release includes three major aspects: (1) S2− liberation directly from river sediment due to the scouring effect [10]; (2) the biodegradation of sulfur-containing high molecular substances such as proteins under the action of native microorganisms [11]; and (3) the bio-reduction of oxidized sulfides as a result of the sulfate-reducing bacteria (SRB) effect [12] (Sorokin et al. 2014).
Inorganic sulfur bio-cycling is an important part of material circulation in nature. In general, S2− is bio-oxidized to high-valence sulfur comprising S0, S2O32− as well as SO32−, and all of them are finally bio-oxidized to SO42− [13,14]. Conversely, SO42− can be bio-reduced to SO32−, S2O32−, and S0, and these low-valence sulfurs are further bio-reduced to S2−. Sulfate-oxidizing bacteria (SOB) are responsible for the bio-oxidation of sulfur, whereas SRB is in charge of the bio-reduction of sulfur [12]. These two paths, as important bio-geochemical processes in nature, maintain the balance of S2− redox and enable its concentration to remain at a low level in native rivers [15,16]. Some studies showed that in a heavily polluted river, dissolved oxygen was often depleted, and the ability of water bodies’ re-oxygenation was greatly weakened, which led to the metabolic block in the bio-oxidation pathway of sulfur [17]. On the other hand, the activities of SRB are significantly enhanced in an oxygen-free environment, which accelerates the bio-reduction process of sulfur, resulting in the large accumulation of S2− and the further exacerbation of river quality [2]. Inoculating special strains, especially mixed culture, has been proven to be a good method to remove pollutants due to the microbial diversity or abundant enzyme system [12,18,19]. Moreover, SOB obtained from the black-odor river could better adapt to the conditions of the original black-odor river, have better survivability in treating the river, and have higher processing efficiency than the SOB obtained from other sources. Therefore, using SOB with mixed culture screened from a heavily polluted river may be an effective measure to eliminate S2− pollution. SOB is a large topic that has attracted attention around the world. To date, many pure SOB have been isolated mainly from sludge [19,20], and these SOB are widely applied in bioleaching and chemical wastewater treatment [21,22]. Gevertz et al. isolated SOB from an oil field in Canada to reduce/cycle the sulfide and elemental sulfur [23]; Brock et al. isolated Sulfolobus as a potential geochemical agent in high-temperature hydrothermal systems [24]; Anandham et al. isolated SOB from rhizosphere soils and discussed the possible thiosulfate oxidation pathways of the SOB and the phylogenetic distribution of the sulfur oxidation gene (soxB) [25]. In comparison, less work has been carried out with the SOB under the condition of mixed culture isolated from a river; the mechanism of S2− removal involved is also less reported.
In this study, three strains with better sulfur oxidation effects were isolated from the Dongsha River, a heavily polluted river located in Beijing, China. These strains, including Citrobacter sp.sp1, Ochrobactrum sp.sp2, and Stenotrophomonas sp.sp3, were used to remove S2− with mixed culture in our previous work [4,26]. Although the consortium in previous work showed the potential to use S2− as electron donors, the mechanism involved in the S2− oxidation process was still unclear. Therefore, the purpose of this article was to investigate the bio-augmentation process of S2− oxidizing and increase the understanding of the involved bio-augmentation mechanism. In order to achieve this purpose, we added a mixed culture microbial consortium (MCMC) into a bio-treatment group and set up a control group (without MCMC) to analyze the effect of the MCMC on the structure of the original microbial community using diversity analysis; infer the sulfate metabolism bacterium (SMB) that might be activated by the MCMC through difference analysis of the bio-treatment group and the control group; explore the mechanism by high throughput sequencing analysis and the correlation analysis between sulfide and SMB; and infer the key role of these microorganisms in each of the sulfur oxidation process. These results will help provide fundamental information for the treatment of heavily polluted rivers in the future and understand the bio-augmentation effect on S2− oxidation.
2. Materials and Methods
2.1. Water Sample
The water sample was collected from the Dongsha River. The characteristics of the water sample were analyzed, and the concentrations of S2−, chemical oxygen demand (COD), ammonia (NH3-H), and total phosphorus (TP) were 20.7 ± 1.2 mg/L, 104.5 ± 8.0 mg/L, 5.3 ± 0.4 mg/L, and 3.0 ± 0.1 mg/L, respectively. The water samples were collected in September, the temperature at the time was 25 °C; the water body of the Dongsha River was stagnant, not flowing, with some algae on the surface and no fish in it.
2.2. The Strains
According to our previous study [4,26], Citrobacter sp. sp1, Ochrobactrum sp. sp2, and Stenotrophomonas sp. sp3 were all isolated from the Dongsha River of Beijing, which was heavily polluted [4], and each of them had been proved to oxidize S2− efficiently. The sequences of these three strains were all uploaded to The National Center for Biotechnology Information (NCBI), and their GenBank accession numbers were MH181794, MH181795, and MH181796, respectively. They had also been deposited in China General Microbiological Culture Collection Center (CGMCC), and the strain numbers were CGMCC No. 24337, 24338, and 18394, respectively.
2.3. The MCMC
The MCMC consisted of Citrobacter sp. sp1, Ochrobactrum sp. sp2, and Stenotrophomonas sp. sp3 in a proportion of 1:1:1, and the initial cell density of each strain was 8.96 × 105 cells/mL (Colony-Forming Units, Cfu). The MCMC was cultured using a liquid medium containing 20 g/L of glucose, 10 g/L of peptone, and 10 g/L of yeast extract with a pH of 7. All experiments were carried out in 250 mL conical flasks containing 150 mL sterilized liquid medium, and the cultivations were conducted on a rotary shaker with 120 rpm at 25 °C. After 48 h, the MCMC was harvested for the following sulfur oxidation experiments.
2.4. Experiment of Sulfur Oxidation
The experiment was divided into two groups. One was the bio-treatment group that was inoculated with 0.1 g/L of the MCMC, while the other was the control group without the MCMC. The sulfur oxidation process was investigated using the water sample from the Dongsha River in a conical flask with 800 mL of working volume. All of the experiments were run on a rotary shaker at 25 °C and 120 rpm. Regularly, samples were taken out for analysis. All experiments were operated in triplicate.
2.4.1. DNA Extraction and PCR Amplification
Microbial DNA was extracted from the samples using the E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, USA) according to the manufacturer’s protocols. The V3-V4 region of the prokaryote 16S ribosomal RNA gene was amplified by the polymerase chain reaction (PCR). The procedure of PCR was as follows: 95 °C for 3 min, followed by 27 cycles at 95 °C for 30 s, 55 °C for 30 s, 72 °C for 45 s, and a final extension at 72 °C for 10 min. The primers for prokaryote were 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′), respectively. The barcode was an eight-base sequence unique to each sample. PCR reactions were performed in triplicate in a 20 μL mixture containing 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, and 10 ng of template DNA.
2.4.2. Illumina MiSeq Sequencing
Amplicons were extracted using 2% agarose gels and purified with an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to the manufacturer’s instructions. The purified amplicons were then quantified using a QuantiFluor™-ST Kit (Promega, Madison, WI, USA). The Purified amplicons were pooled in equimolar and paired-end sequenced (2 × 250) on an Illumina MiSeq platform according to the standard protocols. The raw reads were deposited into Sequence Read Archive (SRA) database (Accession Number: PRJNA394809) of the NCBI.
2.4.3. Processing of Sequencing Data
Raw fastq files were de-multiplexed and quality-filtered using QIIME (version 1.17) with the following conditions: (1) the 300 bp reads were truncated at any site receiving an average quality score <20 over a 50 bp sliding window, discarding the truncated reads that were shorter than 50 bp; (2) exact barcode matching, 2 nucleotide mismatch in primer matching, reads containing ambiguous characters were removed; (3) only sequences that overlap longer than 10 bp were assembled according to their overlap sequence. Reads that could not be assembled were discarded.
Operational Units (OTUs) were clustered with a 97% similarity cutoff using UPARSE (version 7.1 http://drive5.com/uparse/, accessed on 28 January 2017), and chimeric sequences were identified and removed using UCHIME. The taxonomy of each 16S rRNA gene sequence was analyzed using RDP Classifier (http://rdp.cme.msu.edu/, accessed on 28 January 2017) and then compared using the silva 16S rRNA gene reference database (SSU128). The confidence threshold was set at 70% [27].
2.5. Analytical Methods
The concentrations of S2−, S0, S2O32−, SO32−, and SO42− were separately analyzed according to Chinese water and wastewater monitoring and analysis methods, respectively [6]. A Uv-vis spectrophotometry (Uv-1800, Kyoto, Japan) was used to determine the concentrations of S2−, S0, and SO42−; while the concentration of SO32− was measured using an ion chromatograph (ISC-1100, Waltham, USA). Meanwhile, S2O32− was measured according to the titrimetric method.
The relationship between sulfur oxidization and SMB was analyzed using SPSS statistical software (version 17.0, New York, NY, USA). “*” and “**” meant that the correlation was significant at the 0.05 level and 0.01 level, respectively.
3. Results
3.1. Variation of Inorganic Sulfur as Result of MCMC
The concentrations of S2−, S0, S2O32−, SO32−, and SO42− were detected throughout the whole process, and the results are shown in Figure 1. As shown in Figure 1a, for the sample inoculated with the MCMC, the variations of different chemical forms of sulfur were significant during 50 h of the oxidization process. The concentration of S2− gradually decreased as the bio-treatment time was extended, and its final concentration declined to less than 1.0 mg/L. The main reason for the decreasing concentration of S2− was that S2− was oxidized to generate other chemical forms of sulfur such as S0 with the action of MCMC. This resulted in the rapid accumulation of S0 before the first 18 h and the peak concentration reached 0.4 mg/L. Similarly, S2O32− and SO32− appeared at a peak concentration at 18 h and 28 h, respectively. The maximal value of S2O32− arrived at 57.9 mg/L, while for SO32− it was 51.7 mg/L. In the subsequent oxidation process, the concentrations of S0, S2O32−, and SO32− decreased significantly. On the other hand, the concentration of SO42− was clearly raised throughout the process, especially at the initial 18 h, and the final concentration was up to 43.0 mg/L. As for the control without the MCMC, a part of sulfur chemical forms such as S2−, S0, and SO32− were determined (Figure 1b). The concentration of S2− fell initially, and then slightly raised during the later process, while other sulfur chemical forms kept relatively stable in the whole process.
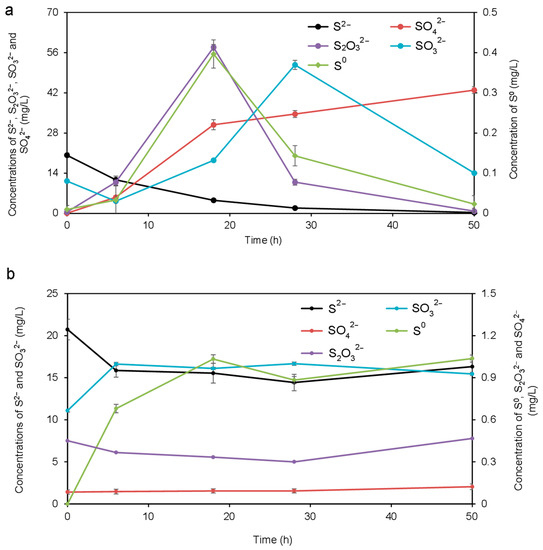
Figure 1.
Variation of inorganic sulfur during the whole oxidization with the (a) bio-treatment group and (b) the control group.
3.2. Variation of Community Structure as a Result of MCMC
Inoculation with special strains to augment bio-reaction occurs mainly through two processes: (1) the formation of stable dominant microorganisms from inoculation in the whole process; and (2) the variation of community structure due to the simulation of original microorganisms by inoculation [28,29]. A comparison of the microbial diversity between the bio-treatment group and the control group was evaluated, as shown in Table 1. In general, different samples showed a relatively similar trend in terms of richness. Furthermore, as indicated by Ace, Chao, Shannon, and Coverage indices implied that there were similar diversity dynamics of the microbial community in the two groups.

Table 1.
Comparison of microbial diversity between the bio-treatment group and the control group.
The difference of community in two groups with or without the cultured MCMC was revealed using PCoa as shown in Figure 2. With regard to principal component 1 (PC1), all samples obtained from the control groups clustered on the left side of the figure, while the samples from the bio-treatment groups covered both the left and right sides. For PC2, each prokaryote sample in the bio-treatment groups was more widely dispersed than other samples in the control groups.
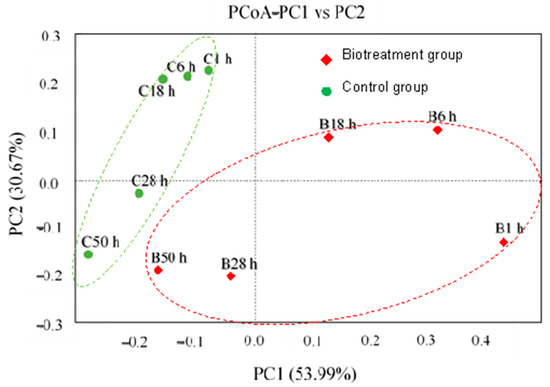
Figure 2.
PCoa analysis of two groups with or without the MCMC.
As shown in Figure 2, the addition of sulfur-oxidizing microorganisms resulted in a significant difference between the two groups. The first 18 h of the bio-reaction process in the bio-treatment group were significantly altered by the MCMC; but for the control group, there was no obvious difference. As the reaction progressed, the differences between the two groups gradually decreased, and at 50 h, the samples in the group with the MCMC (B50 h) tended to be similar to those in the control group (C50 h).
3.3. Analysis of Sulfides Metabolism Bacterium
The differences in microbial abundance between the bio-treatment group and the control group during the reaction were investigated to extrapolate the potential SMB involved in the inorganic sulfides’ bio-oxidation process (only species with mean sums in the top 10 showed). After the addition of the MCMC, Enterococcaceae only showed up in the bio-treatment group after the reaction started. Throughout the reaction process, the abundance of Enterococcaceae, Flavobacteriaceae, Comamonadaceae, Methylophilaceae, Caulobacteraceae, Rhodobacteraceae, and Burkholderiaceae in the bio-treatment group showed significant differences compared with the control group. Most microorganisms play a role in sulfur oxidation, but it cannot be excluded that some microorganisms are associated with sulfur reduction. Moreover, the abundance of Enterococcaceae, Comamonadaceae, Flavobacteriaceae, Caulobacteraceae, Rhodobacteraceae, Sphingomonadaceae, Burkholderiaceae, and Bacteriovoracaceae varied considerably over the reaction time. This suggested that these microorganisms play different roles in the different valence states of sulfates for sulfur oxidation.
After 6 h of the reaction (Figure 3a), in addition to Enterococcaceae and Flavobacteriaceae, the abundance of Caulobacteraceae also showed significant differences between the two groups, and at this time, only the abundance of Enterococcaceae was significantly higher in the bio-treatment group with the MCMC than in the control group. During the first 28 h, the abundance of Flavobacteriaceae, Sphingomonadaceae, and Cryomorphaceae in the control group was consistently higher than the corresponding abundance in the bio-treatment group. As the reaction continued, during the period of 18–50 h, the abundance of 14 microorganisms, including Caulobacteraceae, Rhodobacteraceae, and Sphingomonadaceae, was found to change obviously with the reaction process in both groups (Figure 3b–d). In addition, taxonomically, Citrobacter sp.sp1, Ochrobactrum sp.sp2, and Stenotrophomonas spsp3. were classified as Enterobacteriaceae, Brucellaceae, and Xanthomonadaceae, respectively (the sum of means outside the top 10 species).
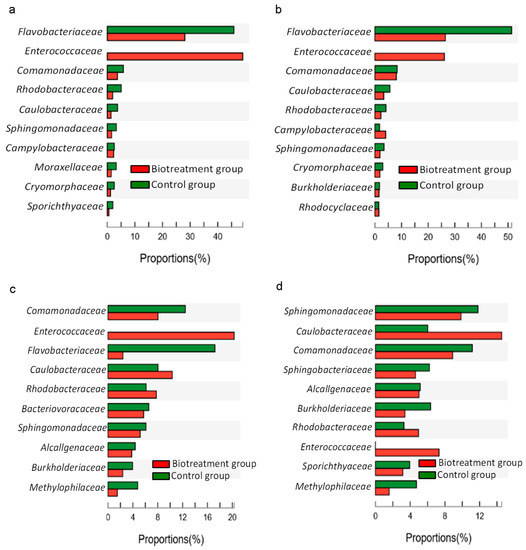
Figure 3.
Abundance difference between the bio-treatment group and the control group ((a–d) Prokaryote difference at 6 h, 18 h, 28 h, and 50 h, respectively).
Differences in S2−, S0, S2O32−, and SO42− concentrations were obvious in the two groups at 18 h, while SO32− concentration showed a great difference at 28 h (Figure 1). Therefore, the samples with different groups at 18 h and 28 h were compared, respectively, to identify the potential functional SMB. Eight prokaryotes at the family level, including Flavobacteriaceae, Enterococcaceae, Caulobacteraceae, Rhodobacteraceae, Campylobacteraceae, Sphingomonadaceae, Cryomorphaceae, and Burkholderiaceae, showed significant abundance differences (p < 0.05) at 18 h. Besides Flavobacteriaceae, Enterococcaceae, Caulobacteraceae, Rhodobacteraceae, Sphingomonadaceae, and Burkholderiaceae, four prokaryotes (family level), including Comamonadaceae, Bacteriovoracaceae, Alcaligenaceae, and Methylophilaceae, also showed significant differences in abundance (p < 0.05) at 28 h. In addition, there were other non-SMB bacteria (Moraxellaceae, Sporichthyaceae, and Sphingobacteriaceae) that differed significantly over time in the control and experimental groups (Figure 3). Table 2 summarizes the relative microorganisms associated with sulfur metabolism in the reported literature. Flavobacteriaceae, Enterococcaceae, Sphingomonadaceae and Cryomorphaceae are SRB; Bacteriovoracaceae, Rhodobacteraceae, Campylobacteraceae, Comamonadaceae, Alcaligenaceae, and Methylophilaceae are SOB; Caulobacteraceae and Burkholderiaceae belong to SRB and SOB.

Table 2.
SMB associated with sulfur metabolism from the literature.
3.4. Identification of SOB and Their Performance
The Pearson correlation analysis between the above series of sulfides and SMB was shown in Table 3. Nine prokaryotes showed significant correlations with some sulfide species in the present study, and a positive correlation (r = 0.900, p < 0.05) was found between Enterococcaceae and S2−. Meanwhile, Enterococcaceae showed a strong negative correlation with both S0 (r = −0.969, p < 0.01) and SO42− (r = −0.977, p < 0.01). Comamonadaceae, Burkholderiaceae, Alcaligenaceae, Methylophilaceae, and Caulobacteraceae showed significant correlations with both S2− and SO42−, respectively; their correlation indices indicated that the S2− oxidation and SO42− accumulation mainly depended on these microorganisms. Furthermore, Comamonadaceae, Alcaligenaceae, Methylophilaceae, and Caulobacteraceae also showed strong positive correlations to S0, which could be inferred that these four prokaryotes played important roles in the production of S0 as well. In addition, there was a notable correlation between Campylobacteraceae and S2O32− (r = 0.809, p < 0.05), demonstrating that it was attributed to S2O32− generation via S4 Intermediate (S4I) pathway [13]. While Bacteriovoracaceae and Rhodobacteraceae showed a significant correlation with SO32−, suggesting that Bacteriovoracaceae and Rhodobacteraceae were associated with SO32− accumulation.

Table 3.
Pearson correlations between series of sulfides and SMB (family level).
Compared with other microorganisms, Enterococcaceae has the highest positive correlation with S2−. The relationship between these SMBs was also observed in Table 3. A competitive relationship was found between Enterococcaceae and five SOB, including Comamonadaceae, Burkholderiaceae, Alcaligenaceae, Methylophilaceae, and Caulobacteraceae, based on significantly negative correlations. Moreover, the correlation indices of these four SOBs showed a significant relationship between Burkholderiaceae, Alcaligenaceae, Methylophilaceae, and Caulobacteraceae, while Comamonadaceae only had a notable relationship with Methylophilaceae. Meanwhile, there was a significant relationship between Bacteriovoracaceae and Rhodobacteraceae. The bio-oxidation of S2− is the result of the combined action of the SMBs, but the contribution of different microorganisms varies significantly. As discussed above, it was prudent to conclude that Comamonadaceae, Burkholderiaceae, Alcaligenaceae, Methylophilaceae, Caulobacteraceae, Campylobacteraceae, Bacteriovoracaceae, and Rhodobacteraceae were the main SOB involved in this inorganic sulfur oxidation processes (Figure 4a).
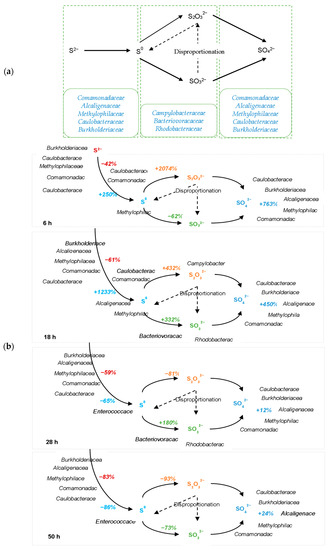
Figure 4.
Inorganic sulfur-oxidizing process (a) inorganic sulfur-oxidizing pathway and involving SOB; (b) performance of SOB and sulfates’ oxidizing process during each period.
The detailed S2− oxidation process and the participation of the SOB could be summarized in Figure 4b. Figure 4b is based on the microbial diversity and the microorganisms’ differences along with the abundance difference and the sulfide changes through Pearson correlation analysis to establish correlations between the microorganisms and the sulfur oxidation process and to speculate on possible pathways and possible key microorganisms for each pathway. The percentages marked in this figure represent the percentage ratio of the changes in inorganic sulfides’ concentration at this instant compared to the previous instant, and the microorganisms next to inorganic sulfides indicate that they might play a major role in that inorganic sulfide change. Throughout the experiment, the S2− was oxidized to other inorganic sulfates mainly through the effect of Comamonadaceae, Burkholderiaceae, Alcaligenaceae, Methylophilaceae, and Caulobacteraceae, resulting in a continuous decrease in S2− concentration in the system. In the first 18 h, The concentration of S0 accumulated rapidly, which was mainly due to the action of Comamonadaceae, Alcaligenaceae, Methylophilaceae, and Caulobacteraceae. In particular, the average generation rate of S0 reached 100% per hour during 6–18 h, which was significantly higher than other inorganic sulfur concentration changes. However, after 18 h, S0 was oxidized to other inorganic sulfides by Campylobacteraceae, Bacteriovoracaceae, and Rhodobacteraceae. At the same time, the concentration of S2O32− accumulated rapidly under the effect of Campylobacteraceae; but the consumption of S2O32− in the last 28 h was higher than the consumption of other inorganic sulfides, which was mainly due to the action of Burkholderiaceae, Methylophilaceae, and Bacteriovoracaceae. The production of SO32− occurs mainly during 6–28 h, which was associated with Bacteriovoracaceae and Rhodobacteraceae, and the generation of SO32− was the dominant sulfides oxidization process during 18–28 h. In contrast to the change in S2− concentration, the concentration of SO42− kept increasing throughout the experiment, this was promoted by Comamonadaceae, Burkholderiaceae, Alcaligenaceae, Methylophilaceae, and Caulobacteraceae; the generation of SO42− was the main sulfide oxidization process in the first 6 h.
4. Discussion
The results of variation of inorganic sulfur indicated that unstable lower-valence sulfur ion species including S2−, S0, S2O32−, and SO32− were consumed to generate stable higher-valence SO42−, and sulfur-oxidizing became the main metabolic path of inorganic sulfur bio-conversion. Moreover, both sulfur oxidation and sulfur reduction processes were present simultaneously in the bio-treatment group and the control group, which led to different forms of sulfur ions being detected during the reaction. In the control group, the concentration of SO32− increased in the first 8 h and the concentration of S0 increased in the first 20 h, both of them remained stable in the subsequent reaction stages; the concentration of S2− first decreased then remained stable after 30 h; the change in the concentration of SO42− and S2O32− was negligible during the reaction. This indicated that although the sulfur oxidation process was produced in the control group at the early stage of the reaction, it was not obvious compared with the bio-treatment group. It could be concluded that the inorganic sulfur-oxidizing process was efficiently augmented by MCMC.
From Table 1, the Ace indices in both groups showed an increase followed by a decrease. However, from 28 h, the Ace indices in the group with the MCMC were higher than the control group, which might be due to the activation of some indigenous microorganisms by sulfur-oxidizing composite microorganisms so that these microorganisms could use the substrate and proliferate rapidly. The Shannon indices in both groups showed similar trends, except at 18 h, indicating that the biodiversity was similar most of the time, but at 18 h the bio-treatment group was higher than that in the control group. The difference at 18 h was probably due to the gradual enrichment of a few inorganic SOB in the system, and thus the removal of S2− might be achieved mainly by the action of these microorganisms [35,36]. The Coverage index reached above 0.997 in both groups, indicating good sequence detection in the samples. In addition, the PCoa in Figure 2 demonstrated that the bacterial communities were most affected by the mixed culture stimulation, with 53.99% of PC1 at the phylum level. That is, the MCMC did not become the dominant microorganism, it just simulated the structure change of the original microorganisms to enhance the sulfur oxidation process. From an ecological point of view, higher microbial community diversity can be considered an augmentation of functional redundancy [37]. Microbial communities with greater evenness are likely to be more functional, which is a key factor in maintaining the functional stability of ecosystems [37]. However, in this study, the new microbial community was shaped after inoculating the MCMC, some of the abundance of the original SRBs was reduced and some of the original SOBs were activated. Furthermore, the bio-augmentation group was generally less diverse and homogeneous than the control group, which seemed to activate the key for inorganic sulfur oxidation and enabled the microbes to cope with the substrate change with the Gibbs free energy changes during the reaction [38,39]. This feature facilitated the optimization of bacterial communities for inorganic sulfur oxidation [40].
From the analysis of sulfide metabolism bacteria, the differences in microbial abundance between the two groups could imply that the MCMC in the bio-treatment group activated the potential sulfur-oxidizing microorganisms. The analysis of sulfide metabolism bacteria revealed that Enterobacteriaceae, Brucellaceae, and Xanthomonadaceae also showed significant differences in abundance between the two groups as the response progressed (mean sums outside the top 10 species). The eight microorganisms (Flavobacteriaceae, Enterococcaceae, Caulobacteraceae, Rhodobacteraceae, Campylobacteraceae, Sphingomonadaceae, Cryomorphaceae, and Burkholderiaceae) have demonstrated the ability to metabolize inorganic sulfur (Table 2). For instance, Flavobacteriaceae, Enterococcaceae, Sphingomonadaceae, Cryomorphaceae, and Caulobacteraceae were often thought to be associated with SRB because of the presence of thioredoxin reductase, sulfur reduction protein DsrE or sulfite reductase [30,32], and they could use SO42− as the substrate as well. Furthermore, Bacteriovoracaceae, Rhodobacteraceae, Campylobacteraceae, Burkholderiaceae, Comamonadaceae, Alcaligenaceae, and Methylophilaceae usually represented SOB in other studies [19,33,41]. The reduced sulfides such as S2−, S0, and S2O32− could be used as electron donors. Interestingly, sometimes Burkholderiaceae and Caulobacteraceae could also play opposite roles, depending on the circumstance they are located in [34,42]. Moreover, from the literature, the non-SMB bacteria with a significant abundance change: Moraxellaceae and Sphingobacteriaceae are aerobic bacteria and Sporichthyaceae is facultative anaerobes bacteria, these microorganisms might play a supporting role in the sulfur oxidation process [43,44,45]. It could be inferred that the abundance differences between these non-SMB in the bio-treatment group and the control group were mainly due to the reproduction of these microorganisms being inhibited in the bio-treatment group, where more oxygen was consumed as the sulfur oxidation process was accelerated by the MCMC.
Moreover, the results of the identification and the performance of SOB suggest that the addition of mixed culture could accelerate the inorganic sulfur oxidization reactions by stimulating SMB. The roles played by these SMB remain unclear. The Pearson correlation analysis in Table 3 indicates that the abundance of Enterococcaceae tends to decrease with the removal of S2− and the accumulation of SO42−, suggesting that Enterococcaceae could either grow or obtain energy by reducing SO42− as an electron acceptor [12,16]. Sulfur redox is a dynamic and reversible process, where S2− is eventually oxidized to SO42− by SOB, while SO42− is also reduced to S2− by SRB during the reaction. The production of S2− during the inorganic sulfur oxidation process is mainly derived from the disproportionation of S2O32− [19]. From the Pearson analysis above, Enterococcaceae has the highest positive correlation with S2−. Therefore, it is hypothesized that Enterococcaceae may play an important role in regulating the S2O32−disproportionation reaction, contributing to the production of S2−. In addition, some literature studies indicated that both Comamonadaceae and Alcaligenaceae could participate in the oxidation of S2− with increasing SO42− concentration [8,19,32]. However, a further study has shown that Alcaligenaceae was also able to oxidize S2O32− through aerobic and anaerobic [19]. Campylobacteraceae was proved that could utilize S0 as an electron acceptor and form polysulfide [46]. It should be noted that Enterococcaceae, Burkholderiaceae, and Caulobacteraceae acted as SOB during the process of this work. These results of the action of Enterococcaceae, Burkholderiaceae, and Caulobacteraceae were different from those described by Llorens-Marès et al. and NCBI (Table 3) [34]. This might be due to the fact that this experiment was operated under aeration, whereas other results obtained in the literature under natural conditions might create an anaerobic or anoxic environment [2,12,34]. The dissolved oxygen (DO) in water samples in this study might be higher than DO in the environment under natural conditions. The application of sufficient oxygen might facilitate the oxidative conversion of S2− to SO42−, and the activation of the associated inorganic sulfur oxidases. Some limitations of this study were that some of the pathways of sulfide oxidation regulated by SOB were not clear. For instance, S2O32− could be generated by S0 oxidation and/or SO32− disproportionation [12,47]. It should be noted that this work only demonstrated that Campylobacteraceae was associated with the production of S2O32−, but there was little evidence as to which pathway was primarily or actually regulated by the Campylobacteraceae.
5. Conclusions
Considerable amounts of S2− are emitted from many industrial and agricultural activities. The slow inorganic sulfur-oxidizing process under natural conditions enables S2− to accumulate massively and induces natural water bodies pollution, even making water blacken. The common occurrence of water pollution has become a serious problem around the world. Biological desulfurization is a highly effective, economically viable alternative method, which has indeed been a hot topic in modern environmental science research. However, to date, there is little information about the fundamental research work of mixed culture for S2− removal in a heavily polluted river.
This study reported that mixed culture with Citrobacter sp.sp1, Ochrobactrum sp.sp2, and Stenotrophomonas sp.sp3 had nearly 98% of S2− removal efficiency within 50 h. The mechanism involved in bio-augmentation could be summarized as follows. The native microbial community dynamics were stimulated and simplified by inoculated mixed culture to cope with the sulfides change. Enterococcaceae, Comamonadaceae, Burkholderiaceae, Alcaligenaceae, Methylophilaceae, Caulobacteraceae, Campylobacteraceae, Bacteriovoracaceae, and Rhodobacteraceae performed as SOB for accelerating inorganic sulfur-oxidizing progress in this experiment. Comamonadaceae, Burkholderiaceae, Alcaligenaceae, Methylophilaceae, and Caulobacteraceae played important roles in S2− oxidization and SO42− accumulation across the whole process. Except for Burkholderiaceae, the other four SOB also made contributions to S0 generation in the first 18 h, but Enterococcaceae was in charge of the oxidation of S0 to other sulfides after 18 h. Campylobacteraceae took effect on S2O32− accumulation from 6 h to 18 h. Bacteriovoracaceae and Rhodobacteraceae took effect when SO32− increased from 6 h to 28 h. This study made a high-efficiency microbial consortium to treat the black-odor water bodies and found that the microbial consortium could enhance the sulfur oxidation process by changing the structure of the original microorganisms in the black-odor water body. This study also demonstrated that the mixed culture microbial consortium might have the potential to be used to remove S2− and clarify water in the actual black-odor water bodies and the wastewaters with high concentrations of S2−.
Author Contributions
Conceptualization, C.S. and X.L.; methodology, C.S. and X.L.; software, C.S. and Y.S.; validation, Y.S., H.G. and X.L.; formal analysis, C.S.; investigation, C.S.; resources, C.S.; data curation, C.S.; writing—original draft preparation, C.S.; writing—review and editing, Y.S.; visualization, Y.S.; supervision, X.L. and P.L.; project administration, X.L.; funding acquisition, X.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Foundation of Beijing, China, grant number 8182058 and the Central Level, Scientific Research Institutes for Basic R&D Special Fund Business, grant number 2019YSKY003.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Semrany, S.; Favier, L.; Djelal, H.; Taha, S.; Amrane, A. Bioaugmentation: Possible solution in the treatment of Bio-Refractory Organic Compounds (Bio-ROCs). Biochem. Eng. J. 2012, 69, 75–86. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Fang, Y.; Huang, R. Analysis on the formation condition of the algae-induced odorous black water agglomerate. Saudi J. Biol. Sci. 2014, 21, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tang, Y.; Kou, Z.; Teng, X.; Cai, W.; Hu, J. Shift of Sediments Bacterial Community in the Black-Odor Urban River during In Situ Remediation by Comprehensive Measures. Water 2019, 11, 2129. [Google Scholar] [CrossRef]
- Song, C.; Liu, X.; Song, Y.; Liu, R.; Gao, H.; Han, L.; Peng, J. Key blackening and stinking pollutants in Dongsha River of Beijing: Spatial distribution and source identification. J. Environ. Manag. 2017, 200, 335–346. [Google Scholar] [CrossRef]
- Sheng, Y.; Qu, Y.; Ding, C.; Sun, Q.; Mortimer, R.J.G. A combined application of different engineering and biological techniques to remediate a heavily polluted river. Ecol. Eng. 2013, 57, 1–7. [Google Scholar] [CrossRef]
- Ministry of Environment and Protection of the People’s Republic of China (MEP). Chinese Water and Wastewater Monitoring and Analysis Methods, 4th ed.; China Environmental Science Press: Beijing, China, 2002.
- Liu, C.; Shen, Q.; Zhou, Q.; Fan, C.; Shao, S. Precontrol of algae-induced black blooms through sediment dredging at appropriate depth in a typical eutrophic shallow lake. Ecol. Eng. 2015, 77, 139–145. [Google Scholar] [CrossRef]
- Xia, F.F.; Zhang, H.T.; Wei, X.M.; Su, Y.; He, R. Characterization of H2S removal and microbial community in landfill cover soils. Environ. Sci. Pollut. Res. 2015, 22, 18906–18917. [Google Scholar] [CrossRef]
- Pikaar, I.; Likosova, E.M.; Freguia, S.; Keller, J.; Rabaey, K.; Yuan, Z. Electrochemical abatement of hydrogen sulfide from waste streams. Crit. Rev. Environ. Sci. Technol. 2015, 45, 1555–1578. [Google Scholar] [CrossRef]
- He, D.F.; Chen, R.R.; Zhu, E.H.; Chen, N.; Yang, B.; Shi, H.H.; Huang, M.S. Toxicity bioassays for water from black-odor rivers in Wenzhou. China Environ. Sci. Pollut. Res. 2015, 22, 1731. [Google Scholar] [CrossRef]
- Lu, X.; Feng, Z.; Shang, J.; Fan, C.; Fan, J. Black water bloom induced by different types of organic matters and forming mechanisms of major odorous compounds. Environ. Sci. 2012, 33, 3152–3159. [Google Scholar] [PubMed]
- Sorokin, D.Y.; Berben, T.; Melton, E.D.; Overmars, L.; Vavourakis, C.D.; Muyzer, G. Microbial diversity and biogeochemical cycling in soda lakes. Extremophiles 2014, 18, 791. [Google Scholar] [CrossRef]
- Rohwerder, T.; Sand, W. Oxidation of inorganic sulfur compounds in Acidophilic Prokaryotes. Eng. Life. Sci. 2007, 7, 301–309. [Google Scholar] [CrossRef]
- Rzhepishevska, O.; Valdes, J.; Marcinkeviciene, L.; Gallardo, C.; Meskys, R.; Bonnefoy, V.; Holmes, D.; Dopson, M. Regulation of a novel Acidithiobacillus caldus gene cluster involved in metabolism of reduced inorganic sulfur compounds. Appl. Environ. Microbiol. 2007, 73, 7367. [Google Scholar] [CrossRef]
- Sakurai, H.; Ogawa, T.; Shiga, M.; Inoue, K. Inorganic sulfur oxidizing system in green sulfur bacteria. Photosyn. Res. 2010, 104, 163. [Google Scholar] [CrossRef]
- Tian, H.; Gao, P.; Chen, Z.; Li, Y.; Li, Y.; Wang, Y.; Zhou, J.; Li, G.; Ma, T. Compositions and abundances of sulfate-reducing and sulfur-oxidizing microorganisms in water-flooded petroleum reservoirs with different temperatures in China. Front. Microbiol. 2017, 8, 143. [Google Scholar] [CrossRef]
- Liu, J.; Jiang, T.; Huang, R.; Wang, D.; Zhang, J.; Qian, S.; Yin, D.; Chen, H. A simulation study of inorganic sulfur cycling in the water level fluctuation zone of the Three Gorges Reservoir, China and the implications for mercury methylation. Chemosphere 2017, 166, 31. [Google Scholar] [CrossRef]
- Kiani, M.H.; Ahmadi, A.; Zilouei, H. Biological removal of sulfur and ash from fine-grained high pyritic sulfur coals using a mixed culture of mesophilic microorganisms. Fuel 2014, 131, 89–95. [Google Scholar] [CrossRef]
- Luo, J.; Tian, G.; Lin, W. Enrichment, isolation and identification of sulfur-oxidizing bacteria from sulfide removing bioreactor. J. Environ. Sci. 2013, 25, 1393–1399. [Google Scholar] [CrossRef]
- Liu, Y.G.; Zhou, M.; Zeng, G.M.; Wang, X.; Li, X.; Fan, T.; Xu, W.H. Bioleaching of heavy metals from mine tailings by indigenous sulfur-oxidizing bacteria: Effects of substrate concentration. Bioresour. Technol. 2008, 99, 4124–4129. [Google Scholar] [CrossRef]
- Aguilar, J.R.P.; Cabriales, J.J.P.A.; Vega, M.A.M. Identification and characterization of sulfur-oxidizing bacteria in an artificial wetland that treats wastewater from a tannery. Int. J. Phytoremediation 2008, 10, 359–370. [Google Scholar] [CrossRef]
- Chen, P.; Yan, L.; Leng, F.; Nan, W.; Yue, X.; Zheng, Y.; Feng, N.; Li, H. Bioleaching of realgar by Acidithiobacillus ferrooxidans using ferrous iron and elemental sulfur as the sole and mixed energy sources. Bioresour. Technol. 2011, 102, 3260–3267. [Google Scholar] [CrossRef] [PubMed]
- Gevertz, D.; Telang, A.J.; Voordouw, G.; Jenneman, G.E. Isolation and characterization of strains CVO and FWKO B, two novel nitrate-reducing, sulfide-oxidizing bacteria isolated from oil field brine. Appl. Environ. Microb. 2000, 66, 6. [Google Scholar] [CrossRef] [PubMed]
- Brock, T.D.; Brock, K.M.; Belly, R.T.; Weiss, R.L. Sulfolobus: A new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 1972, 84, 54–68. [Google Scholar] [CrossRef] [PubMed]
- Anandham, R.; Indiragandhi, P.; Madhaiyan, M.; Ryu, K.Y.; Jee, H.J.; Sa, T.M. Chemolithoautotrophic oxidation of thiosulfate and phylogenetic distribution of sulfur oxidation gene (soxB) in rhizobacteria isolated from crop plants. Res. Microbiol. 2008, 159, 579–589. [Google Scholar] [CrossRef]
- Zhang, L.; Song, C.; Xu, Y.; Shi, Y.; Liu, X. Isolation, Characterization and S2--oxidation Metabolic Pathway of a Sulfur-oxidizing Strain from a Black-odor River in Beijing. Water Sci. Technol. Water Supply. 2022, 22, 4. [Google Scholar] [CrossRef]
- Amato, K.R.; Yeoman, C.J.; Kent, A.; Righini, N.; Carbonero, F.; Estrada, A.; Gaskins, H.R.; Stumpf, R.M.; Yildirim, S.; Torralba, M. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 2013, 7, 1344–1353. [Google Scholar] [CrossRef]
- Chen, J.; Zhan, P.; Koopman, B.; Fang, G.; Shi, Y. Bioaugmentation with Gordonia strain JW8 in treatment of pulp and paper wastewater. Clean Technol. Environ. Policy 2012, 14, 899–904. [Google Scholar] [CrossRef]
- Wang, J.H.; He, H.Z.; Wang, M.Z.; Wang, S.; Zhang, J.; Wei, W.; Xu, H.X.; Lv, Z.M.; Shen, D.S. Bioaugmentation of activated sludge with Acinetobacter sp. TW enhances nicotine degradation in a synthetic tobacco wastewater treatment system. Bioresour. Technol. 2013, 142, 445. [Google Scholar] [CrossRef]
- Delmont, T.; Eren, A.; Vineis, J.; Post, A. Genome reconstructions indicate the partitioning of ecological functions inside a phytoplankton bloom in the Amundsen Sea, Antarctica. Front Microbiol. 2015, 6, 1090. [Google Scholar] [CrossRef]
- Portela, C.A.F.; Smart, K.F.; Tumanov, S.; Cook, G.M.; Villasbôas, S. Global metabolic response of enterococcus faecalis to oxygen. J. Bacteriol. 2014, 196, 2012–2022. [Google Scholar] [CrossRef]
- Johnson, S.S.; Chevrette, M.G.; Ehlmann, B.L.; Benison, K.C. Insights from the Metagenome of an Acid Salt Lake: The Role of Biology in an Extreme Depositional Environment. PLoS ONE 2015, 10, e0122869. [Google Scholar] [CrossRef]
- Carbajal-Rodríguez, I.; Stoveken, N.; Satola, B. Aerobic degradation of mercaptosuccinate by the gram-negative bacterium Variovorax paradoxus strain B4. J. Bacteriol. 2011, 193, 527–539. [Google Scholar] [CrossRef]
- Llorens-Marès, T.; Yooseph, S.; Goll, J.; Hoffman, J.; Vila-Costa, M.; Borrego, C.M.; Dupont, C.L.; Casamayor, E.O. Connecting biodiversity and potential functional role in modern euxinic environments by microbial metagenomics. ISME J. 2015, 9, 1648. [Google Scholar] [CrossRef]
- Balcke, G.U.; Turunen, L.P.; Geyer, R.; Wenderoth, D.F.; Schlosser, D. Chlorobenzene biodegradation under consecutive aerobic–anaerobic conditions. FEMS Microbiol. Ecol. 2004, 49, 109–120. [Google Scholar] [CrossRef]
- Lin, C.W.; Lin, C.Y. MTBE biodegradation and degrader microbial community dynamics in MTBE, BTEX, and heavy metal-contaminated water. INT Biodeter. Biodegr. 2007, 59, 97–102. [Google Scholar] [CrossRef]
- Guo, X.; Wang, C.; Sun, F.; Zhu, W.; Wu, W. A comparison of microbial characteristics between the thermophilic and mesophilic anaerobic digesters exposed to elevated food waste loadings. Bioresour. Technol. 2014, 152, 420–428. [Google Scholar] [CrossRef]
- Hattori, S. Syntrophic acetate-oxidizing microbes in methanogenic environments. Microbes Environ. 2008, 23, 118. [Google Scholar] [CrossRef]
- Xin, B.; Zhang, D.; Zhang, X.; Xi, Y.; Wu, F.; Chen, S.; Li, L. Bioleaching mechanism of Co and Li from spent lithium-ion battery by the mixed culture of acidophilic sulfur-oxidizing and iron-oxidizing bacteria. Bioresour. Technol. 2009, 100, 6163–6169. [Google Scholar] [CrossRef]
- Zhao, Y.; Fang, Y.; Jin, Y.; Huang, J.; Bao, S.; Fu, T.; He, Z.; Wang, F.; Zhao, H. Potential of duckweed in the conversion of wastewater nutrients to valuable biomass: A pilot-scale comparison with water hyacinth. Bioresour. Technol. 2014, 163, 82–91. [Google Scholar] [CrossRef]
- Zhuang, R.; Lou, Y.; Qiu, X.; Zhao, Y.; Dong, Q.; Yan, X.; He, X.; Shen, Q.; Qian, L. Identification of a yeast strain able to oxidize and remove sulfide high efficiently. Appl. Microbiol. Biotechnol. 2017, 101, 391–400. [Google Scholar] [CrossRef]
- Pawar, S.S.; Niel, E.W.J. Evaluation of assimilatory sulfur metabolism in Caldicellulosiruptor saccharolyticus. Bioresour. Technol. 2014, 169, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Frank, J.F.; Hassan, A.N. Microorganisms associated with milk. In Encyclopedia of Dairy Sciences, 1st ed.; Roginski, H., Ed.; Werribee: Melbourne, Australia, 2002; pp. 1786–1796. [Google Scholar] [CrossRef]
- Brady, M.T.; Marcon, M.J. Less Commonly Encountered Nonenteric Gram-Negative Bacilli. In Principles and Practice of Pediatric Infectious Disease, 3rd ed.; Long, S.S., Ed.; W.B. Saunders: London, UK, 2008; Part III; pp. 828–831. [Google Scholar] [CrossRef]
- Tamura, T. The Family Sporichthyaceae. In The Prokaryotes, 4th ed.; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 883–888. [Google Scholar] [CrossRef]
- Roalkvam, I.; Drønen, K.; Stokke, R.; Daae, F.L.; Dahle, H.; Steen, I.H. Physiological and genomic characterization of Arcobacter anaerophilus IR-1 reveals new metabolic features in Epsilonproteobacteria. Front Microbiol. 2015, 6, 987. [Google Scholar] [CrossRef] [PubMed]
- Zopfi, J.; Ferdelman, T.G.; Fossing, H.; Amend, J.P.; Edwards, K.J.; Lyons, T.W. Distribution and fate of sulfur intermediates—Sulfite, tetrathionate, thiosulfate, and elemental sulfur—In marine sediments. In Sulfur Biogeochemistry-Past and Present; Amed, J.P., Edwards, K.J., Lyons, T.W., Eds.; Geological Society of America: Washington, DC, USA, 2004; Volume 379, pp. 97–116. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).