Aspergillus sclerotiorum Whole-Cell Biocatalysis: A Sustainable Approach to Produce 3-Hydroxy-phenazine 1-Carboxylic Acid from Phenazine 1-Carboxylic Acid
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Chemicals and Media
2.2. Isolation and Identification of Fungus Strain
2.3. Whole-Cell Biotransformation of PCA by Aspergillus sclerotiorum
2.4. Isolation and Extraction of the Target Compound
2.5. The Effect of Various Factors on the Whole-Cell Biotransformation of PCA
2.6. Time Course of Whole-Cell Biotransformation of PCA and the Production of 3-OH-PCA
2.7. The Reusability of Fungus Pellets for Fed-Batch Fermentation
2.8. Analytical Methods for PCA Biotransformation
2.9. Statistical Analysis
3. Results and Discussion
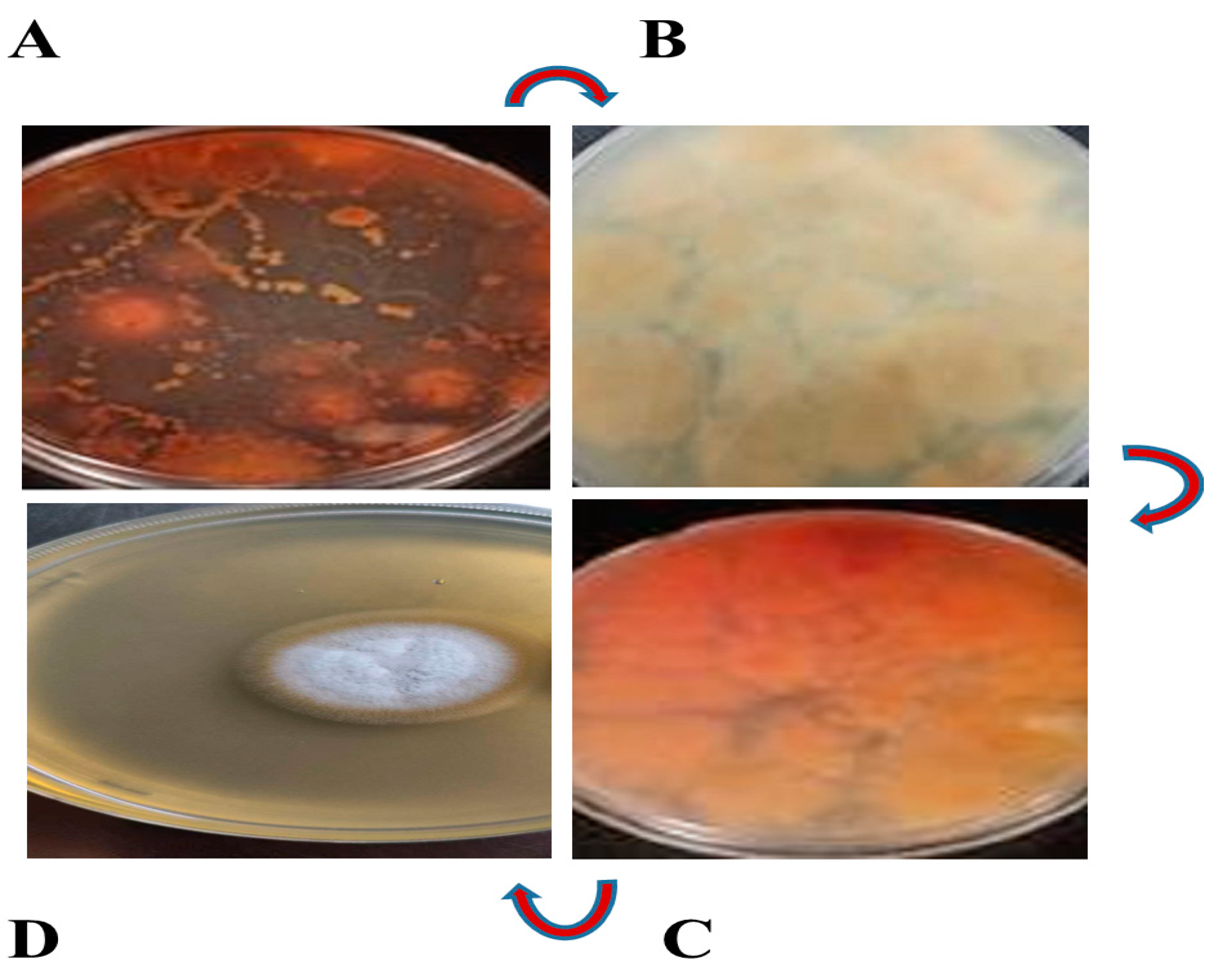
3.1. Fungus Isolate Is Responsible for Red Pigment Production
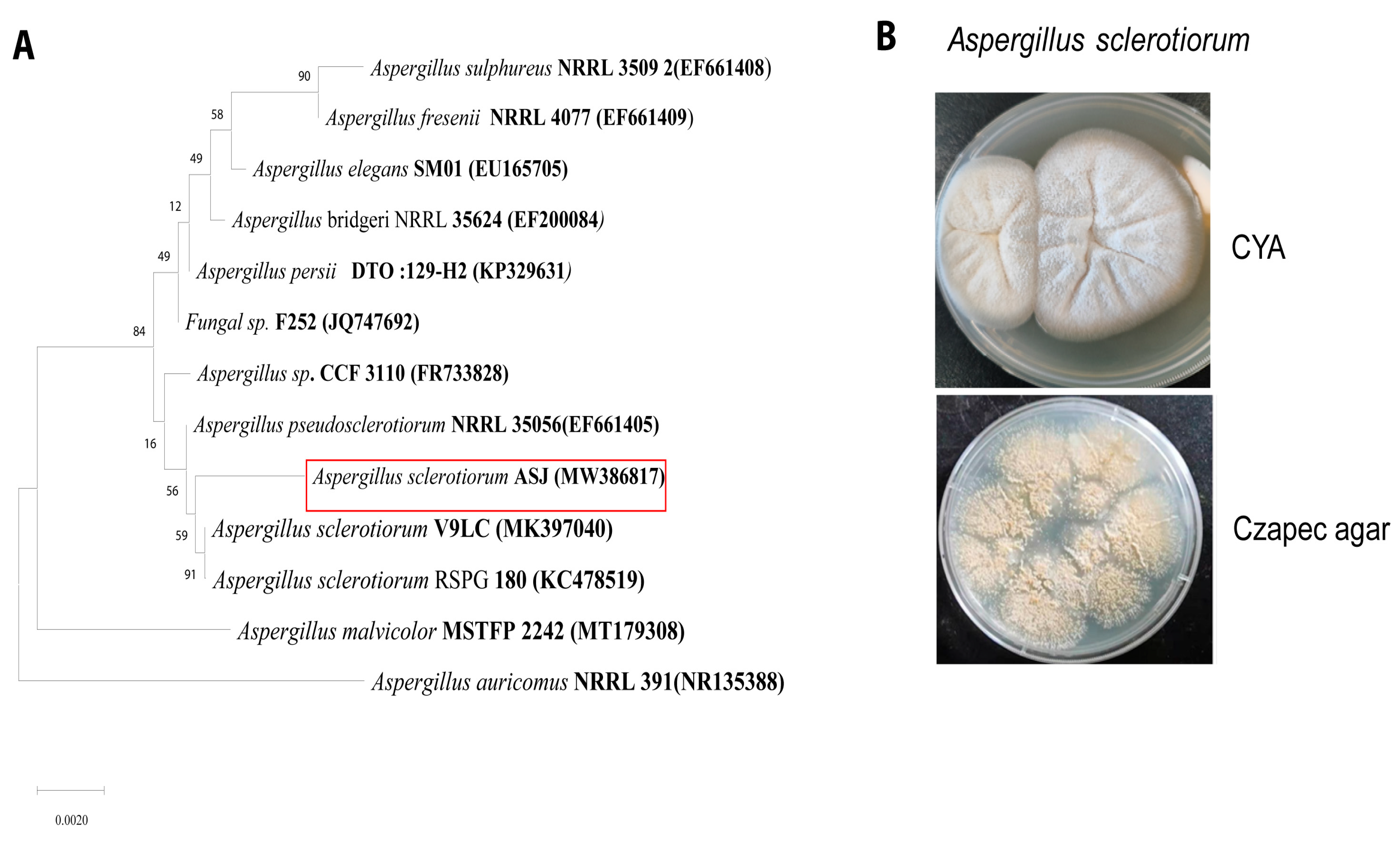
3.2. Identification of Fungus Strain as Aspergillus sclerotiorum
3.3. Whole-Cell Biocatalysis of Phenazine 1-Carboxylic Acid by Aspergillus sclerotiorum
3.4. Purification and Structure Elucidation of the Biotransformation Product
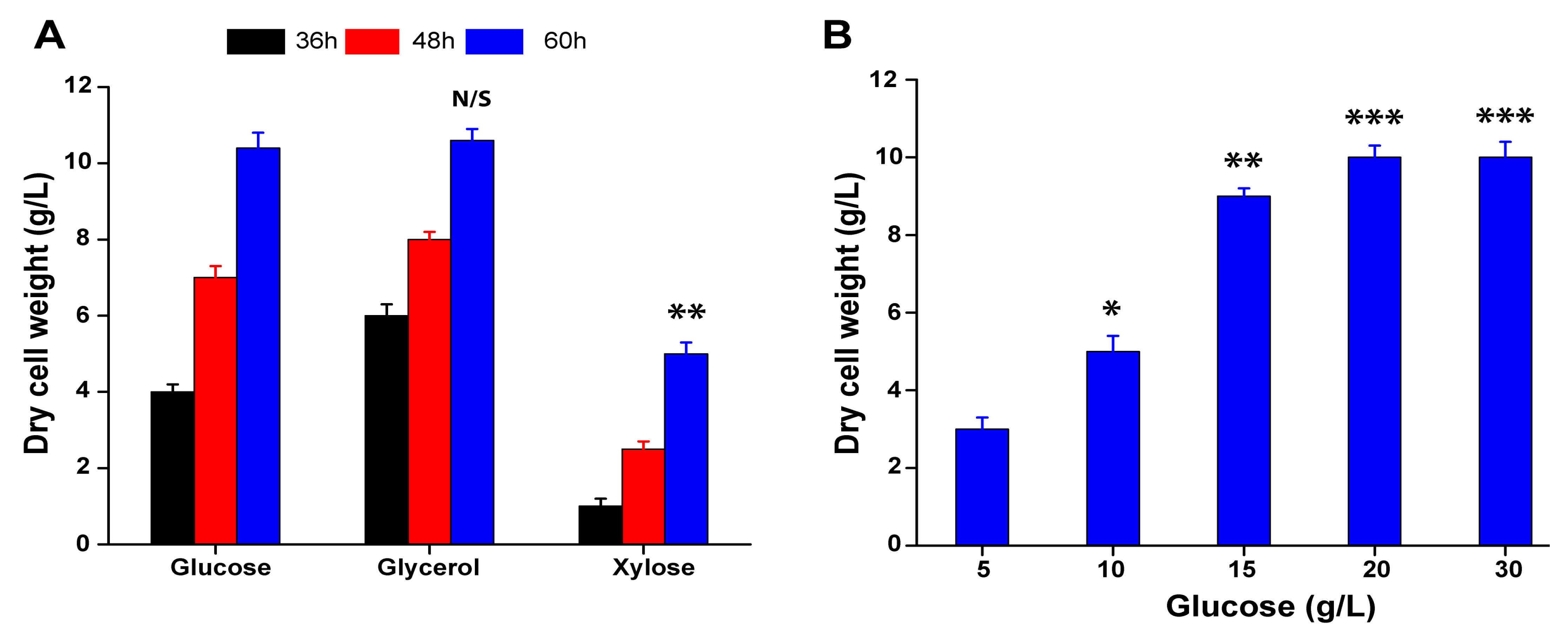
3.5. Whole Cell Biomass Preparation of A. sclerotiorum from Glucose
3.6. Biocatalytic Production of 3-OH-PCA under Optimized Conditions
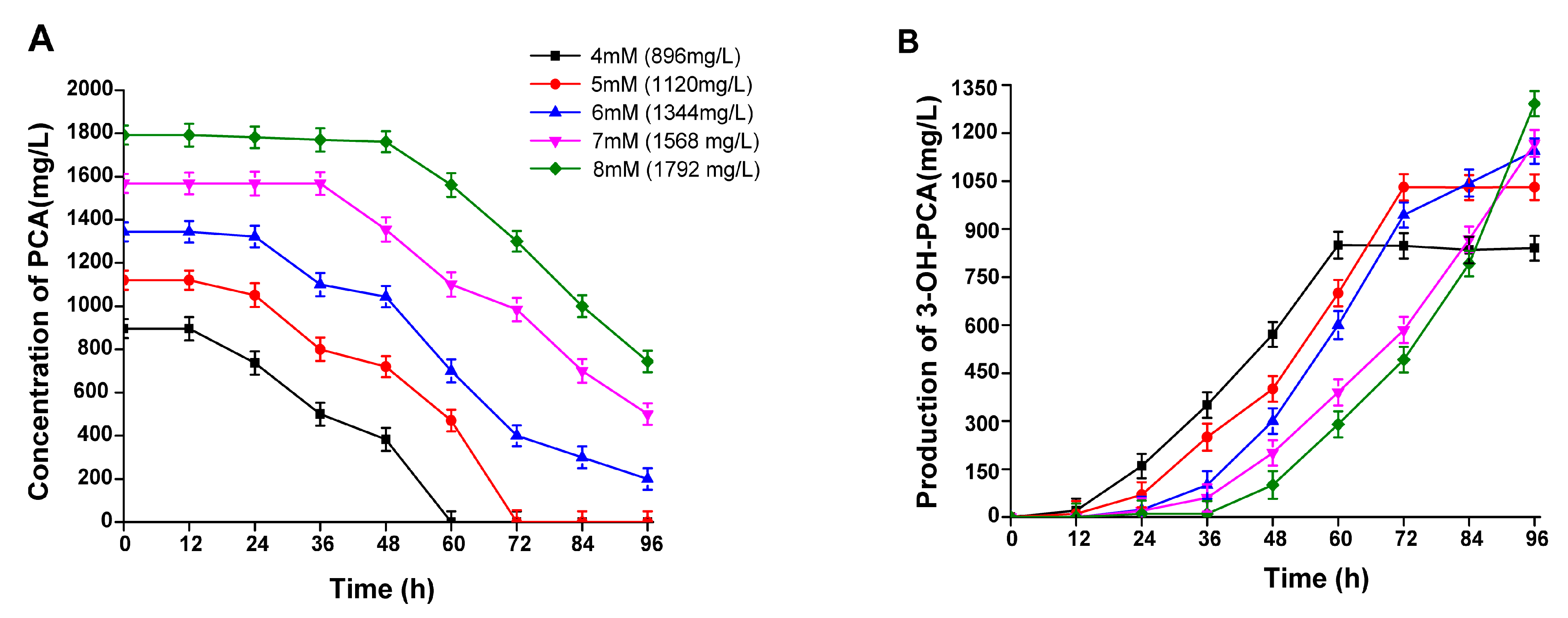
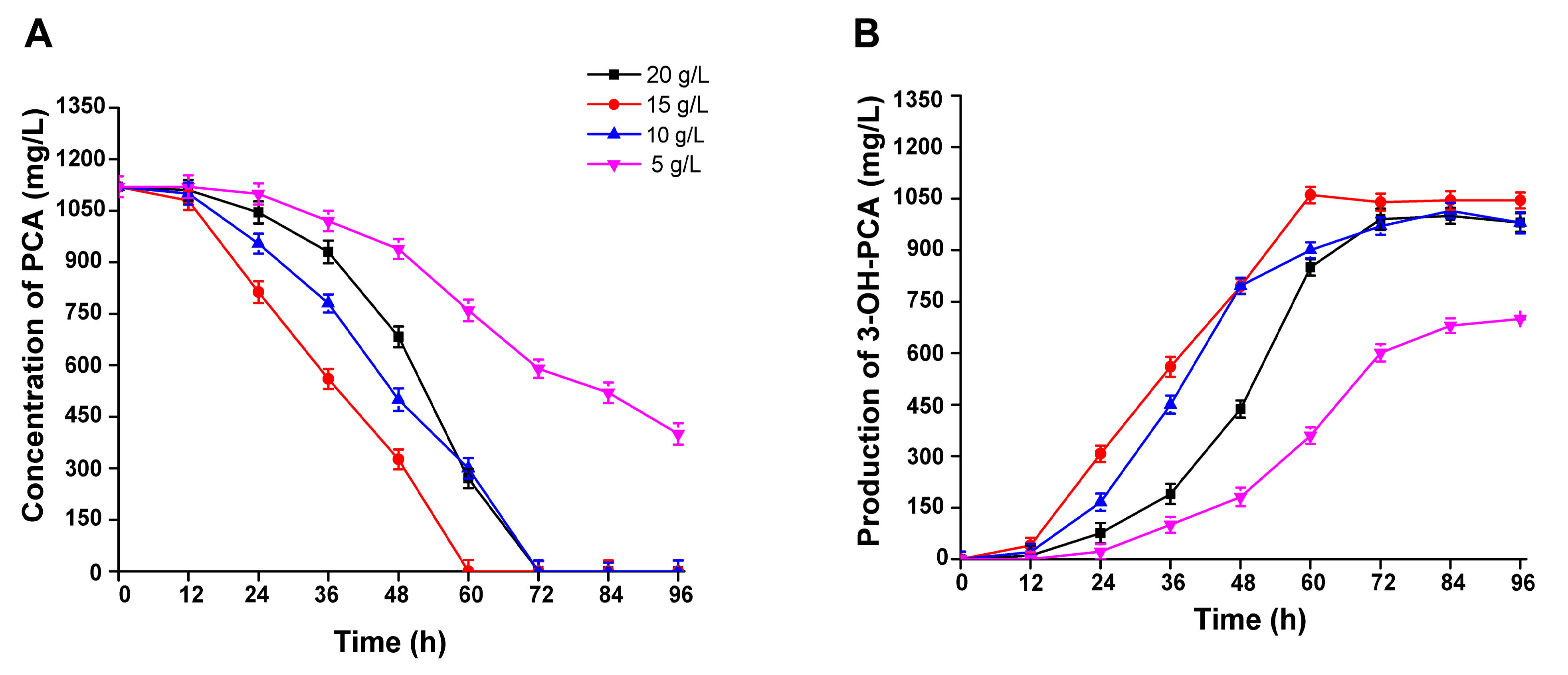
3.7. The Effect of PCA Concentration on the Biocatalytic Production of 3-OH-PCA
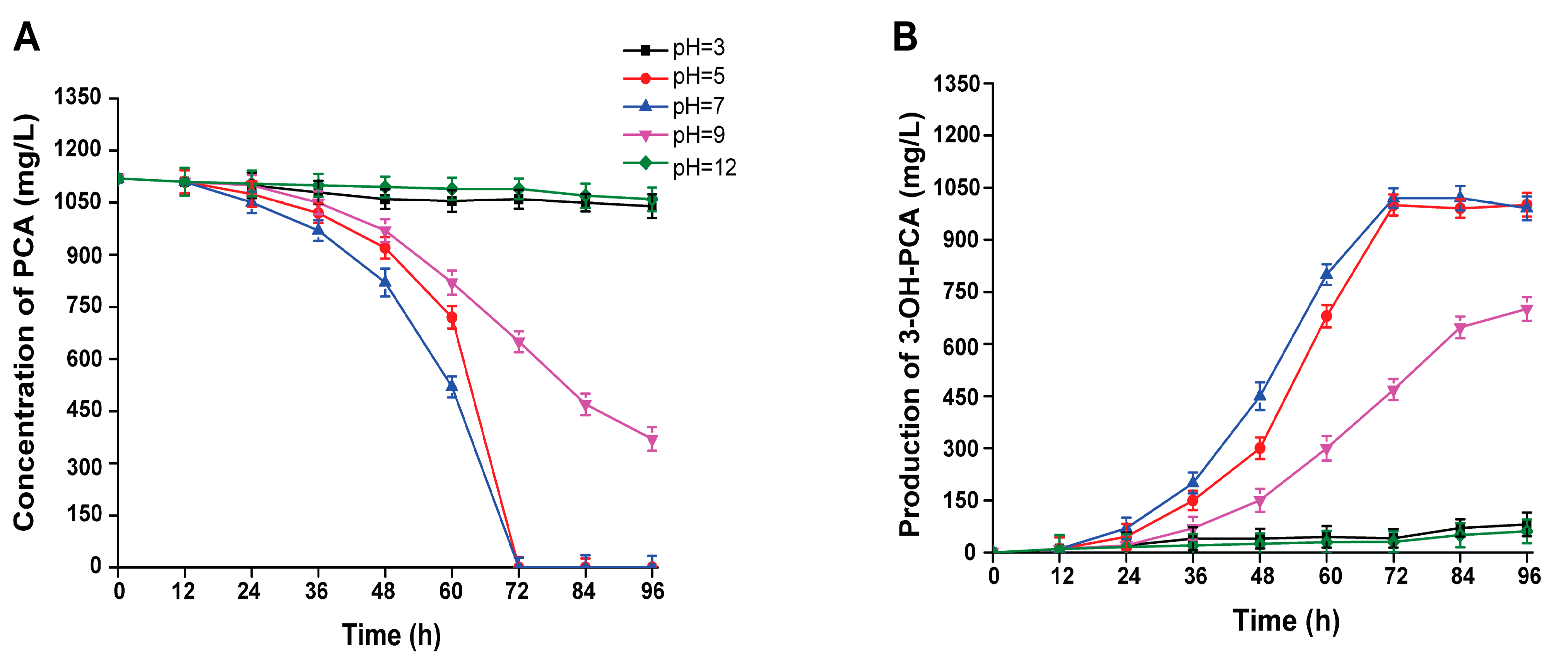
3.8. Effect of Initial pH on the Biotransformation of PCA
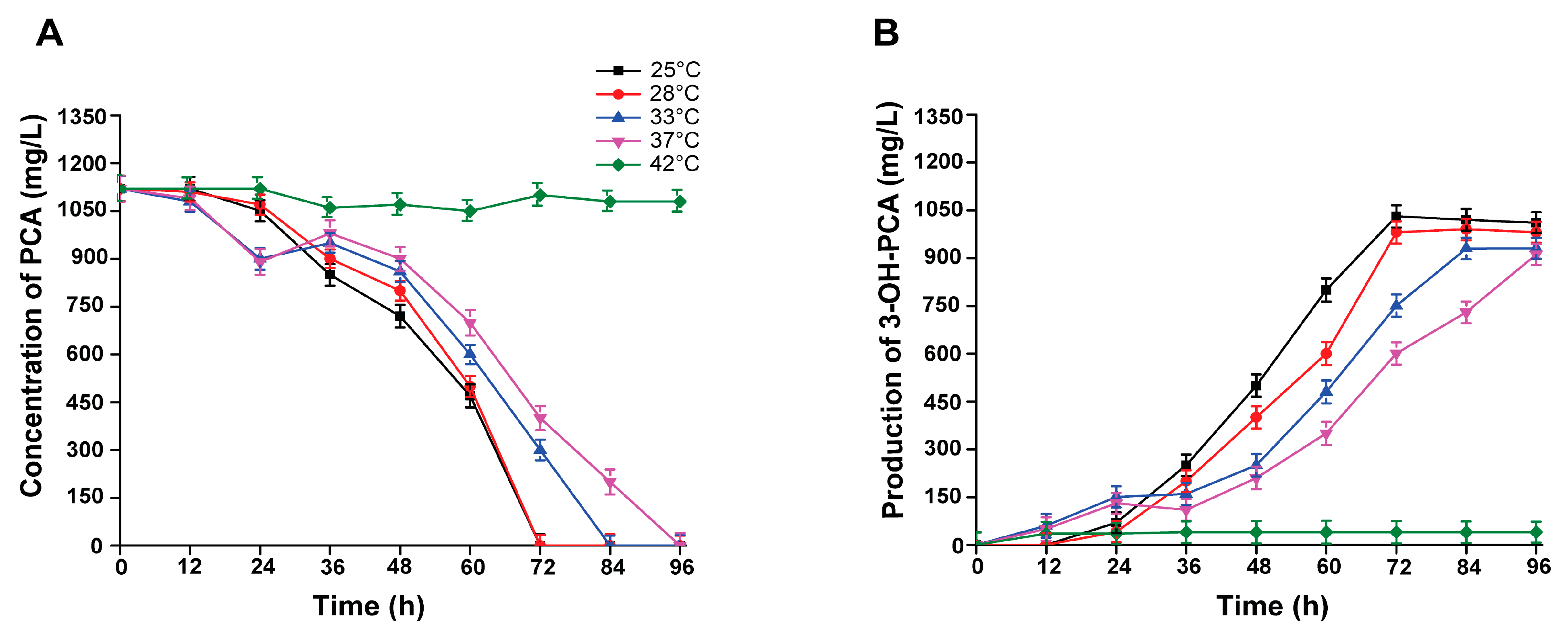
3.9. Effect of Incubation Temperature on the Biotransformation of PCA
3.10. Effect of Agitation Rate(rpm) on the Biotransformation of PCA
3.11. Fungus Biomass Optimization and Its Effect on the Longevity on the Biotransformation of PCA
3.12. Time Course of 3-OH-PCA Production in One-Pot Whole-Cell Biotransformation of PCA
3.13. Repetition and Continuous Utilization of Fungus Pellets for Fed-Batch Fermentation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alvarez-Fitz, P.; Alvarez, L.; Marquina, S.; Luna-Herrera, J.; Navarro-García, V.M. Enzymatic reduction of 9-methoxytariacuripyrone by Saccharomyces cerevisiae and its antimycobacterial activity. Molecules 2012, 17, 8464–8470. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wu, Q.; Zhu, D.; Ma, Y. Biotransformation Enables Innovations Toward Green Synthesis of Steroidal Pharmaceuticals. ChemSusChem 2022, 15, e202102399. [Google Scholar] [CrossRef]
- Liu, J.-H.; Yu, B.-Y. Biotransformation of bioactive natural products for pharmaceutical lead compounds. Curr. Org. Chem. 2010, 14, 1400–1406. [Google Scholar] [CrossRef]
- Alcántara, A. Biotransformations in drug synthesis: A green and powerful tool for medicinal chemistry. J. Med. Chem. Drug Des. 2018, 1, 1–7. [Google Scholar]
- Shah, S.A.; Tan, H.L.; Sultan, S.; Faridz, M.A.; Shah, M.A.; Nurfazilah, S.; Hussain, M. Microbial-catalyzed biotransformation of multifunctional triterpenoids derived from phytonutrients. Int. J. Mol. Sci. 2014, 15, 12027–12060. [Google Scholar] [CrossRef]
- Clouthier, C.M.; Pelletier, J.N. Expanding the organic toolbox: A guide to integrating biocatalysis in synthesis. Chem. Soc. Rev. 2012, 41, 1585–1605. [Google Scholar] [CrossRef]
- de Carvalho, C.C. Whole cell biocatalysts: Essential workers from nature to the industry. Microb. Biotechnol. 2017, 10, 250–263. [Google Scholar] [CrossRef]
- Bilal, M.; Yue, S.J.; Hu, H.B.; Wang, W.; Zhang, X.H. Adsorption/desorption characteristics, separation and purification of phenazine-1-carboxylic acid from fermentation extract by macroporous adsorbing resins. J. Chem. Technol. Biotechnol. 2018, 93, 3176–3184. [Google Scholar] [CrossRef]
- Lee, W.-H.; Kim, M.-D.; Jin, Y.-S.; Seo, J.-H. Engineering of NADPH regenerators in Escherichia coli for enhanced biotransformation. Appl. Microbiol. Biotechnol. 2013, 97, 2761–2772. [Google Scholar] [CrossRef] [PubMed]
- Verho, R.; Londesborough, J.; Penttila, M.; Richard, P. Engineering redox cofactor regeneration for improved pentose fermentation in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2003, 69, 5892–5897. [Google Scholar] [CrossRef]
- Pfruender, H.; Amidjojo, M.; Kragl, U.; Weuster-Botz, D. Efficient whole-cell biotransformation in a biphasic ionic liquid/water system. Angew. Chem. Int. Ed. 2004, 43, 4529–4531. [Google Scholar] [CrossRef] [PubMed]
- Piel, J. Approaches to capturing and designing biologically active small molecules produced by uncultured microbes. Annu. Rev. Microbiol. 2011, 65, 431–453. [Google Scholar] [CrossRef] [PubMed]
- Otten, S.L.; Rosazza, J.P. Microbial transformations of natural antitumor agents. 17. Conversions of lapachol by Cunninghamella echinulata. J. Nat. Prod. 1981, 44, 562–568. [Google Scholar] [CrossRef]
- Huang, Z.; Bonsall, R.F.; Mavrodi, D.V.; Weller, D.M.; Thomashow, L.S. Transformation of Pseudomonas fluorescens with genes for biosynthesis of phenazine-1-carboxylic acid improves biocontrol of rhizoctonia root rot and in situ antibiotic production. FEMS Microbiol. Ecol. 2004, 49, 243–251. [Google Scholar] [CrossRef]
- Yang, M.M.; Mavrodi, D.V.; Mavrodi, O.V.; Bonsall, R.F.; Parejko, J.A.; Paulitz, T.C.; Thomashow, L.S.; Yang, H.T.; Weller, D.M.; Guo, J.H. Biological control of take-all by fluorescent Pseudomonas spp. from Chinese wheat fields. Phytopathology 2011, 101, 1481–1491. [Google Scholar] [CrossRef]
- Mavrodi, D.V.; Mavrodi, O.V.; Parejko, J.A.; Bonsall, R.F.; Kwak, Y.-S.; Paulitz, T.C.; Thomashow, L.S.; Weller, D.M. Accumulation of the antibiotic phenazine-1-carboxylic acid in the rhizosphere of dryland cereals. Appl. Environ. Microbiol. 2012, 78, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Price-Whelan, A.; Dietrich, L.E.; Newman, D.K. Rethinking’secondary’metabolism: Physiological roles for phenazine antibiotics. Nat. Chem. Biol. 2006, 2, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-J.; Wang, W.; Jin, Y.; Hu, H.-B.; Zhang, X.-H.; Xu, Y.-Q. Isolation, identification, and degradation characteristics of phenazine-1-carboxylic acid–degrading strain Sphingomonas sp. DP58. Curr. Microbiol. 2007, 55, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Hu, H.-B.; Wang, W.; Huang, X.-Q.; Zhang, X.-H. Novel three-component phenazine-1-carboxylic acid 1, 2-dioxygenase in Sphingomonas wittichii DP58. Appl. Environ. Microbiol. 2017, 83, e00133-17. [Google Scholar] [CrossRef]
- Costa, K.C.; Moskatel, L.S.; Meirelles, L.A.; Newman, D.K. PhdA catalyzes the first step of phenazine-1-carboxylic acid degradation in Mycobacterium fortuitum. J. Bacteriol. 2018, 200, e00763-17. [Google Scholar] [CrossRef]
- Costa, K.C.; Bergkessel, M.; Saunders, S.; Korlach, J.; Newman, D.K. Enzymatic degradation of phenazines can generate energy and protect sensitive organisms from toxicity. MBio 2015, 6, e01520-15. [Google Scholar] [CrossRef]
- Hill, J.C.; Johnson, G.T. Microbial transformation of phenazines by Aspergillus sclerotiorum. Mycologia 1969, 61, 452–467. [Google Scholar] [CrossRef]
- Moree, W.J.; Phelan, V.V.; Wu, C.-H.; Bandeira, N.; Cornett, D.S.; Duggan, B.M.; Dorrestein, P.C. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc. Natl. Acad. Sci. USA 2012, 109, 13811–13816. [Google Scholar] [CrossRef] [PubMed]
- Chin-A-Woeng, T.F.; Thomas-Oates, J.E.; Lugtenberg, B.J.; Bloemberg, G.V. Introduction of the phzH gene of Pseudomonas chlororaphis PCL1391 extends the range of biocontrol ability of phenazine-1-carboxylic acid-producing Pseudomonas spp. strains. Mol. Plant-Microbe Interact. 2001, 14, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Barbosa Coitinho, L.; Fumagalli, F.; da Rosa-Garzon, N.G.; da Silva Emery, F.; Cabral, H. Lapachol biotransformation by filamentous fungi yields bioactive quinone derivatives and lapachol-stimulated secondary metabolites. Prep. Biochem. Biotechnol. 2019, 49, 459–463. [Google Scholar] [CrossRef] [PubMed]
- de Matos, I.L.; Nitschke, M.; Porto, A.L.M. Regioselective and chemoselective biotransformation of 2′-hydroxychalcone derivatives by marine-derived fungi. Biocatal. Biotransform. 2023, 41, 46–56. [Google Scholar] [CrossRef]
- Wierckx, N.; Elink Schuurman, T.D.; Blank, L.M.; Ruijssenaars, H.J. Whole-cell biocatalytic production of 2, 5-furandicarboxylic acid. In Microorganisms in Biorefineries; Springer: Berlin/Heidelberg, Germany, 2014; pp. 207–223. [Google Scholar]
- Rajesh, R.O.; Godan, T.K.; Rai, A.K.; Sahoo, D.; Pandey, A.; Binod, P. Biosynthesis of 2, 5-furan dicarboxylic acid by Aspergillus flavus APLS-1: Process optimization and intermediate product analysis. Bioresour. Technol. 2019, 284, 155–160. [Google Scholar] [CrossRef]
- Deng, R.-X.; Yue, S.-J.; Wang, W.; Hu, H.-B.; Zhang, X.-H. Identification, biological evaluation, and improved biotransformation of a phenazine antioxidant using Streptomyces lomondensis S015 whole cells. Process Biochem. 2023, 125, 154–161. [Google Scholar] [CrossRef]
- Yue, S.J.; Song, C.; Li, S.; Huang, P.; Guo, S.Q.; Hu, H.B.; Wang, W.; Zhang, X.H. Synthesis of cinnabarinic acid by metabolically engineered Pseudomonas chlororaphis GP72. Biotechnol. Bioeng. 2019, 116, 3072–3083. [Google Scholar] [CrossRef]
- Klich, M.A. Identification of Common Aspergillus Species; CBS: New York, NY, USA, 2002. [Google Scholar]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Consortium, F.B.; List, F.B.C.A.; Bolchacova, E. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal identification using molecular tools: A primer for the natural products research community. J. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gao, Y.; Yin, Y.; Cai, M.; Zhou, X.; Zhang, Y. Effective pH pretreatment and cell disruption method for real-time intracellular enzyme activity assay of a marine fungus covered with pigments. Prep. Biochem. Biotechnol. 2017, 47, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Fan, D.; Su, C.; Pan, L.; He, Q.; Li, Z.; Liu, C. Biotransformation of sulfamethoxazole by a novel strain, Nitratireductor sp. GZWM139: Characterized performance, metabolic mechanism and application potential. J. Hazard. Mater. 2023, 441, 129861. [Google Scholar] [CrossRef]
- Supramani, S.; Jailani, N.; Ramarao, K.; Zain, N.A.M.; Klaus, A.; Ahmad, R.; Wan, W.A.A.Q.I. Pellet diameter and morphology of European Ganoderma pfeifferi in a repeated-batch fermentation for exopolysaccharide production. Biocatal. Agric. Biotechnol. 2019, 19, 101118. [Google Scholar] [CrossRef]
- Birhanli, E.; Yesilada, O. Increased production of laccase by pellets of Funalia trogii ATCC 200800 and Trametes versicolor ATCC 200801 in repeated-batch mode. Enzym. Microb. Technol. 2006, 39, 1286–1293. [Google Scholar] [CrossRef]
- Liu, Y.; Yue, S.; Bilal, M.; Jan, M.; Wang, W.; Hu, H.; Zhang, X. Development of Artificial Synthetic Pathway of Endophenazines in Pseudomonas chlororaphis P3. Biology 2022, 11, 363. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Zhang, P.; Bilal, M.; Wang, W.; Hu, H.; Zhang, X. Enhanced biosynthesis of phenazine-1-carboxamide by engineered Pseudomonas chlororaphis HT66. Microb. Cell Factories 2018, 17, 117. [Google Scholar] [CrossRef]
- Hansen, G.M.; Laird, T.S.; Woertz, E.; Ojala, D.; Glanzer, D.; Ring, K.; Richart, S.M. Aspergillus sclerotiorum fungus is lethal to both Western drywood (Incisitermes minor) and Western subterranean (Reticulitermes hesperus) termites. Fine Focus 2016, 2, 23–38. [Google Scholar] [CrossRef]
- Zhou, Q.; Su, J.; Jiang, H.; Huang, X.; Xu, Y. Optimization of phenazine-1-carboxylic acid production by a gacA/qscR-inactivated Pseudomonas sp. M18GQ harboring pME6032Phz using response surface methodology. Appl. Microbiol. Biotechnol. 2010, 86, 1761–1773. [Google Scholar] [CrossRef]
- Kobayashi, M.; Nagasawa, T.; Yamada, H. Enzymatic synthesis of acrylamide: A success story not yet over. Trends Biotechnol. 1992, 10, 402–408. [Google Scholar] [CrossRef]
- Dejaegher, B.; Vander Heyden, Y. Ruggedness and robustness testing. J. Chromatogr. A 2007, 1158, 138–157. [Google Scholar] [CrossRef] [PubMed]
- Şahin, E. First green synthesis of (R)-2-methyl-1-phenylpropan-1-ol using whole-cell Lactobacillus paracasei BD101 biotransformation. Biocatal. Biotransform. 2020, 38, 138–143. [Google Scholar] [CrossRef]
- Salvi, N.A.; Chattopadhyay, S. Laboratory scale-up synthesis of chiral carbinols using Rhizopus arrhizus. Tetrahedron Asymmetry 2016, 27, 188–192. [Google Scholar] [CrossRef]
- Xiao, Z.; Lv, C.; Gao, C.; Qin, J.; Ma, C.; Liu, Z.; Liu, P.; Li, L.; Xu, P. A novel whole-cell biocatalyst with NAD+ regeneration for production of chiral chemicals. PLoS ONE 2010, 5, e8860. [Google Scholar] [CrossRef]
- de Oliveira, J.R.; Seleghim, M.H.R.; Porto, A.L.M. Biotransformation of methylphenylacetonitriles by Brazilian marine fungal strain Aspergillus sydowii CBMAI 934: Eco-friendly reactions. Mar. Biotechnol. 2014, 16, 156–160. [Google Scholar] [CrossRef]
- Şahin, E.; Serencam, H.; Dertli, E. Whole cell application of Lactobacillus paracasei BD101 to produce enantiomerically pure (S)-cyclohexyl (phenyl) methanol. Chirality 2019, 31, 211–218. [Google Scholar] [CrossRef]
- Yılmaz, D.; Şahin, E.; Dertli, E. Highly enantioselective production of chiral secondary alcohols using Lactobacillus paracasei BD 101 as a new whole cell biocatalyst and evaluation of their antimicrobial effects. Chem. Biodivers. 2017, 14, e1700269. [Google Scholar] [CrossRef] [PubMed]
- Şahin, E. Production of (R)-1-(1,3-benzodioxol-5-yl) ethanol in high enantiomeric purity by Lactobacillus paracasei BD 101. Chirality 2018, 30, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Zilbeyaz, K.; Kurbanoglu, E.B.; Kilic, H. Preparation of Enantiomerically Pure (S)-(−)-1-(1’-naphthyl)-ethanol by the Fungus Alternaria alternata. Chirality 2016, 28, 669–673. [Google Scholar] [CrossRef]
- Parshikov, I.A.; Sutherland, J.B. Biotransformation of steroids and flavonoids by cultures of Aspergillus niger. Appl. Biochem. Biotechnol. 2015, 176, 903–923. [Google Scholar] [CrossRef] [PubMed]
- Parsons, J.F.; Greenhagen, B.T.; Shi, K.; Calabrese, K.; Robinson, H.; Ladner, J.E. Structural and functional analysis of the pyocyanin biosynthetic protein PhzM from Pseudomonas aeruginosa. Biochemistry 2007, 46, 1821–1828. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.-X.; Zhang, Z.-J.; Liu, W.-F.; Dong, Z.-Y.; Tao, Y. Enhanced production of N-acetyl-D-neuraminic acid by multi-approach whole-cell biocatalyst. Appl. Microbiol. Biotechnol. 2013, 97, 4775–4784. [Google Scholar] [CrossRef] [PubMed]
- Knapp, J.S.; Zhang, F.m.; Tapley, K.N. Decolourisation of Orange II by a wood-rotting fungus. J. Chem. Technol. Biotechnol. Int. Res. Process Environ. Clean Technol. 1997, 69, 289–296. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jan, M.; Yue, S.-J.; Deng, R.-X.; Nie, Y.-F.; Zhang, H.-Y.; Hao, X.-R.; Wang, W.; Hu, H.-B.; Zhang, X.-H. Aspergillus sclerotiorum Whole-Cell Biocatalysis: A Sustainable Approach to Produce 3-Hydroxy-phenazine 1-Carboxylic Acid from Phenazine 1-Carboxylic Acid. Fermentation 2023, 9, 579. https://doi.org/10.3390/fermentation9060579
Jan M, Yue S-J, Deng R-X, Nie Y-F, Zhang H-Y, Hao X-R, Wang W, Hu H-B, Zhang X-H. Aspergillus sclerotiorum Whole-Cell Biocatalysis: A Sustainable Approach to Produce 3-Hydroxy-phenazine 1-Carboxylic Acid from Phenazine 1-Carboxylic Acid. Fermentation. 2023; 9(6):579. https://doi.org/10.3390/fermentation9060579
Chicago/Turabian StyleJan, Malik, Sheng-Jie Yue, Ru-Xiang Deng, Yan-Fang Nie, Hong-Yan Zhang, Xiang-Rui Hao, Wei Wang, Hong-Bo Hu, and Xue-Hong Zhang. 2023. "Aspergillus sclerotiorum Whole-Cell Biocatalysis: A Sustainable Approach to Produce 3-Hydroxy-phenazine 1-Carboxylic Acid from Phenazine 1-Carboxylic Acid" Fermentation 9, no. 6: 579. https://doi.org/10.3390/fermentation9060579
APA StyleJan, M., Yue, S.-J., Deng, R.-X., Nie, Y.-F., Zhang, H.-Y., Hao, X.-R., Wang, W., Hu, H.-B., & Zhang, X.-H. (2023). Aspergillus sclerotiorum Whole-Cell Biocatalysis: A Sustainable Approach to Produce 3-Hydroxy-phenazine 1-Carboxylic Acid from Phenazine 1-Carboxylic Acid. Fermentation, 9(6), 579. https://doi.org/10.3390/fermentation9060579






