Abstract
Pichia kudriavzevii is one of the major non-Saccharomyces cerevisiae yeasts in Chinese baijiu brewing, which has shown a substantially higher tolerance to acid, heat, and ethanol. Exploring the mechanism of P. kudriavzevii could have a positive effect on the artificially controlled production of baijiu. In this study, an efficient acetic-acid-tolerant P. kudriavzevii strain, Y2, was isolated from the yellow water of strong-flavored baijiu brewing waste, and its molecular mechanism of acetic acid tolerance was investigated through a comparative transcriptomic analysis. The strain Y2 could tolerate 12 g/L of acetic acid. According to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, differentially expressed genes (DEGs) were mainly enriched in oxidative phosphorylation, the citrate cycle, glycolysis/gluconeogenesis, and carbon metabolism under low (AL group) and high (AH group) concentrations of acetic acid. However, the DEG enrichment was more profound in the AH group when compared to the control. Compared with the AL group, the expression of genes related to oxidative phosphorylation was more significantly upregulated, while in terms of the TCA cycle, phosphoenolpyruvate carboxykinase was significantly upregulated in both the AH and AL groups and was positively correlated with tolerance to acetic acid. This was followed by citrate synthase, isocitrate dehydrogenase, malate dehydrogenase, and succinate dehydrogenase. These results illustrated a possible mechanism of acid tolerance by regulating the metabolism-related pathways in P. kudriavzevii and provided a basis for the further investigation of the acid tolerance mechanism.
1. Introduction
Baijiu is a long-established alcoholic beverage, being the most distinctive of traditional Chinese fermented foods, and it is also one of the six most famous distilled spirits in the world [1,2]. In China, the consumption of baijiu had reached over 13 billion liters by 2016, and its market value can reach about USD97 billion [3]. Chinese baijiu can be divided into 12 types of flavors according to the origin, climate, raw materials, and brewing methods. Strong-flavored, Maotai-flavored, and light-flavored baijiu are the three most popular types among consumers [4,5]. The brewing of baijiu is often carried out in a harsh environment with high heat, acidity, and ethanol [6,7]. Among these, acetic acid and lactic acid are the two main organic acids in baijiu, and the levels of both can significantly affect the aroma of baijiu [8,9]. In addition, baijiu production involves complex microbial processes, where diverse microbial communities are one of the important factors affecting the flavor of baijiu in addition to the raw materials and production processes [2,5,10]. However, under an open-fermentation mode, the original balance of the microbial community can be easily disrupted. For example, a hot environment in summer can cause an excessive accumulation of organic acids such as lactic acid and acetic acid, which can seriously inhibit the growth of the functional yeast flora in the system [11]. In addition, the utilization of nutrients and the accumulation of metabolites during the brewing process can easily lead to changes in the microenvironment, such as low pH, an increased ethanol concentration, and a decreased oxygen content, etc. These conditions can subject the brewing microorganisms to a variety of stresses and limit their growth; however, there exists a small number of microorganisms with good tolerance that can adapt to this extreme environment and grow and multiply in large numbers [12]. Saccharomyces cerevisiae has long been widely regarded as the main fermentation driver for baijiu production. However, with the gradual deepening of research on the microbial community of baijiu brewing, it has been found that non-Saccharomyces-cerevisiae yeast species (abnormal Wickham yeast, Candida, Pichia, etc.) are also indispensable core microbes. In particular, these non-Saccharomyces-cerevisiae yeast species have a good tolerance to stresses such as high temperature, high acidity, and high osmotic pressure in the baijiu fermentation environment [13], and they play an irreplaceable role in stabilizing the composition and metabolic activity of the bacterial flora in the fermentation system. Therefore, by exploring the microbial communities in the baijiu brewing environment and uncovering highly resistant non-Saccharomyces-cerevisiae yeast species, finding solutions to maintain the brewing microecological balance can help to improve the quality of baijiu.
Pichia kudriavzevii is one of the core non-saccharomyces yeasts in baijiu brewing engineering [14,15], which is widely found in various spontaneous fermentations, soils, and fruits in nature, and it has an excellent tolerance to high-temperature, low-pH, and high-ethanol environments [16]. P. kudriavzevii is widely used in the food fermentation industries, such as Chinese baijiu, coffee fermentation, fermented milk, and wine [17]. It has been found that P. kudriavzevii is the main functional yeast in the fermentation process of Chinese baijiu brewing, including Maotai-flavored, strong-flavored, chi-flavored, and light-flavored baijiu [11]. During baijiu brewing, P. kudriavzevii can effectively degrade urea, thus reducing the amount of ethyl carbamate (EC) in Maotai-flavored baijiu production, ensuring the safety of the base liquor [18]. In addition, P. kudriavzevii also plays a regulatory role in fungal succession in brewing microecology, and its metabolite phenylethanol significantly inhibits the growth of other fungi in the community [19]. P. kudriavzevii is the yeast of choice for coffee fermentation because of its wide growth temperature range (25–40 °C) and its ability to survive under multiple stresses of 1–3% acetic acid, 2–10% ethanol, and 5–50% glucose and fructose [20,21]. Coffee fermented with P. kudriavzevii has prominent tastes of malt, berry, and honey flavors on the sensory level, and therefore P. kudriavzevii can be applied to specialty coffee production and in the enhancement of its unique flavor [22]. With reference to its tolerance to low pH, P. kudriavzevii not only has a strong acid tolerance and ability to degrade lactic acid by itself but also its co-culturing with S. cerevisiae can significantly increase the consumption of lactic acid [23]. In addition, during bioethanol fermentation, P. kudriavzevii can tolerate acetic acid and formic acid concentrations of up to 8–10 g/L and 2 g/L, respectively [24], and it can even grow under conditions with a pH as low as 2.0 [25]. However, the molecular mechanisms of the acid tolerance associated with P. kudriavzevii have not been sufficiently studied, and in-depth studies on its genome and metabolic pathways are needed.
In this study, a Pichia kudriavzevii strain, Y2, was isolated from yellow water—a waste product of strong-flavored baijiu production, and its acetic acid tolerance and three enzymatic activities (α-amylase, cellulase, and lipase) were studied. Later, the molecular mechanism of acid tolerance by P. kudriavzevii was then investigated by transcriptomic analysis, providing a theoretical basis for the study of the regulatory mechanisms of the acid tolerance of P. kudriavzevii and the control of the artificial production of strong-flavored baijiu.
2. Materials and Methods
2.1. Isolation, Purification, and Identification of the Yeast Strain
The yellow-water samples of strong-flavored baijiu brewing were obtained from a baijiu distillery in Yibin, China. A total of 10 mL of the yellow-water sample was added into 90 mL of sterile saline solution (0.9%) and was mixed in a shaker at 37 °C and 180 rpm for 30 min to obtain the mother liquor. The above mother liquor was serially diluted to 10−7, and 50 μL of each dilution was evenly applied on a YDP solid medium and incubated at 37 °C for 2–3 days. The culture was purified through subculturing on YPD, and the purity was monitored under a microscope (WMF-3690, WUMO, Shanghai, China) after staining with lactophenol cotton-blue stain. The pure cultures were then inoculated on YPD slants and stored at 4 °C.
2.2. Screening of Acid-Tolerant Yeast Strains
To assess the acid tolerance potential of the strain Y2, a YPD liquid medium (Sangon Biotech, Shanghai, China) was prepared, and the pH was adjusted using various concentration of acetic acid including 2 g/L, (pH = 3.12), 3 g/L, (pH = 3.03), 4 g/L, (pH = 2.97), 5 g/L, (pH = 2.92), and 6 g/L. (pH = 2.88). Then, the purified strain, activated for 24 h, was inoculated in the medium at a 5% volume fraction of the inoculum. To set a control, the cultures were incubated at 37 °C and 180 rpm for 48 h in two parallel groups. The OD600 was measured using a NanoDrop kit (Yitao Science, Guangzhou, China).
2.3. Molecular Biological Identification of the Yeast Strain
A fungal genomic DNA extraction kit (Solarbio, Beijing, China) was used to extract the acid-tolerant yeast DNA, and the DNA was used as a template for PCR amplification with universal primers ITS1: 5′-TCCGTAGGTGAACCTGCGG-3′ and ITS4: 5′-TCCTCCGCTTATTGATATGC-3′ [26]. The PCR amplification system was as follows: 25 μL: 8.5 μL ddH2O, 12.5 μL of 2×Taq PCR Master Mix, 1 μL of forward and reverse primers, and 2 μL of DNA template. The PCR reaction was initiated at a pre-denaturation step at 94 °C for 5 min, which was followed by 36 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 40 s, and a final extension at 72 °C for 5 min. The PCR amplification products were examined by gel electrophoresis, and the gel bands were observed with a dark box UV analyzer (Qiwei Instrument, Hangzhou, China) and a Tannon-2500B gel-imaging system (Tannon, Shanghai, China). The PCR products were sent to Biotech Bioengineering (Shanghai, China) Co. The sequences were compared with those of existing strains in NCBI (BLAST: Basic Local Alignment Search Tool), and the phylogenetic tree was constructed using MEGA 11 following the neighbor joining method, while the bootstrapping value was set as 1000.
2.4. Determination of Growth Curve of Acid-Tolerant Yeast
The acidity of the YPD liquid medium was adjusted using different concentrations of acetic acid (0 g/L (pH = 6.50), 4 g/L, (pH = 2.97), 8 g/L, (pH = 2.82), 10 g/L, (pH = 2.77), and 12 g/L (pH = 2.73)) with three replicates per group. The yeast seed solution was then inoculated in a 48-well plate at 5% (v/v) inoculum, and the well plates were placed in a MicroScreen-HT real-time microbial growth analysis system (Gering, Tianjin, China) and incubated at 37 °C and 180 rpm with shaking, and the OD600 was measured. The growth curves of the yeast under different acidities were plotted with the fermentation time (x) as the horizontal coordinate and the OD600 (y) as the vertical coordinate.
2.5. Determination of Enzyme Activities
The yeast seed solution was inoculated in 50 mL of YPD liquid medium (without acetic acid) at 5% (v/v) inoculum, incubated at 37 °C and 180 rpm with shaking, and then stopped after reaching the middle of the logarithmic growth period, and the culture was preserved. The samples were treated according to the sample processing method using an LPS activity test kit, a CL activity test kit, and an α-amylase activity test kit (Sangon Biotech, Shanghai, China), and the enzyme activities of lipase (LPS), cellulase (CL), and α-amylase of the acid-tolerant yeast Y2 were measured. The lipase activity (LPS) was calculated using the standard curve y = 0.0117x + 0.0219, with R2 = 0.9992; the cellulase activity (CL) was calculated using the standard curve y = 1.8507x + 0.0837, with R2 = 0.9992; and the α-amylase activity was calculated using the standard curve y = 3.7259x + 0.0076, with R2 = 0.9957.
2.6. RNA Sample Preparation, Extraction, and Sequencing
The pH of the YPD liquid medium (50 mL) was adjusted with 0 g/L (C group, pH = 6.50), 4 g/L (AL group, pH = 2.97), and 8 g/L (AH group, pH = 2.82) of acetic acid, and then the samples were inoculated (5% v/v of the acid tolerant Y2 strain) and incubated at 37 °C with shaking at 180 rpm. According to the growth curves obtained during the previous stage, the strains were collected from each group at the middle of the logarithmic growth period (centrifuged at 3000 rpm for 3 min, and the supernatant discarded), snap-frozen in liquid nitrogen for 10 min, and stored at −80 °C.
The cells for RNA extraction were collected from three parallel groups including a low-acidic-strength group (AL), a high-acidic-strength group (AH) and a control group (C). Total RNA was extracted using Trizol reagent following the kit’s protocol (Invitrogen, Carlsbad, CA, USA). The quality and quantity of RNA were determined, and libraries were constructed, which was followed by sequencing on the Illumina HiSeq X Ten platform, where 150 bp paired-end reads were obtained.
2.7. RNA-Seq Analysis
RNA sequence analysis was performed by Novogene Co., Ltd. (Beijing, China). The DESeq2 R package (1.20.0) was used for differential expression analysis between the two comparative combinations. The clusterProfiler R package (3.8.1) was used to perform gene ontology (GO) enrichment analysis on differentially expressed genes (DEGs), and the gene length deviation was corrected. GO terms corrected for p < 0.05 were considered as significant enrichment terms for differentially expressed genes. Statistical enrichment of differentially expressed genes in the KEGG pathway was analyzed using the clusterProfiler R package (3.8.1).
2.8. Statistical Analysis
All data are shown as the mean ± SD of three replications for each experiment. Tukey’s post hoc test and one-way ANOVA were performed using GraphPad Prism 9.0 (San Diego, CA, USA). p-values of less than 0.05 indicated significant differences.
3. Results
3.1. Isolation, Purification, and Morphological Characterization of the Yeast Strains
The yellow water samples were diluted, spread on YPD solid medium and streaked on plates, and were observed for colony formation and cell morphology. Here, 10 yeast strains were isolated and purified, and they were labelled as Y1 to Y10 in order. Among them, Y2, Y5, and Y7 grew relatively well, and the single colonies of these three strains were round, creamy white, sticky, moist, and with a smooth surface of 1–2 mm in diameter (Figure 1A). The cells were oval or circular in shape under a microscope with a size of (2.5–4.0) μm × (6–10) μm (Figure 1B).
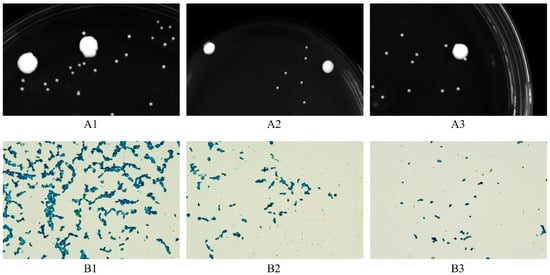
Figure 1.
Morphological characterization of strains. (A) The colony morphology of the strains (A1–A3) in order is shown for strains Y2, Y5, and Y7. (B) Cell morphology of strains under light microscope (×100) after staining with lactophenol cotton-blue stain. (B1–B3) The cell morphology of strains Y2, Y5, and Y7 in order.
3.2. Screening of Acid-Tolerance Potential of the Yeast Strains
The three strains, namely Y2, Y5, and Y7, which showed stronger growth activity, were inoculated in YPD liquid media with different acidities, and the results showed that there were significant differences in the OD600 of the three yeast strains at different acetic acid concentrations (Figure 2A). Under lower concentrations of acetic acid (2 and 3 g/L) stress, the OD600 of strain Y5 was significantly higher than that of strain Y2 and strain Y7 after 48 h. However, when the acetic acid concentration was increased, strain Y2 showed the highest growth activity with a significantly higher OD600 when compared to the other two strains. This indicated that strain Y2 had a better acetic acid tolerance than Y5 and Y7. In addition, strain Y2 was the most adaptable under 5 g/L acetic acid stress and had the maximum OD600 value. To sum up, the three strains had different adaptabilities under different acetic acid concentrations, and the growth of strain Y2 was significantly better than that of the other two strains under higher acetic acid concentrations. Therefore, strain Y2 was selected as the target for the subsequent study of the acid tolerance mechanism.
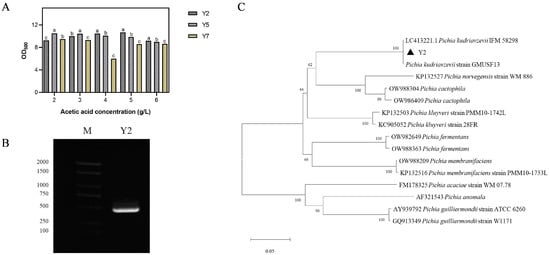
Figure 2.
Screening and molecular identification of acid-tolerant yeast strains. (A) The growth of strains Y2, Y5, and Y7 under different concentrations of acetic acid stress (OD600), respectively, with different lowercase letters indicating significance. (B) Electrophoresis bands at 1% (w/v) agarose gel of the PCR amplification product of strain Y2, lane M (Maker). (C) Phylogenetic tree constructed based on strain Y2.
3.3. Molecular Identification of Acid-Tolerant Yeast Strain Y2
The ITS1 and ITS4 primers were used for PCR amplification. The PCR amplification products of strain Y2 were subjected to gel electrophoresis and showed a clear band of 500 bp (Figure 2B), which was purified and sent for sequencing. The sequencing analysis showed that the sequence similarity between strain Y2 and LC413221.1 Pichia kudriavzevii IFM 58298 was 99.6%, and both showed up on the same clade (Figure 2C). Therefore, strain Y2 was identified as Pichia kudriavzevii.
3.4. Analysis of Acetic Acid Resistance of Acid-Tolerant Yeast
The Pichia kudriavzevii Y2 strain was exposed to various concentration of acetic acid including 0 g/L (control), 4 g/L, 8 g/L, 10 g/L, and 12 g/L, and it was found that, at an acetic acid concentration of 4 g/L, strain Y2 proliferated rapidly during the logarithmic growth period from 2 h to 28 h and then entered into a stable period with OD600 values reaching about 9.0. However, the growth rate of the strain was slowed down significantly after the acetic acid concentration increased. Under the conditions of 8 g/L, 10 g/L, and 12 g/L acetic acid, the yeast growth was faster at an acetic acid concentration of 12 g/L. However, there was no significant difference in the growth of strain Y2 after 24 h, with all samples entering the stable phase after about 54 h with OD600 values at about 9.0, which were not significantly different from those at the 4 g/L concentration. In addition, the acclimatization period for strain growth at the 4 g/L concentration was significantly shorter than that of the other three concentrations (Figure 3). These results indicated that strain Y2 could tolerate up to a 12 g/L concentration of acetic acid but adapted relatively better at lower concentrations and entered the logarithmic phase more quickly, suggesting that high concentrations of acetic acid inhibited the strain growth to a certain extent.
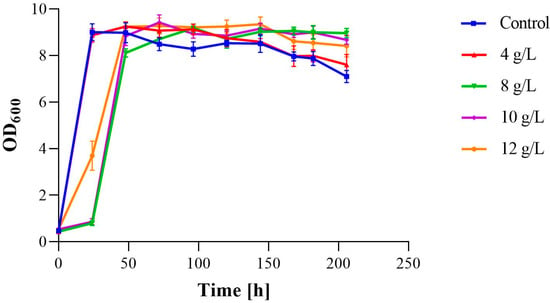
Figure 3.
Biomass of Pichia kudriavzevii Y2 during fermentation (control) and its growth under different concentrations of acetic acid stress (4 g/L, 8 g/L, 10 g/L, and 12 g/L).
3.5. Enzyme Activities under Acid Stress
Samples were collected at the middle of the logarithmic growth period, and the enzyme activity levels of lipase (LPS), cellulase (CL), and α-amylase were determined separately. The strain showed 2.64 U/g mass, 0.058 U/104 cell, and 0.076 U/g mass of lipase, cellulase, and α-amylase activities, respectively.
3.6. Transcriptome Analysis of Acid-Tolerant Yeast under Acetic Acid Stress
The mechanism of tolerance to acetic acid in strain Y2 under different concentrations of acetic acid stress was investigated through an acid-stress-responsive transcriptome analysis. Accordingly, a total of 674,794,634 raw sequencing reads were generated, and 648,168,878 clean reads were left after quality control filtering (Table S1). Principal component analysis (PCA) was performed on the gene expression values (FPKM) of all the samples to assess inter-group differences and within-group sample duplication. The results showed that the samples were dispersed between the C, AL, and AH groups, but they were clustered together within the groups, which indicated that there was a significant difference in the FPKM between the three groups of samples (Figure 4A). The combined change in the first and second principal components was 82.77% (PC1: 45.04%, PC2: 27.73%), which indicated a difference between the control, AL, and AH groups.
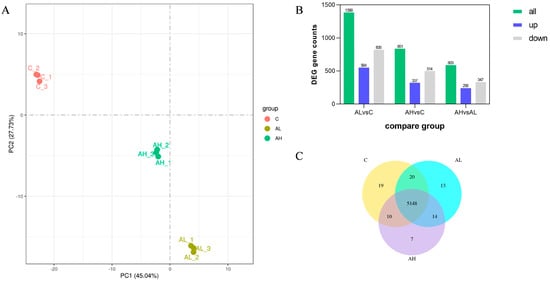
Figure 4.
Transcriptome analysis of Pichia kudriavzevii Y2. C: control group, AL: low-concentration acetic acid group (4 g/L), AH: high-concentration acetic acid group (8 g/L). (A) Principal component analysis results. The abscissa is the first principal component, and the ordinate is the second principal component. (B) Statistical histogram of the number of differentially compared genes in combination. Blue and grey represent upregulated and downregulated differential genes, respectively, and the numbers on the bars represent the number of differential genes. (C) Venn diagram of coexpression.
3.7. Identification of Differentially Expressed Genes
Differentially expressed genes (DEGs) were screened with absolute values of log2 (fold change) ≥ 1 and padj ≤ 0.05. In contrast to the C group, 1399 DEGs and 851 DEGs were identified under 4 g/L (AL group) and 8 g/L acetic acid (AH group) stresses, respectively. Additionally, 603 DEGs were identified in the AL vs. AH group (Figure 4B). Among these, 564 genes were downregulated and 835 genes were upregulated in the AL vs. C group; 337 genes were upregulated and 514 genes were downregulated in the AH vs. C group; and 256 genes were upregulated and 347 genes were downregulated in the AH vs. AL group, respectively. Among these nineteen, thirteen, and seven DEGs were found to be unique to the C, AL, and AH groups, respectively, while there were five thousand one hundred and forty-eight differentially expressed genes common to all three groups (Figure 4C).
3.8. GO Enrichment Analysis of DEGs
GO function enrichment analysis was performed on the DEGs with padj < 0.05. The results showed that the DEGs between AH, AL, and C groups were mainly enriched in biological processes. In addition, the DEGs of the AH vs. C group were also enriched in molecular function and cellular component processes (Figure 5). Among these, the significantly enriched entries in biological processes were shown to be involved in ribonucleoprotein complex biogenesis (GO:0022613), ribosome biogenesis (GO:0042254), and oxidation–reduction processes (GO:0055114). Cellular components were significantly enriched in preribosome (GO:0030684) and membrane (GO:0016020) processes. The entries that were significantly enriched in molecular function processes were related to oxidoreductase activity (GO:0016491) (Tables S2–S4). Notably, the DEGs of both the AH vs. C group and the AL vs. C group were significantly enriched in oxidation–reduction processes (GO:0055114) and oxidoreductase activity (GO:0016491). However, the AH vs. C group had a higher significance level. Therefore, strain Y2 may enhance tolerance to acetic acid by enhancing redox intensity in the presence of elevated acetic acid concentrations.
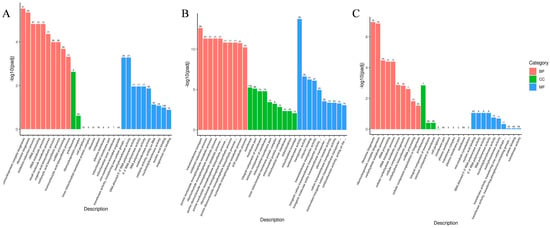
Figure 5.
GO (gene ontology) enrichment analysis. The horizontal coordinate is the GO term, the vertical coordinate is the significance level of GO term enrichment, expressed by -log10 (padj), and different colors indicate different functional classifications, respectively. C: control group, AL: low-concentration acetic acid group (4 g/L), AH: high-concentration acetic acid group (8 g/L). (A) AL vs. C; (B) AH vs. C; (C) AH vs. AL.
3.9. KEGG Enrichment Analysis of DEGs under Acetic Acid Stress
Further KEGG enrichment analysis of the DEGs showed that the DEGs were mainly enriched in the broad category of metabolism, where most of the differential genes were annotated to the carbohydrate and amino acid metabolism pathways. In addition, the DEGs were also involved in genetic-information-processing-, environmental-information-processing-, cellular-processes-, and organismal-systems-related pathways (Figure 6B). Especially, in the AH vs. C group compared with the AL vs. C group, the DEGs were more effective in terms of oxidative phosphorylation (ppa00190), the citrate cycle (TCA cycle) (ppa00020), glycolysis/gluconeogenesis (ppa00010), carbon metabolism (ppa01200), valine, leucine, and isoleucine biosynthesis (ppa00290), and glycine, serine, and threonine metabolism (ppa00260) enrichment in these pathways, with the former being more significant (Figure 6A, Tables S5–S7).
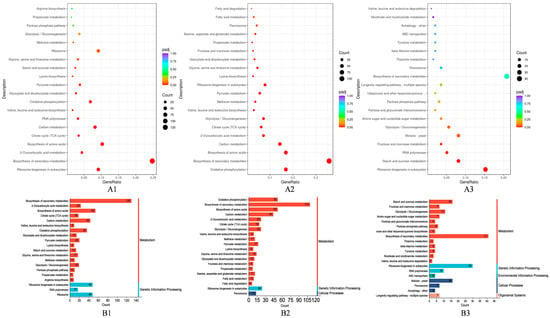
Figure 6.
KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis. C: control group, AL: low-concentration acetic acid group (4 g/L), AH: high-concentration acetic acid group (8 g/L). (A) KEGG enrichment scatter plot (A1–A3) in the order of AL vs. C, AH vs. C, and AH vs. AL. The horizontal coordinate is the ratio of the number of differential genes annotated to the total number of differential genes on the KEGG pathway, and the vertical coordinate is the ratio of the KEGG pathway. (B) KEGG pathway classification chart (B1–B3) in the order of AL vs. C, AH vs. C, and AH vs. AL. The horizontal coordinate is the number of differential genes annotated to the KEGG pathway, and the vertical coordinate is the KEGG pathway and its functional classification.
In addition, the genes related to complex I (NADH dehydrogenase), complex II (succinate dehydrogenase), complex III (cytochrome c reductase), and complex IV (cytochrome c oxidase) of the electron transport chain (oxidative phosphorylation) were significantly more upregulated in the AH vs. C group than in the AL vs C group. At the same time, multiple subunits of F-type H+-transporting ATPases were significantly upregulated (alpha (1.54-/1.08-fold), delta (1.74-/1.29-fold), and OSCP (oligomycin-sensitivity-conferring protein) (1.80-/1.24-fold)), and the increase in the acetate concentration increased the H+ transport and the degree of upregulation (Table 1).

Table 1.
DEGs related to oxidative phosphorylation.
In the citrate cycle (TCA cycle), phosphoenolpyruvate carboxykinase was the most significantly upregulated (8197501, 4.22-/3.56-fold), and the key enzymes citrate synthase (8197246, 2.99-/2.74-fold), isocitrate dehydrogenase (8198516 (3.03-/2.08-fold) and 8200845 (2.71-/1.89-fold)), and malate dehydrogenase (8198787, 2.97-/2.75-fold) were also significantly upregulated. In addition, genes related to succinate dehydrogenase (8197074 (2.14-/1.37-fold), 8198288 (1.93-/1.28-fold), and 8199556 (1.75-/1.36-fold)) were also significantly upregulated, which was consistent with the results of oxidative phosphorylation (Table 2).

Table 2.
DEGs related to citrate cycle (TCA cycle).
In short, the acid-tolerant yeast Y2 regulates the TCA cycle and the oxidative phosphorylation pathway mainly by upregulating the gene expression levels of key enzymes in response to acetic acid stress (Figure 7).
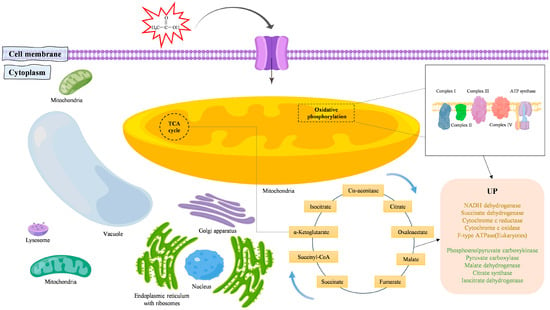
Figure 7.
Diagram of the Pichia kudriavzevii Y2 acid resistance mechanism.
4. Discussion
Baijiu brewing is generally carried out in an environment of multiple stresses conferred by high temperature, high acidity, and high levels of ethanol. While acetic acid is one of the main organic acids present in baijiu, changes in its content will not only significantly affect the flavor of baijiu but also affect the microbial community in the baijiu brewing process. If the acetic acid content is excessive, the growth of functional yeast flora will be inhibited to a certain extent, thus affecting the quality of the baijiu.
The current study focused on acetic acid stress with the intent of isolating an acid-tolerant strain followed by elucidating the acid tolerance mechanism. Here, a newly isolated Pichia kudriavzevii strain, Y2, was studied for its excellent acetic acid tolerance. Through transcriptome analysis, its acid tolerance mechanism was explored, which could have a positive effect on the artificially controlled production of baijiu. Pichia kudriavzevii is one of the core functional microorganisms in brewing. It is the main functional yeast in the brewing process of various aromatic baijiu products, and it has a regulatory role in the brewing microenvironment and the microbial community succession in the brewing environment. In addition, Pichia kudriavzevii can grow under multiple stresses of high temperature and high acidity [20,27]. In a previous report, the strain XDNZ_PK05 (Pichia kudriavzevii) was screened in spirits of Maotai-flavored baijiu under the conditions of double stresses of high acidity and high temperature. It was not only tolerant to high temperature (45 °C) but also grew normally under 5 g/L acetic acid and 70 g/L lactic acid concentrations [11], which indicated that P. kudriavzevii has a high resistance to high temperature and high acidity. In this study, the Pichia kudriavzevii strain Y2, isolated from the yellow water of Maotai-flavored baijiu under acid stress conditions, also showed a high acetic acid tolerance and could grow normally under a 12 g/L acetic acid concentration stress.
To further investigate the mechanism of acetate tolerance in the acid-tolerant strain Y2, a transcriptomic analysis of yeast RNA samples was performed. Based on the results of the KEGG pathway analysis, DEGs were mainly enriched in oxidative phosphorylation (ppa00190), the citrate cycle (TCA cycle) (ppa00020), glycolysis/gluconeogenesis (ppa00010), carbon metabolism (ppa01200), valine, leucine, and isoleucine biosynthesis (ppa00290), and glycine, serine, and threonine metabolism (ppa00260) pathways. Among these, the oxidative phosphorylation pathway is the process of aerobic biologically driven ATP synthesis, while the citrate cycle (TCA cycle) and glycolysis/gluconeogenesis belong to two pathways involved in carbohydrate metabolism. It can be concluded that the metabolism-related pathways of the acid-tolerant yeast Y2 were significantly changed under the different concentrations of acetic acid stress, and the changes were more obvious under higher concentrations of acetic acid. In addition, the possible mechanism of acid tolerance involved increasing intracellular ATP synthesis at the cost of improved carbohydrate metabolism and amino acid metabolism.
In most cells, mitochondrial oxidative phosphorylation is central to metabolism and is the main source of ATP (>95%), accounting for more than 95% of the total oxygen consumption. Any perturbation of the oxidative phosphorylation function has a direct impact on cellular metabolism [28]. An enhanced oxidative phosphorylation and an increased respiratory capacity were reported in Streptomyces albulus under low-pH stress. Among them, genes related to succinate-Q reductase and cytochrome C oxidase were significantly upregulated, while the δ-subunit (atpH), β-subunit (atpF), and C subunit (atpE) of ATP synthase were downregulated, which may be aimed at limiting H+ influx into the cell under acidic conditions, thus preventing the acidification of the intracellular environment [29]. Under acetic acid stress, the permeability of Saccharomyces cerevisiae’s membrane increases, and V-ATPase acts as a proton pump to pump H+ out of the cell and into subcellular vesicles to restore homeostasis in vivo. However, this process requires a large consumption of ATP [30]. For Saccharomyces cerevisiae, ATP is mainly produced by glycolysis, the TCA cycle, and oxidative phosphorylation [31]. In addition, during brewing, major genes in the NADH/NADPH regeneration reaction are upregulated when under a variety of stress conditions, thus enabling a normal biosynthetic pathway for amino acids, lipids, and nucleotides in Saccharomyces cerevisiae [32]. In our study, DEGs were significantly enriched in the oxidative phosphorylation pathway under acetic acid stress, and the enrichment was more pronounced at higher acetic acid concentrations. In addition, four protein complexes (NADH dehydrogenase, succinate dehydrogenase, cytochrome C reductase, and cytochrome C oxidase) and F-type H+-transporting ATPases (alpha (1.54-/1.08-fold), delta (1.74-/1.29-fold), and OSCP (1.80-/1.24-fold)) in the respiratory chain of the acid-tolerant yeast Y2 were significantly upregulated. The upregulation was greater in the higher-acetic-acid-concentration group. These results suggested that under acetic acid stress, the acid-tolerant yeast Y2 had an enhanced oxidative phosphorylation ability, an increased respiratory capacity, and an increased translocation of H+, which maintained homeostasis inside and outside the cell, which may be the mechanism of action by which the strain Y2 tolerated the low pH.
Mitochondria are organelles that carry out the most efficient metabolic pathway, which, in addition to oxidative phosphorylation, includes the TCA cycle, which provides energy to the whole cell in the form of adenosine triphosphate (ATP) [33]. The TCA cycle is a series of chemical reactions that generate energy through the oxidation of acetyl coenzyme a (CoA) from carbohydrates, fatty acids, and proteins [34]. According to the results of the transcriptome analysis, the DEGs in Saccharomyces cerevisiae were mainly involved in carbohydrate metabolism and amino acid metabolism under formic acid (Fa), acetic acid (Aa), and mixed Fa–Aa stress conditions [35]. The carbohydrate metabolic processes mainly include the glycolytic pathway, the TCA cycle, and the pentose phosphate pathway. It has been shown that the TCA cycle serves as the main pathway for energy supply and substance oxidation in Streptomyces albulus under low-pH stress, and the key genes of citrate synthase, α-ketoglutarate dehydrogenase complex, and succinate dehydrogenase are significantly upregulated [29], which indicates that under a low-pH environment, the bacterium obtains a large amount of energy to resist acid stress by enhancing the TCA cycle. In addition, for acetic-acid-resistant Escherichia coli, the key enzymes in the TCA cycle, phosphoenolpyruvate carboxykinase, malate dehydrogenase, and isocitrate dehydrogenase, all showed a decreasing and then increasing trend with increasing the mass concentration of exogenous sodium acetate [36]. Among them, isocitate dehydrogenase is the first enzyme after the branch of the TCA cycle and the glyoxylate branch, and it is a rate-limiting enzyme of the TCA cycle; malate dehydrogenase catalyzes the mutual interconversion of malate and oxaloacetate. The TCA cycle not only provides energy for the growth of the bacterium but is also an important source of intermediates for biosynthesis. When these intermediate products of the TCA pathway are reduced due to their participation in amino acid synthesis, they need to be replenished through a compensatory pathway to ensure the smooth operation of the TCA cycle. Phosphoenolpyruvate carboxykinase catalyzes the reaction of phosphoenolpyruvate with CO2 to produce oxaloacetic acid, which is the main intracellular recharge pathway in Escherichia coli. It is speculated that a mechanism such as that of Acetobacter may have been used to solve the problem of cytosolic acidification, that is, the flow of acetic acid through the TCA cycle and not to glyoxylic acid, which is the main mechanism of Acetobacter to alleviate the internal accumulation of acetic acid [37]. The addition of 1% acetic acid was reported to promote the expression of the key enzymes of the intracellular TCA cycle metabolism in Acetobacter pasteurianus, especially aconitase (1.80-fold), citrate synthetase (1.90-fold), fumarate synthetase (2.14-fold), malate dehydrogenase (2.00-fold), succinate dehydrogenase (2.69-fold), and malate quinone oxidoreductase (1.63-fold) [38]. Acetic acid may convert intracellular acetic acid to acetyl coenzyme A through the succinate coenzyme A pathway, which further enters the TCA cycle and intensifies the energy metabolism of the intracellular TCA cycle, allowing Acetobacter pasteurianus to produce more NADH and ATP to meet the energy requirements of the cell during growth [39,40]. Our results showed that the acid-tolerant yeast Y2 significantly enriched the DEGs in the citrate cycle (TCA cycle) of the carbohydrate metabolism pathway under acetic acid stress, and the enrichment was more pronounced in the AH vs. C group. The expression of the key enzymes of the TCA cycle metabolism was also significantly upregulated. Phosphoenolpyruvate carboxykinase was the most significantly upregulated (8197501, 4.22-/3.56-fold) enzyme, while citrate synthase (8197246, 2.99-/2.74-fold), isocitrate dehydrogenase (8198516, 3.03-/2.08-fold and 8200845, 2.71-/1.89-fold), and malate dehydrogenase (8198787, 2.97-/2.75-fold) were also significantly upregulated. In addition, succinate dehydrogenase (8197074, 2.14-/1.37-fold; 8198288, 1.93-/1.28-fold; and 8199556, 1.75-/1.36-fold) were also significantly upregulated. The above results indicate that the possible acid tolerance mechanism of strain Y2 is to enhance the TCA cycle by increasing the expression of key enzyme-related genes in the TCA cycle, thus increasing the energy supply of the cell and resisting acid stress.
5. Conclusions
In this study, the acid-tolerant yeast Pichia kudriavzevii Y2 was studied, and its acid tolerance molecular mechanism was explored by transcriptomic analysis. It was shown that the DEGs were mainly enriched in oxidative phosphorylation and the citrate cycle (TCA cycle), and they were upregulated the expression levels of key enzymes in these pathways. It was concluded that the acid stress was alleviated through the improved synthesis of ATP via the upregulated metabolism of carbohydrate and amino acids. This study provided a molecular basis for understanding the acid tolerance mechanism of P. kudriavzevii, which may be exploited to further improve the quality of Chinese baijiu through microflora regulation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation9060540/s1, Table S1: Sample sequencing data quality summary; Table S2: Significant terms in GO enrichment analysis (AL vs. C); Table S3: Significant terms in GO enrichment analysis (AH vs. C); Table S4: Significant terms in GO enrichment analysis (AH vs. AL); Table S5: Significant pathways in KEGG enrichment analysis (AL vs. C); Table S6: Significant pathways in KEGG enrichment analysis (AH vs. C); Supplementary Table S7: Significant pathways in KEGG enrichment analysis (AH vs. AL).
Author Contributions
N.W.: conceptualization, methodology, investigation, writing—review and editing, funding acquisition. P.Z.: methodology, writing—original draft. X.Z. (Xiaoli Zhou): data curation, writing—review and editing. J.Z.: conceptualization, investigation. Y.M.: data curation, formal analysis. C.L.: visualization, investigation, supervision. T.W.: visualization, supervision. H.L.: methodology, conceptualization. X.W.: supervision. H.W.: visualization, investigation. X.Z. (Xudong Zhao): visualization, investigation. M.A.M.: conceptualization, methodology, data curation, software, writing—review and editing. H.Z.: conceptualization, methodology, data curation, software, writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by grants from the Wuliangye Industry University research cooperation project (grant no. CXY2021ZR009) and the Talent Introduction Project of Sichuan University of Science & Engineering (grant no. 2019RC30).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw sequence data reported in this paper were deposited in the genome sequence archive (Genomics, Proteomics & Bioinformatics 2017) in the National Genomics Data Center (Nucleic Acids Res 2021), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences, under accession number CRA009003 and are publicly accessible at https://bigd.big.ac.cn/gsa (accessed on 27 October 2022).
Acknowledgments
The authors would like to give special thanks to the Wuliangye Industry University research cooperation project (grant no. CXY2021ZR009) for financial support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kang, J.; Sun, Y.; Huang, X.; Ye, L.; Chen, Y.; Chen, X.; Zheng, X.; Han, B.Z. Unraveling the microbial compositions, metabolic functions, and antibacterial properties of Huangshui, a byproduct of Baijiu fermentation. Food Res. Int. 2022, 157, 111320. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.; Cao, X.; Cheng, J.; Li, L.; Zhang, T.; Wu, Q.; Xiang, P.; Shen, C.; Li, Q. Chinese Baijiu: The Perfect Works of Microorganisms. Front. Microbiol. 2022, 13, 919044. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Sun, B. Effect of Fermentation Processing on the Flavor of Baijiu. J. Agric. Food Chem. 2018, 66, 5425–5432. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.W.; Han, B.Z. Baijiu (白酒), Chinese liquor: History, classification and manufacture. J. Ethn. Foods 2016, 3, 19–25. [Google Scholar] [CrossRef]
- Zou, W.; Zhao, C.; Luo, H. Diversity and Function of Microbial Community in Chinese Strong-Flavor Baijiu Ecosystem: A Review. Front. Microbiol. 2018, 9, 671. [Google Scholar] [CrossRef]
- Wang, S.; Wu, Q.; Nie, Y.; Wu, J.; Xu, Y. Construction of Synthetic Microbiota for Reproducible Flavor Compound Metabolism in Chinese Light-Aroma-Type Liquor Produced by Solid-State Fermentation. Appl. Environ. Microbiol. 2019, 85, 3090. [Google Scholar] [CrossRef]
- Xu, Y.; Sun, B.; Fan, G.; Teng, C.; Xiong, K.; Zhu, Y.; Li, J.; Li, X. The brewing process and microbial diversity of strong flavour Chinese spirits: A review. J. Inst. Brew. 2017, 123, 5–12. [Google Scholar] [CrossRef]
- Yan, Q.; Zhang, K.; Zou, W.; Hou, Y. Three main flavour types of Chinese Baijiu: Characteristics, research, and perspectives. J. Inst. Brew. 2021, 127, 317–326. [Google Scholar] [CrossRef]
- Song, Z.W.; Du, H.; Zhang, M.H.; Nie, Y.; Xu, Y. Schizosaccharomyces pombe Can Reduce Acetic Acid Produced by Baijiu Spontaneous Fermentation Microbiota. Microorganisms 2019, 7, 606. [Google Scholar] [CrossRef]
- You, L.; Zhao, D.; Zhou, R.; Tan, Y.; Wang, T.; Zheng, J. Distribution and function of dominant yeast species in the fermentation of strong-flavor baijiu. World J. Microbiol. Biotechnol. 2021, 37, 26. [Google Scholar] [CrossRef]
- Lin, L.C.; Bai, R.; Gao, Y.; Mu, J.Z.; Lu, J.; Li, C.W.; Zhang, C.Y. Screening of a robust high-tolerance Pichia kudriavzevii strain and its application in Baijiu fermentation. Food Ferment. Ind. 2023, 49, 60–67. [Google Scholar] [CrossRef]
- Di Martino, C.; Testa, B.; Letizia, F.; Iorizzo, M.; Lombardi, S.J.; Ianiro, M.; Di Renzo, M.; Strollo, D.; Coppola, R. Effect of exogenous proline on the ethanolic tolerance and malolactic performance of Oenococcus oeni. J. Food Sci. Technol. 2020, 57, 3973–3979. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Bae, J.H.; Ko, H.J.; Lee, S.H.; Sung, B.H.; Han, J.I.; Sohn, J.H. Low-pH production of d-lactic acid using newly isolated acid tolerant yeast Pichia kudriavzevii NG7. Biotechnol. Bioeng. 2018, 115, 2232–2242. [Google Scholar] [CrossRef] [PubMed]
- Ciani, M.; Canonico, L.; Oro, L.; Comitini, F. Footprint of nonconventional yeasts and their contribution in alcoholic fermentations. Biotechnol. Prog. Beverage Consum. 2020, 19, 435–465. [Google Scholar] [CrossRef]
- Du, H.; Song, Z.; Zhang, M.; Nie, Y.; Xu, Y. The deletion of Schizosaccharomyces pombe decreased the production of flavor-related metabolites during traditional Baijiu fermentation. Food Res. Int. 2021, 140, 109872. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Ko, H.J.; Jeong, H.; Lee, S.H.; Ko, H.J.; Bae, J.H.; Sung, B.H.; Han, J.I.; Sohn, J.H. Draft Genome Sequence of a Multistress-Tolerant Yeast, Pichia kudriavzevii NG7. Genome Announc. 2018, 6, e01515-17. [Google Scholar] [CrossRef] [PubMed]
- Bourdichon, F.; Casaregola, S.; Farrokh, C.; Frisvad, J.C.; Gerds, M.L.; Hammes, W.P.; Harnett, J.; Huys, G.; Laulund, S.; Ouwehand, A.; et al. Food fermentations: Microorganisms with technological beneficial use. Int. J. Food Microbiol. 2012, 154, 87–97. [Google Scholar] [CrossRef]
- Du, H.; Song, Z.; Xu, Y. Ethyl Carbamate Formation Regulated by Lactic Acid Bacteria and Nonconventional Yeasts in Solid-State Fermentation of Chinese Moutai-Flavor Liquor. J. Agric. Food Chem. 2018, 66, 387–392. [Google Scholar] [CrossRef]
- Zhang, H.; Du, H.; Xu, Y. Volatile Organic Compound-Mediated Antifungal Activity of Pichia spp. and Its Effect on the Metabolic Profiles of Fermentation Communities. Appl. Environ. Microbiol. 2021, 87, 2992. [Google Scholar] [CrossRef]
- Elhalis, H.; Cox, J.; Frank, D.; Zhao, J. Microbiological and biochemical performances of six yeast species as potential starter cultures for wet fermentation of coffee beans. LWT 2021, 137, 110430. [Google Scholar] [CrossRef]
- Chagas Junior, G.C.A.; Ferreira, N.R.; Andrade, E.H.A.; Nascimento, L.D.D.; Siqueira, F.C.; Lopes, A.S. Profile of Volatile Compounds of On-Farm Fermented and Dried Cocoa Beans Inoculated with Saccharomyces cerevisiae KY794742 and Pichia kudriavzevii KY794725. Molecules 2021, 26, 344. [Google Scholar] [CrossRef] [PubMed]
- Shankar, S.R.; Sneha, H.P.; Prakash, I.; Khan, M.; HN, P.K.; Om, H.; Basavaraj, K.; Murthy, P.S. Microbial ecology and functional coffee fermentation dynamics with Pichia kudriavzevii. Food Microbiol. 2022, 105, 104012. [Google Scholar] [CrossRef]
- Deng, N.; Du, H.; Xu, Y. Cooperative Response of Pichia kudriavzevii and Saccharomyces cerevisiae to Lactic Acid Stress in Baijiu Fermentation. J. Agric. Food Chem. 2020, 68, 4903–4911. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.J.; Ahn, T.Y.; Sohn, J.H. Analysis of microbial diversity in makgeolli fermentation using PCR-DGGE. J. Life Sci. 2012, 22, 232–238. [Google Scholar] [CrossRef]
- Ruyters, S.; Mukherjee, V.; Verstrepen, K.J.; Thevelein, J.M.; Willems, K.A.; Lievens, B. Assessing the potential of wild yeasts for bioethanol production. J. Ind. Microbiol. Biotechnol. 2015, 42, 39–48. [Google Scholar] [CrossRef]
- Duan, Y.; Wu, F.; He, D.; Xu, R.; Feng, H.; Chen, T.; Liu, G.; Wang, W. Seasonal Variation of Airborne Fungi of the Tiantishan Grottoes and Western Xia Museum, Wuwei, China. Sci. Cold Arid. Reg. 2020, 13, 522–532. [Google Scholar] [CrossRef]
- Díaz-Nava, L.; Aguilar-Uscanga, M.; Ortiz-Muñiz, B.; Montes-García, N.; Domínguez, J.; Gómez-Rodríguez, J. Acetic acid-tolerant native yeast Pichia kudriavzevii ITV-S42 isolated from sweet sorghum juice for ethanol production. Sugar Tech 2022, 24, 576–584. [Google Scholar] [CrossRef]
- Wilson, D.F.; Harrison, D.K.; Vinogradov, S.A. Oxygen, pH, and mitochondrial oxidative phosphorylation. J. Appl. Physiol. 2012, 113, 1838–1845. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, K.F.; Pan, L.; Chen, X.S. Global gene transcriptome analysis of the acid stress response of Streptomyces albulus M-Z18. Food Ferment. Ind. 2022, 48, 1–10. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, Y.; Cai, W.; Zeng, J.; Liu, N.; Wan, Y.; Fu, G. Research progress of anti-environmental factor stress mechanism and anti-stress tolerance way of Saccharomyces cerevisiae during the brewing process. Crit. Rev. Food Sci. Nutr. 2022, 1–16. [Google Scholar] [CrossRef]
- Snoek, T.; Verstrepen, K.J.; Voordeckers, K. How do yeast cells become tolerant to high ethanol concentrations? Curr. Genet. 2016, 62, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Liu, Z.L. Mechanisms of ethanol tolerance in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2010, 87, 829–845. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, P.; Schnellmann, R.G. Mitochondrial energetics in the kidney. Nat. Rev. Nephrol. 2017, 13, 629–646. [Google Scholar] [CrossRef]
- Choi, I.; Son, H.; Baek, J.H. Tricarboxylic Acid (TCA) Cycle Intermediates: Regulators of Immune Responses. Life 2021, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Xie, C.-Y.; Yang, B.-X.; Gou, M.; Xia, Z.-Y.; Sun, Z.-Y.; Tang, Y.-Q. The response mechanisms of industrial Saccharomyces cerevisiae to acetic acid and formic acid during mixed glucose and xylose fermentation. Process. Biochem. 2020, 91, 319–329. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Zhang, P.H.; Zhou, C.; Dong, M.M. Effect of Different Concentration Exogenous Sodium Acetate on Metabolism and the Key Enzyme Activity in the Acetate-Tolerant Escherichia coli DA19. J. Microbiol. 2016, 36, 15–20. [Google Scholar] [CrossRef]
- Trček, J.; Mira, N.P.; Jarboe, L.R. Adaptation and tolerance of bacteria against acetic acid. Appl. Microbiol. Biotechnol. 2015, 99, 6215–6229. [Google Scholar] [CrossRef]
- Yin, H.; Zhang, R.; Chang, Y.; Zheng, Y.; Wang, M. Effects of TCA cycle metabolism on the acetic acid fermentation of Acetobacter pasteurianus. Food Sci. 2017, 38, 82–86. [Google Scholar] [CrossRef]
- Mullins, E.A.; Francois, J.A.; Kappock, T.J. A specialized citric acid cycle requiring succinyl-coenzyme A (CoA):acetate CoA-transferase (AarC) confers acetic acid resistance on the acidophile Acetobacter aceti. J. Bacteriol. 2008, 190, 4933–4940. [Google Scholar] [CrossRef]
- Fukaya, M.; Takemura, H.; Tayama, K.; Okumura, H.; Kawamura, Y.; Horinouchi, S.; Beppu, T. The aarC gene responsible for acetic acid assimilation confers acetic acid resistance on Acetobacter aceti. J. Ferment. Bioeng. 1993, 76, 270–275. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).